Aberrantly Expressed tRNA-Val Fragments Can Distinguish Canine Hepatocellular Carcinoma from Canine Hepatocellular Adenoma
Highlights
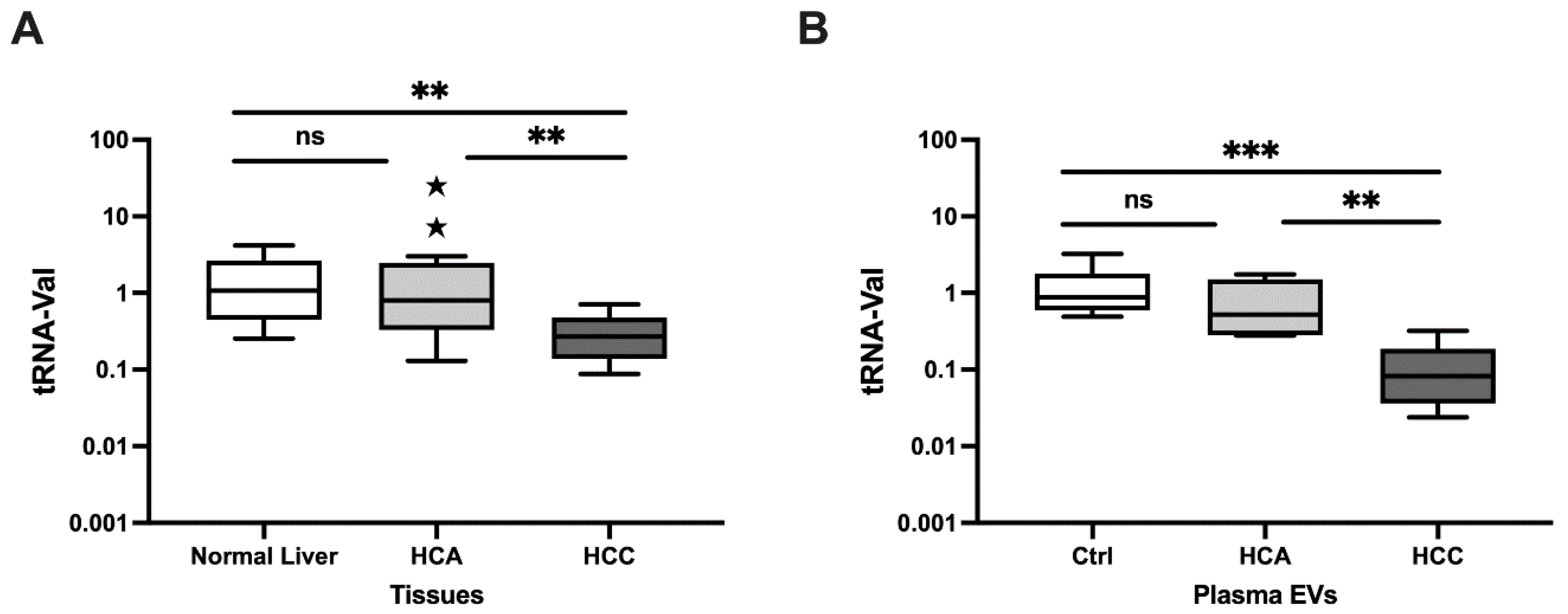
- tRNA-Val fragments distinguish canine HCC from HCA using tissues, plasma EVs, and cell lines.
- tRNA-Val is significantly downregulated in HCC versus HCA and normal tissues (AUC = 1.00 and 0.950).
- tRNA-Val is linked to DNA repair, mRNA processing, and protein degradation pathways.
- tRNA-Val is a promising biomarker for differentiating HCC and HCA.
- Insights into tRNA-Val's functional roles may contribute to understanding tumorigenesis and highlight therapeutic targets in both veterinary and human oncology.
- Research involving dogs provides insights applicable to human HCC.
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Cell Lines and Cell Culture
2.3. Isolation of EVs
2.4. RNA Extraction
2.5. qRT-PCR Analysis for tRNA-Val
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment and Protein-Protein Interaction (PPI) Analysis
2.7. Statistical Analysis
3. Results
3.1. Expression of tRNA-Val Using qRT-PCR
3.1.1. Relative Expression in Clinical Tissue Samples
3.1.2. Relative Expression in Plasma EVs
3.1.3. Relative Expression in Canine HCC Cell Lines
3.2. Diagnostic Value of tRNA-Val
3.3. Predicted Targets for tRNA-Val and Enrichment Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zheng, J.; Sadot, E.; Vigidal, J.A.; Klimstra, D.S.; Balachandran, V.P.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; et al. Characterization of hepatocellular adenoma and carcinoma using microRNA profiling and targeted gene sequencing. PLoS ONE 2018, 13, e0200776. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.K.; Hurvitz, A.I.; Lieberman, P.H.; Johnson, G.F. Canine hepatocellular carcinoma. Vet. Pathol. 1981, 18, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Van Sprundel, R.G.; van den Ingh, T.S.; Guscetti, F.; Kershaw, O.; Kanemoto, H.; van Gils, H.M.; Rothuizen, J.; Roskams, T.; Spee, B. Classification of primary hepatic tumours in the dog. Vet. J. 2013, 197, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.K.; Hurvitz, A.I.; Lieberman, P.H. Canine hepatic neoplasms: A clinicopathologic study. Vet. Pathol. 1980, 17, 553–564. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Jeannot, E.; Nhieu, J.T.; Scoazec, J.Y.; Guettier, C.; Rebouissou, S.; Bacq, Y.; Leteurtre, E.; Paradis, V.; Michalak, S.; et al. Genotype-phenotype correlation in hepatocellular adenoma: New classification and relationship with HCC. Hepatology 2006, 43, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Bioulac-Sage, P.; Laumonier, H.; Couchy, G.; Le Bail, B.; Sa Cunha, A.; Rullier, A.; Laurent, C.; Blanc, J.F.; Cubel, G.; Trillaud, H.; et al. Hepatocellular adenoma management and phenotypic classification: The Bordeaux experience. Hepatology 2009, 50, 481–489. [Google Scholar] [CrossRef]
- Zhu, L.; Ge, J.; Li, T.; Shen, Y.; Guo, J. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett. 2019, 452, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Gong, M.; Deng, Y.; Xiang, Y.; Ye, D. The role and mechanism of action of tRNA-derived fragments in the diagnosis and treatment of malignant tumors. Cell Commun. Signal 2023, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Wang, M.; Zhang, J.; Chen, Y.; Jiang, P.; Zhu, T.; Zhang, X. Emerging roles of tRNA-derived fragments in cancer. Mol. Cancer 2023, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, D.; Zhang, L.; Wang, J.; Ding, Y.; Sun, Z.; Wang, W. 5′-tiRNA-Gln inhibits hepatocellular carcinoma progression by repressing translation through the interaction with eukaryotic initiation factor 4A-I. Front. Med. 2023, 17, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulou, K.; Magkou, P.; Dreyer, T.; Dorn, J.; Obermayr, E.; Mahner, S.; van Gorp, T.; Braicu, I.; Magdolen, V.; Zeillinger, R.; et al. tRNA-derived small RNA 3′U-tRF(ValCAC) promotes tumour migration and early progression in ovarian cancer. Eur. J. Cancer 2023, 180, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Li, H.; Wu, H.; Du, F.; Xie, X.; Zeng, S.; Zhang, Z.; Dong, K.; Shang, L.; Jing, C.; et al. A novel 3′tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death Dis. 2022, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Ma, S.; Liang, B.; Ju, S. Serum hsa_tsr016141 as a Kind of tRNA-Derived Fragments Is a Novel Biomarker in Gastric Cancer. Front. Oncol. 2021, 11, 679366. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, C.; Zhu, Z.; Yang, F.; Wang, X.; Jiang, P.; Yan, F. A 5′-tRNA Derived Fragment NamedtiRNA-Val-CAC-001 Works as a Suppressor in Gastric Cancer. Cancer Manag. Res. 2022, 14, 2323–2337. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, B.; Wang, J.; Tang, L.; Hu, Q.; Wang, J.; Chen, H.; Zheng, J.; Yan, F.; Chen, H. tRNA-Derived Fragment tRF-Glu-TTC-027 Regulates the Progression of Gastric Carcinoma via MAPK Signaling Pathway. Front. Oncol. 2021, 11, 733763. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, Z.; Yu, X.; Ruan, Y.; Shen, Y.; Shao, Y.; Zhang, X.; Ye, G.; Guo, J. The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem Cell Res. Ther. 2021, 12, 418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bi, Z.; Dong, X.; Yu, M.; Wang, K.; Song, X.; Xie, L.; Song, X. tRNA-derived fragments: tRF-Gly-CCC-046, tRF-Tyr-GTA-010 and tRF-Pro-TGG-001 as novel diagnostic biomarkers for breast cancer. Thorac. Cancer 2021, 12, 2314–2323. [Google Scholar] [CrossRef]
- Tong, L.; Zhang, W.; Qu, B.; Zhang, F.; Wu, Z.; Shi, J.; Chen, X.; Song, Y.; Wang, Z. The tRNA-Derived Fragment-3017A Promotes Metastasis by Inhibiting NELL2 in Human Gastric Cancer. Front. Oncol. 2020, 10, 570916. [Google Scholar] [CrossRef]
- Mo, D.; Jiang, P.; Yang, Y.; Mao, X.; Tan, X.; Tang, X.; Wei, D.; Li, B.; Wang, X.; Tang, L.; et al. A tRNA fragment, 5′-tiRNA(Val), suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019, 457, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Sahlolbei, M.; Fattahi, F.; Vafaei, S.; Rajabzadeh, R.; Shiralipour, A.; Madjd, Z.; Kiani, J. Relationship Between Low Expressions of tRNA-Derived Fragments with Metastatic Behavior of Colorectal Cancer. J. Gastrointest. Cancer 2022, 53, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Lai, Y.C.; Husna, A.A.; Chen, H.W.; Tanaka, Y.; Kawaguchi, H.; Hatai, H.; Miyoshi, N.; Nakagawa, T.; Fukushima, R.; et al. Aberrantly expressed snoRNA, snRNA, piRNA and tRFs in canine melanoma. Vet. Comp. Oncol. 2020, 18, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.N.; Rahman, M.M.; Husna, A.A.; Nozaki, N.; Yamato, O.; Miura, N. YRNA and tRNA fragments can differentiate benign from malignant canine mammary gland tumors. Biochem. Biophys. Res. Commun. 2024, 691, 149336. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Zheng, B.; Song, X.; Wang, L.; Zhang, Y.; Tang, Y.; Wang, S.; Li, L.; Wu, Y.; Song, X.; Xie, L. Plasma exosomal tRNA-derived fragments as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 2022, 12, 1037523. [Google Scholar] [CrossRef]
- Xi, J.; Zeng, Z.; Li, X.; Zhang, X.; Xu, J. Expression and Diagnostic Value of tRNA-Derived Fragments Secreted by Extracellular Vesicles in Hypopharyngeal Carcinoma. Onco Targets Ther. 2021, 14, 4189–4199. [Google Scholar] [CrossRef]
- Lin, C.; Zheng, L.; Huang, R.; Yang, G.; Chen, J.; Li, H. tRFs as Potential Exosome tRNA-Derived Fragment Biomarkers for Gastric Carcinoma. Clin. Lab. 2020, 66, 961. [Google Scholar] [CrossRef]
- Husna, A.A.; Rahman, M.M.; Lai, Y.C.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Identification of melanoma-specific exosomal miRNAs as the potential biomarker for canine oral melanoma. Pigment. Cell Melanoma Res. 2021, 34, 1062–1073. [Google Scholar] [CrossRef]
- Gustafson, D.L.; Duval, D.L.; Regan, D.P.; Thamm, D.H. Canine sarcomas as a surrogate for the human disease. Pharmacol. Ther. 2018, 188, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Gardner, H.L.; Fenger, J.M.; London, C.A. Dogs as a Model for Cancer. Annu. Rev. Anim. Biosci. 2016, 4, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Ushio, N.; Rahman, M.M.; Katanoda, Y.; Ogihara, K.; Naya, Y.; Moriyama, A.; Iwanaga, T.; Saitoh, Y.; Sogawa, T.; et al. Aberrant expression of microRNAs and the miR-1/MET pathway in canine hepatocellular carcinoma. Vet. Comp. Oncol. 2018, 16, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Neo, S.; Ishizuka, C.; Kato, T.; Segawa, K.; Kawarai, S.; Ogihara, K.; Hisasue, M.; Tsuchiya, R. Identification of cell surface antigen expression in canine hepatocellular carcinoma cell lines. J. Vet. Med. Sci. 2013, 75, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Hino, Y.; Rahman, M.M.; Lai, Y.C.; Husna, A.A.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Hypoxic miRNAs expression are different between primary and metastatic melanoma cells. Gene 2021, 782, 145552. [Google Scholar] [CrossRef] [PubMed]
- Husna, A.A.; Rahman, M.M.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Long non-coding RNA and transfer RNA-derived small fragments in exosomes are potential biomarkers for canine oral melanoma. Vet. Comp. Oncol. 2022, 20, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Weng, Q.; Wang, Y.; Xie, Y.; Yu, X.; Zhang, S.; Ge, J.; Li, Z.; Ye, G.; Guo, J. Extracellular vesicles-associated tRNA-derived fragments (tRFs): Biogenesis, biological functions, and their role as potential biomarkers in human diseases. J. Mol. Med. 2022, 100, 679–695. [Google Scholar] [CrossRef]
- Suresh, P.S.; Thankachan, S.; Venkatesh, T. Landscape of Clinically Relevant Exosomal tRNA-Derived Non-coding RNAs. Mol. Biotechnol. 2023, 65, 300–310. [Google Scholar] [CrossRef]
- Wang, J.; Ma, G.; Li, M.; Han, X.; Xu, J.; Liang, M.; Mao, X.; Chen, X.; Xia, T.; Liu, X.; et al. Plasma tRNA Fragments Derived from 5′ Ends as Novel Diagnostic Biomarkers for Early-Stage Breast Cancer. Mol. Ther. Nucleic Acids 2020, 21, 954–964. [Google Scholar] [CrossRef]
- Majumder, A.; Syed, K.M.; Mukherjee, A.; Lankadasari, M.B.; Azeez, J.M.; Sreeja, S.; Harikumar, K.B.; Pillai, M.R.; Dutta, D. Enhanced expression of histone chaperone APLF associate with breast cancer. Mol. Cancer 2018, 17, 76. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, G.; Liu, Y.; Mei, C.; Yao, Y.; Wu, X.; Chen, X.; Ma, W.; Li, K.; Zhang, Z.; et al. A novel long non-coding RNA RP11-286H15.1 represses hepatocellular carcinoma progression by promoting ubiquitination of PABPC4. Cancer Lett. 2021, 499, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhang, Q.; Liu, M.; Yan, M.; Chu, X.; Li, Y. Identification of DNA repair-related genes predicting pathogenesis and prognosis for liver cancer. Cancer Cell Int. 2021, 21, 81. [Google Scholar] [CrossRef] [PubMed]
- Hernanda, P.Y.; Chen, K.; Das, A.M.; Sideras, K.; Wang, W.; Li, J.; Cao, W.; Bots, S.J.; Kodach, L.L.; de Man, R.A.; et al. SMAD4 exerts a tumor-promoting role in hepatocellular carcinoma. Oncogene 2015, 34, 5055–5068. [Google Scholar] [CrossRef] [PubMed]
- Kang, A.R.; An, H.T.; Ko, J.; Choi, E.J.; Kang, S. Ataxin-1 is involved in tumorigenesis of cervical cancer cells via the EGFR-RAS-MAPK signaling pathway. Oncotarget 2017, 8, 94606–94618. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.; Yun, Y.C.; Idris, M.; Cheng, S.; Boot, A.; Iain, T.B.H.; Rozen, S.G.; Tan, P.; Epstein, D.M. A tumor-associated splice-isoform of MAP2K7 drives dedifferentiation in MBNL1-low cancers via JNK activation. Proc. Natl. Acad. Sci. USA 2020, 117, 16391–16400. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Cheng, Y.; Liu, Y.; Chen, L.; Liu, L.; Wei, N.; Xie, Z.; Wu, W.; Feng, Y. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res. 2017, 77, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Huang, W.L.; Xu, Q.G.; Zhang, L.; Sun, S.H.; Zhou, W.P.; Yang, F. Overactivated neddylation pathway in human hepatocellular carcinoma. Cancer Med. 2018, 7, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, J.; Song, Q.; Ding, X.; Sun, F.; Yang, L. UBA3 promotes the occurrence and metastasis of intrahepatic cholangiocarcinoma through MAPK signaling pathway. Acta Biochim. Biophys. Sin. 2024, 56, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Bieberich, E. Synthesis, Processing, and Function of N-glycans in N-glycoproteins. Adv. Neurobiol. 2014, 9, 47–70. [Google Scholar]
- Tu, Y.; Chen, C.; Pan, J.; Xu, J.; Zhou, Z.G.; Wang, C.Y. The Ubiquitin Proteasome Pathway (UPP) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. Int. J. Clin. Exp. Pathol. 2012, 5, 726–738. [Google Scholar] [PubMed]
- Shang, F.; Taylor, A. Roles for the ubiquitin-proteasome pathway in protein quality control and signaling in the retina: Implications in the pathogenesis of age-related macular degeneration. Mol. Asp. Med. 2012, 33, 446–466. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, R.; Yang, J.; Zhao, C.; Liu, W.; Huang, Y.; Lyu, H.; Xiao, S.; Guo, D.; Zhou, C.; et al. The role of N-glycosylation modification in the pathogenesis of liver cancer. Cell Death Dis. 2023, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- DelaCourt, A.; Black, A.; Angel, P.; Drake, R.; Hoshida, Y.; Singal, A.; Lewin, D.; Taouli, B.; Lewis, S.; Schwarz, M.; et al. N-Glycosylation Patterns Correlate with Hepatocellular Carcinoma Genetic Subtypes. Mol. Cancer Res. 2021, 19, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Ma, H.; Roh, Y.S. Ubiquitin pathways regulate the pathogenesis of chronic liver disease. Biochem. Pharmacol. 2021, 193, 114764. [Google Scholar] [CrossRef]
- Oh, J.H.; Cho, J.Y. Comparative oncology: Overcoming human cancer through companion animal studies. Exp. Mol. Med. 2023, 55, 725–734. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, S.; Hasan, M.N.; Arif, M.; Nozaki, N.; Husna, A.A.; Furusawa, Y.; Sogawa, T.; Takahashi, K.; Kuramoto, T.; Noguchi, A.; et al. Aberrantly Expressed tRNA-Val Fragments Can Distinguish Canine Hepatocellular Carcinoma from Canine Hepatocellular Adenoma. Genes 2024, 15, 1024. https://doi.org/10.3390/genes15081024
Hashimoto S, Hasan MN, Arif M, Nozaki N, Husna AA, Furusawa Y, Sogawa T, Takahashi K, Kuramoto T, Noguchi A, et al. Aberrantly Expressed tRNA-Val Fragments Can Distinguish Canine Hepatocellular Carcinoma from Canine Hepatocellular Adenoma. Genes. 2024; 15(8):1024. https://doi.org/10.3390/genes15081024
Chicago/Turabian StyleHashimoto, Saki, MD Nazmul Hasan, Mohammad Arif, Nobuhiro Nozaki, Al Asmaul Husna, Yu Furusawa, Takeshi Sogawa, Kaori Takahashi, Tomohide Kuramoto, Aki Noguchi, and et al. 2024. "Aberrantly Expressed tRNA-Val Fragments Can Distinguish Canine Hepatocellular Carcinoma from Canine Hepatocellular Adenoma" Genes 15, no. 8: 1024. https://doi.org/10.3390/genes15081024
APA StyleHashimoto, S., Hasan, M. N., Arif, M., Nozaki, N., Husna, A. A., Furusawa, Y., Sogawa, T., Takahashi, K., Kuramoto, T., Noguchi, A., Takahashi, M., Yamato, O., Rahman, M. M., & Miura, N. (2024). Aberrantly Expressed tRNA-Val Fragments Can Distinguish Canine Hepatocellular Carcinoma from Canine Hepatocellular Adenoma. Genes, 15(8), 1024. https://doi.org/10.3390/genes15081024







