ARGONAUTE2 Localizes to Sites of Sporocysts in the Schistosome-Infected Snail, Biomphalaria glabrata
Abstract
1. Introduction
2. Materials and Methods
2.1. Biomphalaria glabrata and Schistosoma mansoni Maintenance
2.2. MicroRNA Pathway Gene Identification in Biomphalaria glabrata
2.3. Differential Expression of miRNA Pathway Genes in Biomphalaria glabrata Following Its Exposure to Schistosoma mansoni Miracidia
2.4. Production of a Polyclonal Antibody Specific to Biomphalaria glabrata Ago2
2.5. Western Blot Hybridization Analysis
2.6. Histology and Immunofluoresence to Determine Ago2 Localisation in Biomphalaria glabrata Tissues Following Its Exposure to Schistosoma mansoni Miracidia
2.7. High-Throughput Small RNA Sequencing for Profiling of the microRNA Landscape of Biomphalaria glabrata Post Exposure to Schistosoma mansoni Miracidia
3. Results
3.1. Identification, Annotation, and Expression Analysis of Core Pieces of Protein Machinery of the Biomphalaria glabrata miRNA Pathway
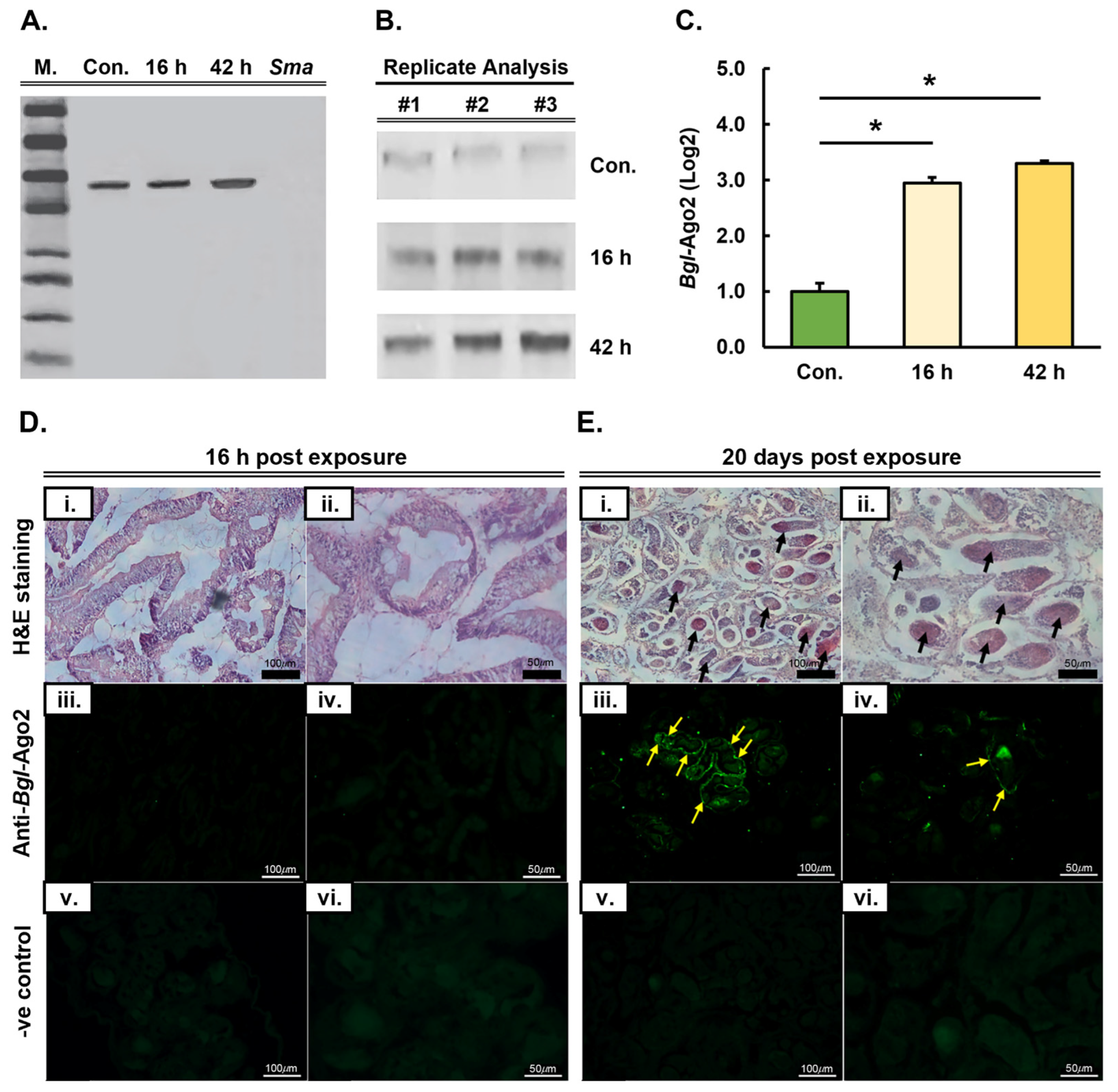
3.2. Expression Analysis and Tissue Localization of the Biomphalaria glabrata Ago2 Protein Post Long-Term Infection by Schistosoma mansoni
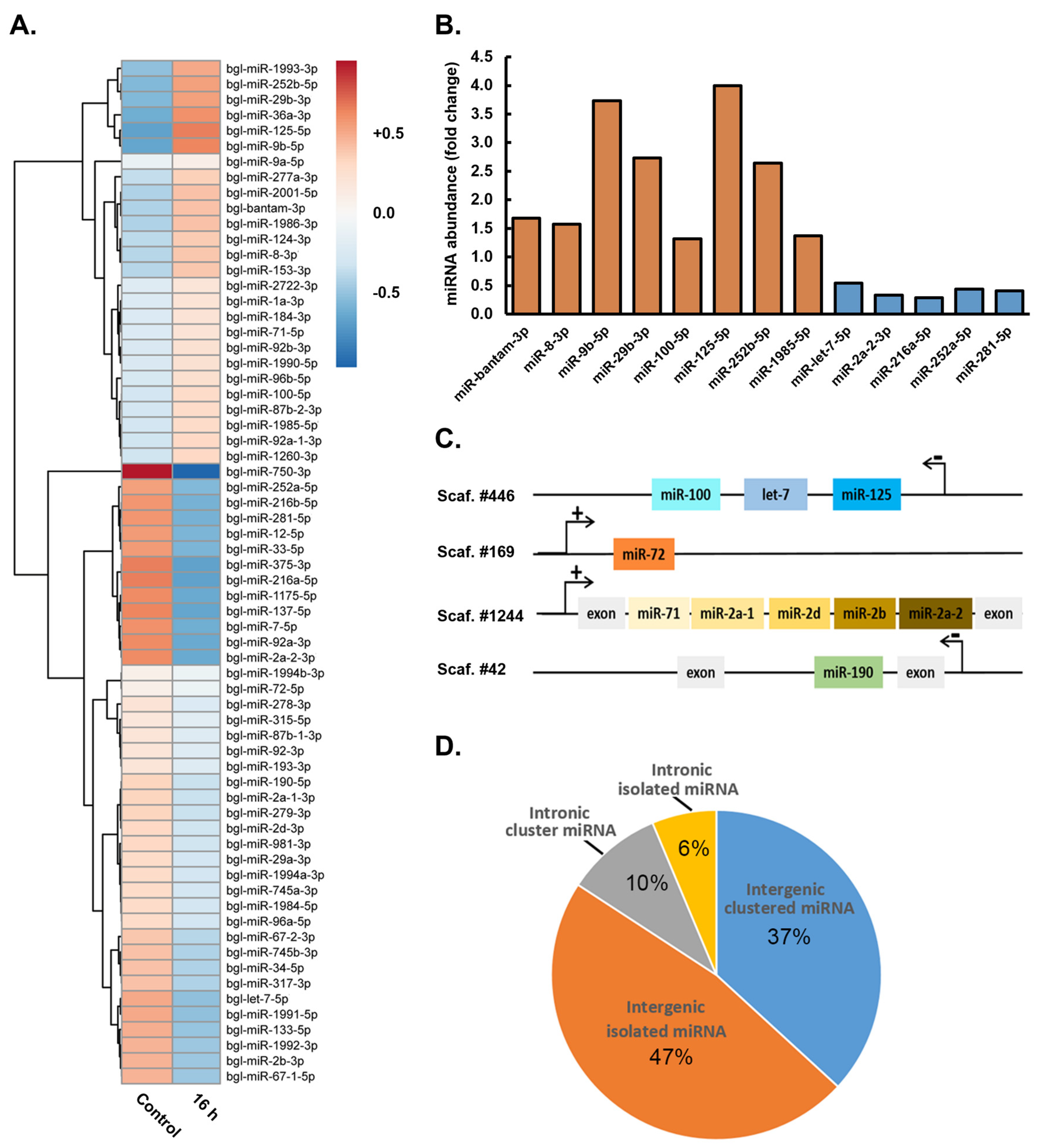
3.3. Demonstration of the Requirement of the miRNA Pathway as Part of the Molecular Response of Biomphalaria glabrata to Exposure to Schistosoma mansoni Miracidia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, J.; Luo, X.; Huang, M.; Liu, G.; You, W.; Ke, C. Identification and characteristics of muscle growth-related microRNA in the Pacific abalone, Haliotis discus hannai. BMC Genom. 2018, 19, 915. [Google Scholar] [CrossRef]
- Walker, S.E.; Spencer, G.E.; Necakov, A.; Carlone, R.L. Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System. Int. J. Mol. Sci. 2018, 19, 2741. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, X.; Feng, Y.; Huang, W.; Wang, W.; Li, L.; Fang, X.; Que, H.; Zhang, G. Identification of conserved and novel microRNAs in the Pacific oyster Crassostrea gigas by deep sequencing. PLoS ONE 2014, 9, e104371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, H.; Kong, L.; Liu, S.; Li, Q. High throughput sequencing of small RNAs transcriptomes in two Crassostrea oysters identifies microRNAs involved in osmotic stress response. Sci. Rep. 2016, 6, 22687. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zheng, Z.; Du, X.; Wang, Q.; Huang, R.; Deng, Y.; Shi, S.; Zhao, X. Identification and characterization of microRNAs in pearl oyster Pinctada martensii by Solexa deep sequencing. Mar. Biotechnol. 2014, 16, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yoshitake, K.; Asaduzzaman, M.; Kinoshita, S.; Watabe, S.; Asakawa, S. Discovery and functional understanding of miRNAs in molluscs: A genome-wide profiling approach. RNA Biol. 2021, 18, 1702–1715. [Google Scholar] [CrossRef]
- Abo-Al-Ela, H.G.; Faggio, C. MicroRNA-mediated stress response in bivalve species. Ecotoxicol. Environ. Saf. 2021, 208, 111442. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, L.; Song, L.; Liu, R.; Zhang, H.; Huang, M.; Chen, H. The identification and characteristics of immune-related microRNAs in haemocytes of oyster Crassostrea gigas. PLoS ONE 2014, 9, e88397. [Google Scholar] [CrossRef]
- Picone, B.; Rhode, C.; Roodt-Wilding, R. Identification and characterization of miRNAs transcriptome in the South African abalone, Haliotis midae. Mar Genomics. 2017, 31, 9–12. [Google Scholar] [CrossRef]
- Chen, X.; Qiu, L.; Si, X.; Zhang, X.; Guo, B.; Liao, Z.; Yan, X.; Qi, P. Exploring the Role of a Novel Interleukin-17 Homolog from Invertebrate Marine Mussel Mytilus coruscus in Innate Immune Response: Is Negative Regulation by Mc-Novel_miR_145 the Key? Int. J. Mol. Sci. 2023, 24, 5928. [Google Scholar] [CrossRef]
- Feng, D.; Li, Q.; Yu, H.; Liu, S.; Kong, L.; Du, S. Integrated analysis of microRNA and mRNA expression profiles in Crassostrea gigas to reveal functional miRNA and miRNA-targets regulating shell pigmentation. Sci. Rep. 2020, 10, 20238. [Google Scholar] [CrossRef] [PubMed]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; do Amaral, L.R.; et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017, 8, 15451. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, J.; Yang, D. miR-96-5p promotes the proliferation and migration of ovarian cancer cells by suppressing Caveolae1. J. Ovarian Res. 2019, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Liang, H.; Uzair-Ur-Rehman; Wang, Y.; Zhang, W.; Zhou, Y.; Chen, S.; Yu, M.; Cui, S.; Liu, M.; et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 2016, 6, 37421. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, F.R.; Portilho, L.G.; Jeremias, W.J.; Babá, É.H.; do Amaral, L.R.; Silva, L.M.; Coelho, P.M.Z.; Caldeira, R.L.; Gomes, M.S. Deep sequencing of small RNAs reveals the repertoire of miRNAs and piRNAs in Biomphalaria glabrata. Mem. Inst. Oswaldo Cruz 2020, 115, e190498. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Smith, M.K.; Bose, U.; Mita, M.; Hall, M.R.; Elizur, A.; Motti, C.A.; Cummins, S.F. Differences in Small Molecule Neurotransmitter Profiles from the Crown-of-Thorns Seastar Radial Nerve Revealed Between Sexes and Following Food-Deprivation. Front. Endocrinol. 2018, 9, 551. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Derrien, B.; Clavel, M.; Baumberger, N.; Iki, T.; Sarazin, A.; Hacquard, T.; Ponce, M.R.; Ziegler-Graff, V.; Vaucheret, H.; Micol, J.L.; et al. A Suppressor Screen for AGO1 Degradation by the Viral F-Box P0 Protein Uncovers a Role for AGO DUF1785 in sRNA Duplex Unwinding. Plant Cell 2018, 30, 1353–1374. [Google Scholar] [CrossRef] [PubMed]
- Krings, W.; Brütt, J.O.; Gorb, S.N. Elemental analyses reveal distinct mineralization patterns in radular teeth of various molluscan taxa. Sci. Rep. 2022, 12, 7499. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Liu, X.; Park, J.K.; Jiang, F.; Liu, Y.; McKearin, D.; Liu, Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 2007, 13, 2324–2349. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Biggar, K.K.; Kornfeld, S.F.; Maistrovski, Y.; Storey, K.B. MicroRNA regulation in extreme environments: Differential expression of microRNAs in the intertidal snail Littorina littorea during extended periods of freezing and anoxia. Genom. Proteom. Bioinform. 2012, 10, 302–309. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Galvin, H.D.; Husain, M. Influenza A virus-induced host caspase and viral PA-X antagonize the antiviral host factor, histone deacetylase 4. J. Biol. Chem. 2019, 294, 20207–20221. [Google Scholar] [CrossRef]
- Menning, M.; Kufer, T.A. A role for the Ankyrin repeat containing protein Ankrd17 in Nod1- and Nod2-mediated inflammatory responses. FEBS Lett. 2013, 587, 2137–2142. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.; Cerullo, A.R.; Parziale, J.; Achrak, E.; Sultana, S.; Ferd, J.; Samad, S.; Deng, W.; Braunschweig, A.B.; Holford, M. Advancing Discovery of Snail Mucins Function and Application. Front. Bioeng. Biotechnol. 2021, 9, 734023. [Google Scholar] [CrossRef]
- Montuori, N.; Ragno, P. Multiple activities of a multifaceted receptor: Roles of cleaved and soluble uPAR. Front. Biosci. 2009, 14, 2494–2503. [Google Scholar] [CrossRef] [PubMed]
- Golias, C.; Tsoutsi, E.; Matziridis, A.; Makridis, P.; Batistatou, A.; Charalabopoulos, K. Leukocyte and endothelial cell adhesion molecules in inflammation focusing on inflammatory heart disease. In Vivo 2007, 21, 757–769. [Google Scholar]
- Glogauer, J.; Sun, C.; Wang, Y.; Glogauer, M. The actin-binding protein Adseverin mediates neutrophil polarization and migration. Cytoskeleton 2021, 78, 206–213. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Senturia, R.; Faller, M.; Yin, S.; Loo, J.A.; Cascio, D.; Sawaya, M.R.; Hwang, D.; Clubb, R.T.; Guo, F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 2010, 19, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.S.; Eckert, D.M.; Bass, B.L. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA 2006, 12, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Fedoroff, N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000, 12, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, F.R.; Silva, L.M.; Jeremias, W.J.; Babá, É.H.; Caldeira, R.L.; Coelho, P.M.Z.; Gomes, M.S. Differential expression of small RNA pathway genes associated with the Biomphalaria glabrata/Schistosoma mansoni interaction. PLoS ONE 2017, 12, e0181483. [Google Scholar] [CrossRef] [PubMed]
- Beezhold, K.J.; Castranova, V.; Chen, F. Microprocessor of microRNAs: Regulation and potential for therapeutic intervention. Mol. Cancer 2010, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Bose, M.; Bhattacharyya, S.N. Target-dependent biogenesis of cognate microRNAs in human cells. Nat. Commun. 2016, 7, 12200. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed]
- Griffin, K.N.; Walters, B.W.; Li, H.; Wang, H.; Biancon, G.; Tebaldi, T.; Kaya, C.B.; Kanyo, J.; Lam, T.T.; Cox, A.L.; et al. Widespread association of the Argonaute protein AGO2 with meiotic chromatin suggests a distinct nuclear function in mammalian male reproduction. Genome Res. 2022, 32, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Rao, L.V.M. The Role of microRNAs in Inflammation. Int. J. Mol. Sci. 2022, 23, 15479. [Google Scholar] [CrossRef]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction Between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 2019, 10, 1053. [Google Scholar] [CrossRef] [PubMed]
- Desseyn, J.L.; Aubert, J.P.; Porchet, N.; Laine, A. Evolution of the large secreted gel-forming mucins. Mol. Biol. Evol. 2000, 17, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Prakobphol, A.; Borén, T.; Ma, W.; Zhixiang, P.; Fisher, S.J. Highly glycosylated human salivary molecules present oligosaccharides that mediate adhesion of leukocytes and Helicobacter pylori. Biochemistry 2005, 44, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Radicioni, G.; Ceppe, A.; Ford, A.A.; Alexis, N.E.; Barr, R.G.; Bleecker, E.R.; Christenson, S.A.; Cooper, C.B.; Han, M.K.; Hansel, N.N.; et al. Airway mucin MUC5AC and MUC5B concentrations and the initiation and progression of chronic obstructive pulmonary disease: An analysis of the SPIROMICS cohort. Lancet Respir. Med. 2021, 9, 1241–1254. [Google Scholar] [CrossRef]
- Roger, E.; Grunau, C.; Pierce, R.J.; Hirai, H.; Gourbal, B.; Galinier, R.; Means, R.; Cesari, I.M.; Cosseau, C.; Mitta, G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata). PLoS Negl. Trop. Dis. 2008, 2, e330. [Google Scholar] [CrossRef]
- Ito, S.; Shimizu, M.; Nagatsuka, M.; Kitajima, S.; Honda, M.; Tsuchiya, T.; Kanzawa, N. High molecular weight lectin isolated from the mucus of the giant African snail Achatina fulica. Biosci. Biotechnol. Biochem. 2011, 75, 20–25. [Google Scholar] [CrossRef]
- Pitt, S.J.; Graham, M.A.; Dedi, C.G.; Taylor-Harris, P.M.; Gunn, A. Antimicrobial properties of mucus from the brown garden snail Helix aspersa. Br. J. Biomed. Sci. 2015, 72, 174–181. [Google Scholar] [CrossRef]
- Montuori, N.; Selleri, C.; Ragno, P. The urokinase-receptor in infectious diseases. Infez. Med. 2012, 20 (Suppl. 2), 13–18. [Google Scholar] [PubMed]
- Lingel, A.; Sattler, M. Novel modes of protein-RNA recognition in the RNAi pathway. Curr. Opin. Struct. Biol. 2005, 15, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Faro, M.J.; Perazzini, M.; Corrêa Ldos, R.; Mello-Silva, C.C.; Pinheiro, J.; Mota, E.M.; de Souza, S.; de Andrade, Z.; Júnior, A.M. Biological, biochemical and histopathological features related to parasitic castration of Biomphalaria glabrata infected by Schistosoma mansoni. Exp. Parasitol. 2013, 134, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Stricker, E.; Hotz-Wagenblatt, A.; Heit-Mondrzyk, A.; Pougialis, G.; Hugo, A.; Kuźmak, J.; Materniak-Kornas, M.; Löchelt, M. Functional Analyses of Bovine Foamy Virus-Encoded miRNAs Reveal the Importance of a Defined miRNA for Virus Replication and Host-Virus Interaction. Viruses 2020, 12, 1250. [Google Scholar] [CrossRef] [PubMed]
- Bennett, V.; Baines, A.J. Spectrin and Ankyrin-Based Pathways: Metazoan Inventions for Integrating Cells Into Tissues. Physiol. Rev. 2001, 81, 1353–1392. [Google Scholar] [CrossRef]
- Huang, H.M.; Bauer, D.C.; Lelliott, P.M.; Greth, A.; McMorran, B.J.; Foote, S.J.; Burgio, G. A novel ENU-induced ankyrin-1 mutation impairs parasite invasion and increases erythrocyte clearance during malaria infection in mice. Sci. Rep. 2016, 6, 37197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Sakashita, K.; Kamijo, T.; Hidaka, E.; Sugane, K.; Kubota, T.; Koike, K. Granulocyte-macrophage colony-stimulating factor induces de novo methylation of the p15 CpG island in hematopoietic cells. Cytokine 2005, 31, 203–212. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, G.; Zhao, T.C. HDAC4: Mechanism of regulation and biological functions. Epigenomics 2014, 6, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, G.Z.; Cingöz, O.; Goff, S.P. NP220 mediates silencing of unintegrated retroviral DNA. Nature 2018, 564, 278–282. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, X.; Lei, X.; Wang, H.; Jiang, J.; Wang, Y.; Bi, K.; Diao, H. Influenza A virus NS1 protein hijacks YAP/TAZ to suppress TLR3-mediated innate immune response. PLoS Pathog. 2022, 18, e1010505. [Google Scholar] [CrossRef]
- Ren, T.; Chen, H.; Liu, X.; Wang, Y.; Fan, A.; Qi, L.; Pan, L.; Bai, W.; Zhang, Y.; Sun, Y. ID1 inhibits foot-and-mouth disease virus replication via targeting of interferon pathways. FEBS J. 2021, 288, 4364–4381. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Ulloa Severino, L.; Panagiotou, T.C.; Moraes, T.F.; Yuen, D.A.; Lavoie, B.D.; Wilde, A. Inhibition of polar actin assembly by astral microtubules is required for cytokinesis. Nat. Commun. 2021, 12, 2409. [Google Scholar] [CrossRef] [PubMed]
- Erba, H.P.; Eddy, R.; Shows, T.; Kedes, L.; Gunning, P. Structure, chromosome location, and expression of the human gamma-actin gene: Differential evolution, location, and expression of the cytoskeletal beta- and gamma-actin genes. Mol. Cell Biol. 1988, 8, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.S.; Lee, S.H.; Sheng, J.; Zhang, Z.; Zhao, W.D.; Wang, D.; Jin, Y.; Charnay, P.; Ervasti, J.M.; Wu, L.G. Actin Is Crucial for All Kinetically Distinguishable Forms of Endocytosis at Synapses. Neuron 2016, 92, 1020–1035. [Google Scholar] [CrossRef]
- Arraes, S.M.; Freitas, M.S.; da Silva, S.V.; de Paula Neto, H.A.; Alves-Filho, J.C.; Auxiliadora Martins, M.; Basile-Filho, A.; Tavares-Murta, B.M.; Barja-Fidalgo, C.; Cunha, F.Q. Impaired neutrophil chemotaxis in sepsis associates with GRK expression and inhibition of actin assembly and tyrosine phosphorylation. Blood 2006, 108, 2906–2913. [Google Scholar] [CrossRef]

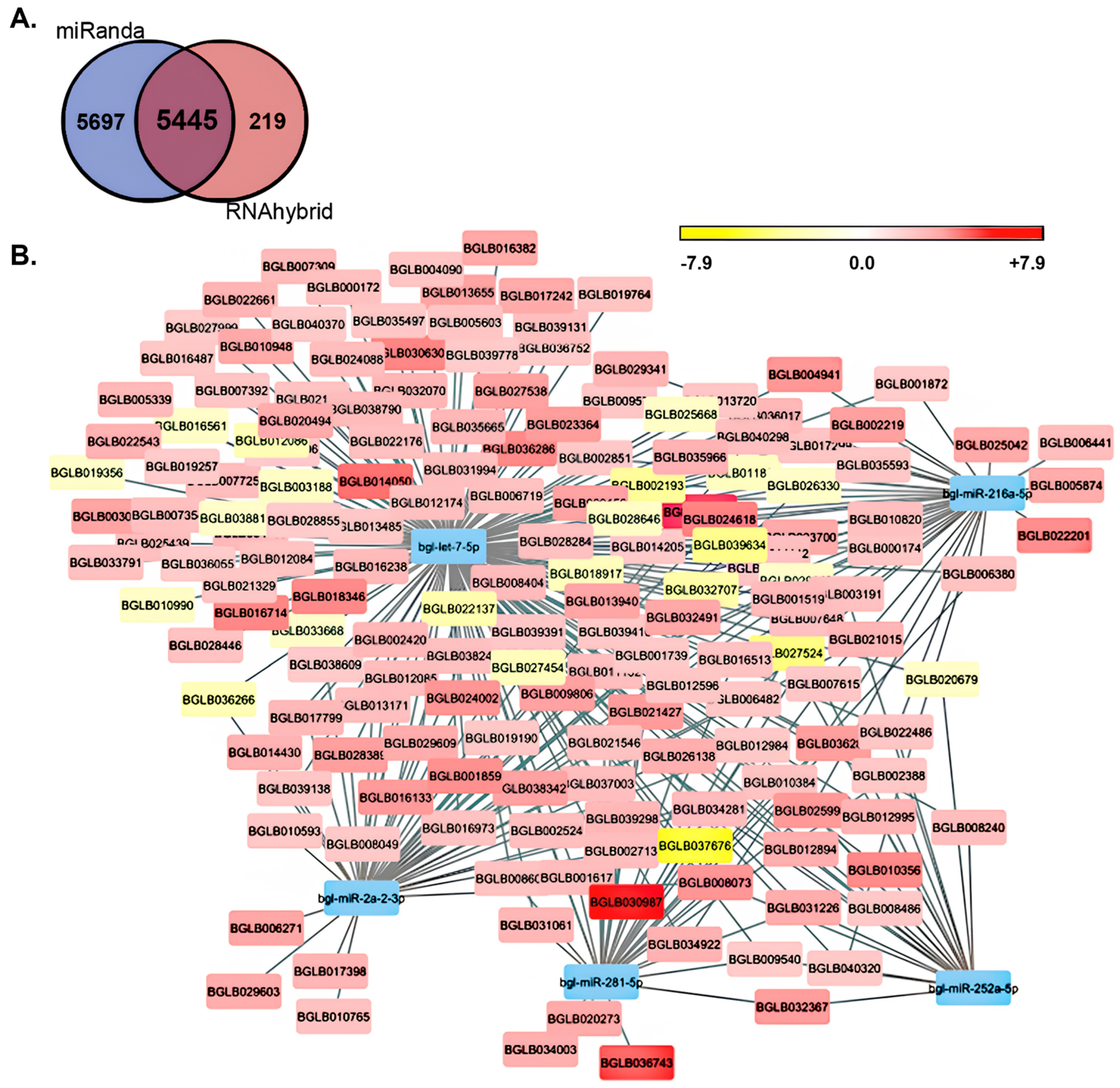
| B. glabrata Gene ID | Expression Fold Change | Targeting miRNA(s) | KEGG Identifier (KO ID) | KEGG Definition |
| BGLB025042 | +13.4 | Bgl-miR-216a-5p | K13908 | mucin-5B |
| BGLB027538 | +11.6 | Bgl-let-7-5p | K03985 | urokinase receptor |
| BGLB029609 | +9.7 | Bgl-let-7-5p Bgl-miR-2a-2-3p | K06787 | endothelial cell adhesion molecule |
| BGLB011132 | +7.3 | Bgl-let-7-5p Bgl-miR-216a-5p | K10380 | ankyrin repeat- domain protein |
| BGLB002524 | +5.9 | Bgl-let-7-5p Bgl-miR-281-5p | K05692 | actin β/γ 1 subunit |
| BGLB012596 | +5.6 | Bgl-let-7-5p Bgl-miR-216a-5p Bgl-miR-2a-2-3p | K11406 | histone deacetylase 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, P.; Fogarty, C.E.; Eamens, A.L.; Duke, M.G.; McManus, D.P.; Wang, T.; Cummins, S.F. ARGONAUTE2 Localizes to Sites of Sporocysts in the Schistosome-Infected Snail, Biomphalaria glabrata. Genes 2024, 15, 1023. https://doi.org/10.3390/genes15081023
Phan P, Fogarty CE, Eamens AL, Duke MG, McManus DP, Wang T, Cummins SF. ARGONAUTE2 Localizes to Sites of Sporocysts in the Schistosome-Infected Snail, Biomphalaria glabrata. Genes. 2024; 15(8):1023. https://doi.org/10.3390/genes15081023
Chicago/Turabian StylePhan, Phong, Conor E. Fogarty, Andrew L. Eamens, Mary G. Duke, Donald P. McManus, Tianfang Wang, and Scott F. Cummins. 2024. "ARGONAUTE2 Localizes to Sites of Sporocysts in the Schistosome-Infected Snail, Biomphalaria glabrata" Genes 15, no. 8: 1023. https://doi.org/10.3390/genes15081023
APA StylePhan, P., Fogarty, C. E., Eamens, A. L., Duke, M. G., McManus, D. P., Wang, T., & Cummins, S. F. (2024). ARGONAUTE2 Localizes to Sites of Sporocysts in the Schistosome-Infected Snail, Biomphalaria glabrata. Genes, 15(8), 1023. https://doi.org/10.3390/genes15081023








