Identification of Autophagy-Related Biomarkers and Diagnostic Model in Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Identification of DEGs and DEAGs
2.3. Biological Functional and Enrichment Analysis of DEAGs
2.4. PPI Network Construction and Hub Gene Identification
2.5. Diagnostic ROC Curve Construction
2.6. Cell Culture and qRT-PCR
2.7. External Dataset Validation
2.8. Exploration of microRNAs Targeting the Hub Genes
3. Results
3.1. Identification of DEAGs
3.2. GO and KEGG Enrichment Analysis of DEAGs
3.3. PPI Network Construction and Hub DEAG Detection
3.4. Diagnostic ROC Model for Hub Genes
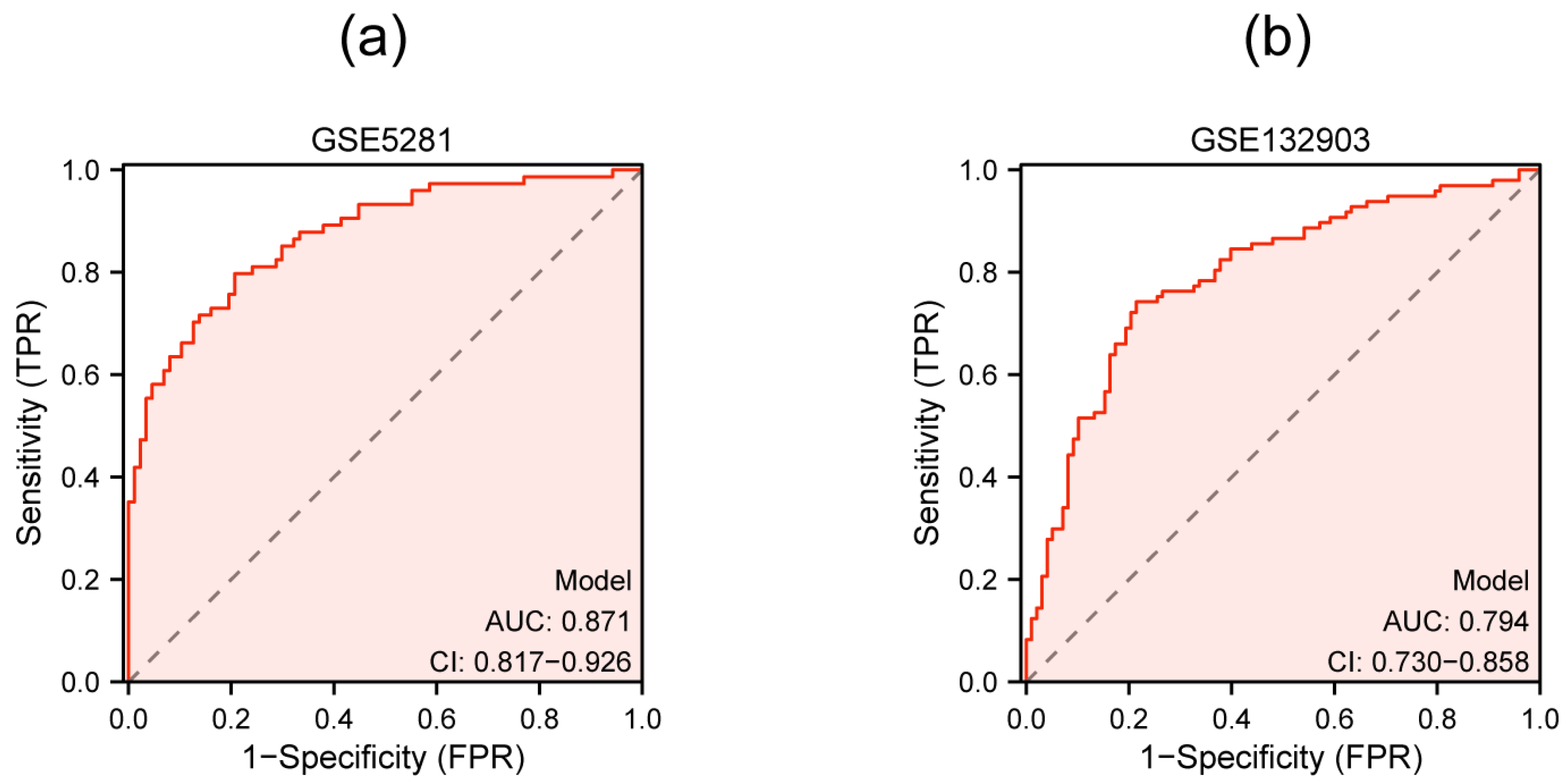
3.5. Validation of Hub Genes by qRT-PCR and External Dataset
3.6. Multigenic Prediction Model Construction and Validation
3.7. Identification of miRNAs Targeting Autophagy-Related Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barker, W.W.; Luis, C.A.; Kashuba, A.; Luis, M.; Harwood, D.G.; Loewenstein, D.; Waters, C.; Jimison, P.; Shepherd, E.; Sevush, S.; et al. Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis. Assoc. Disord. 2002, 16, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Cervellati, C.; Wood, P.L.; Romani, A.; Valacchi, G.; Squerzanti, M.; Sanz, J.M.; Ortolani, B.; Zuliani, G. Oxidative challenge in Alzheimer’s disease: State of knowledge and future needs. J. Investig. Med. Off. Publ. Am. Fed. Clin. Res. 2016, 64, 21–32. [Google Scholar] [CrossRef]
- Kumar, A.; Nisha, C.M.; Silakari, C.; Sharma, I.; Anusha, K.; Gupta, N.; Nair, P.; Tripathi, T.; Kumar, A. Current and novel therapeutic molecules and targets in Alzheimer’s disease. J. Formos. Med. Assoc. Taiwan Yi Zhi 2016, 115, 3–10. [Google Scholar] [CrossRef] [PubMed]
- The Alzheimer’s Association. 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2023, 19, 1598–1695. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Bourdenx, M.; Fujimaki, M.; Karabiyik, C.; Krause, G.J.; Lopez, A.; Martín-Segura, A.; Puri, C.; Scrivo, A.; Skidmore, J.; et al. The different autophagy degradation pathways and neurodegeneration. Neuron 2022, 110, 935–966. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Zhu, X.X.; Tang, D.S.; Lu, J.H. Targeting autophagy in Alzheimer’s disease: Animal models and mechanisms. Zool. Res. 2023, 44, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Pluta, R. The Dual Role of Autophagy in Postischemic Brain Neurodegeneration of Alzheimer’s Disease Proteinopathy. Int. J. Mol. Sci. 2023, 24, 13793. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef]
- Liu, X.; Yamashita, T.; Shang, J.; Shi, X.; Morihara, R.; Huang, Y.; Sato, K.; Takemoto, M.; Hishikawa, N.; Ohta, Y.; et al. Molecular switching from ubiquitin-proteasome to autophagy pathways in mice stroke model. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2020, 40, 214–224. [Google Scholar] [CrossRef]
- Reddy, P.H.; Oliver, D.M. Amyloid Beta and Phosphorylated Tau-Induced Defective Autophagy and Mitophagy in Alzheimer’s Disease. Cells 2019, 8, 488. [Google Scholar] [CrossRef]
- Glatigny, M.; Moriceau, S.; Rivagorda, M.; Ramos-Brossier, M.; Nascimbeni, A.C.; Lante, F.; Shanley, M.R.; Boudarene, N.; Rousseaud, A.; Friedman, A.K.; et al. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr. Biol. CB 2019, 29, 435–448.e438. [Google Scholar] [CrossRef]
- La Barbera, L.; Vedele, F.; Nobili, A.; Krashia, P.; Spoleti, E.; Latagliata, E.C.; Cutuli, D.; Cauzzi, E.; Marino, R.; Viscomi, M.T.; et al. Nilotinib restores memory function by preventing dopaminergic neuron degeneration in a mouse model of Alzheimer’s Disease. Prog. Neurobiol. 2021, 202, 102031. [Google Scholar] [CrossRef]
- Fang, E.F.; Hou, Y.; Palikaras, K.; Adriaanse, B.A.; Kerr, J.S.; Yang, B.; Lautrup, S.; Hasan-Olive, M.M.; Caponio, D.; Dan, X.; et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Moreau, K.; Fleming, A.; Imarisio, S.; Lopez Ramirez, A.; Mercer, J.L.; Jimenez-Sanchez, M.; Bento, C.F.; Puri, C.; Zavodszky, E.; Siddiqi, F.; et al. PICALM modulates autophagy activity and tau accumulation. Nat. Commun. 2014, 5, 4998. [Google Scholar] [CrossRef]
- de Wit, N.M.; den Hoedt, S.; Martinez-Martinez, P.; Rozemuller, A.J.; Mulder, M.T.; de Vries, H.E. Astrocytic ceramide as possible indicator of neuroinflammation. J. Neuroinflamm. 2019, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Qin, Z.H.; Sheng, R. The Multiple Roles of Autophagy in Neural Function and Diseases. Neurosci. Bull. 2024, 40, 363–382. [Google Scholar] [CrossRef]
- Berglund, R.; Guerreiro-Cacais, A.O.; Adzemovic, M.Z.; Zeitelhofer, M.; Lund, H.; Ewing, E.; Ruhrmann, S.; Nutma, E.; Parsa, R.; Thessen-Hedreul, M.; et al. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci. Immunol. 2020, 5, eabb5077. [Google Scholar] [CrossRef]
- Heckmann, B.L.; Teubner, B.J.W.; Boada-Romero, E.; Tummers, B.; Guy, C.; Fitzgerald, P.; Mayer, U.; Carding, S.; Zakharenko, S.S.; Wileman, T.; et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Sci. Adv. 2020, 6, eabb9036. [Google Scholar] [CrossRef]
- Cheng, J.; Liao, Y.; Dong, Y.; Hu, H.; Yang, N.; Kong, X.; Li, S.; Li, X.; Guo, J.; Qin, L.; et al. Microglial autophagy defect causes parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy 2020, 16, 2193–2205. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, J.; Li, Z.; Wang, S.; Yu, G.; Wang, L. Identification ferroptosis-related hub genes and diagnostic model in Alzheimer’s disease. Front. Mol. Neurosci. 2023, 16, 1280639. [Google Scholar] [CrossRef]
- Yan, R.; Wang, W.; Yang, W.; Huang, M.; Xu, W. Mitochondria-Related Candidate Genes and Diagnostic Model to Predict Late-Onset Alzheimer’s Disease and Mild Cognitive Impairment. J. Alzheimer’s Dis. JAD 2024, 99, S299–S315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kiryu, H. Identification of oxidative stress-related genes differentially expressed in Alzheimer’s disease and construction of a hub gene-based diagnostic model. Sci. Rep. 2023, 13, 6817. [Google Scholar] [CrossRef]
- Du, Y.; Chen, X.; Zhang, B.; Jin, X.; Wan, Z.; Zhan, M.; Yan, J.; Zhang, P.; Ke, P.; Huang, X.; et al. Identification of Copper Metabolism Related Biomarkers, Polygenic Prediction Model, and Potential Therapeutic Agents in Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2023, 95, 1481–1496. [Google Scholar] [CrossRef]
- Gu, X.; Lai, D.; Liu, S.; Chen, K.; Zhang, P.; Chen, B.; Huang, G.; Cheng, X.; Lu, C. Hub Genes, Diagnostic Model, and Predicted Drugs Related to Iron Metabolism in Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 949083. [Google Scholar] [CrossRef]
- Qin, Q.; Gu, Z.; Li, F.; Pan, Y.; Zhang, T.; Fang, Y.; Zhang, L. A Diagnostic Model for Alzheimer’s Disease Based on Blood Levels of Autophagy-Related Genes. Front. Aging Neurosci. 2022, 14, 881890. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, W.; Sun, W.; Rao, X.; Chen, X.; Yu, L. A Study on Autophagy Related Biomarkers in Alzheimer’s Disease Based on Bioinformatics. Cell. Mol. Neurobiol. 2023, 43, 3693–3703. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Cui, C.; Zhong, B.; Fan, R.; Cui, Q. HMDD v4.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2024, 52, D1327–D1332. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Tarazona-Sánchez, A.; Braza-Boils, A.; Balaguer, A.; Ferré-González, L.; Cañada-Martínez, A.J.; Baquero, M.; Cháfer-Pericás, C. Plasma microRNAs as potential biomarkers in early Alzheimer disease expression. Sci. Rep. 2022, 12, 15589. [Google Scholar] [CrossRef]
- Garcia, G.; Fernandes, A.; Stein, F.; Brites, D. Protective Signature of IFNγ-Stimulated Microglia Relies on miR-124-3p Regulation From the Secretome Released by Mutant APP Swedish Neuronal Cells. Front. Pharmacol. 2022, 13, 833066. [Google Scholar] [CrossRef]
- Abuelezz, N.Z.; Nasr, F.E.; Abdel Aal, W.M.; Molokhia, T.; Zaky, A. Sera miR-34a, miR-29b and miR-181c as potential novel diagnostic biomarker panel for Alzheimers in the Egyptian population. Exp. Gerontol. 2022, 169, 111961. [Google Scholar] [CrossRef] [PubMed]
- Sha, S.; Shen, X.; Cao, Y.; Qu, L. Mesenchymal stem cells-derived extracellular vesicles ameliorate Alzheimer’s disease in rat models via the microRNA-29c-3p/BACE1 axis and the Wnt/β-catenin pathway. Aging 2021, 13, 15285–15306. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Kim, M.S. Exposure to a mixture of heavy metals induces cognitive impairment: Genes and microRNAs involved. Toxicology 2022, 471, 153164. [Google Scholar] [CrossRef] [PubMed]
- Pulcrano, S.; De Gregorio, R.; De Sanctis, C.; Lahti, L.; Perrone-Capano, C.; Ponti, D.; di Porzio, U.; Perlmann, T.; Caiazzo, M.; Volpicelli, F.; et al. Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation. Int. J. Mol. Sci. 2022, 23, 6961. [Google Scholar] [CrossRef]
- Sedighi, M.; Baluchnejadmojarad, T.; Fallah, S.; Moradi, N.; Afshin-Majd, S.; Roghani, M. The Association Between Circulating Klotho and Dipeptidyl Peptidase-4 Activity and Inflammatory Cytokines in Elderly Patients With Alzheimer Disease. Basic Clin. Neurosci. 2020, 11, 349–357. [Google Scholar] [CrossRef]
- Tasker, R.; Rowlands, J.; Ahmed, Z.; Di Pietro, V. Co-Expression Network Analysis of Micro-RNAs and Proteins in the Alzheimer’s Brain: A Systematic Review of Studies in the Last 10 Years. Cells 2021, 10, 3479. [Google Scholar] [CrossRef] [PubMed]
- Aloi, M.S.; Prater, K.E.; Sopher, B.; Davidson, S.; Jayadev, S.; Garden, G.A. The pro-inflammatory microRNA miR-155 influences fibrillar β-Amyloid(1)(-42) catabolism by microglia. Glia 2021, 69, 1736–1748. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lin, M.; Ma, J.; Liu, W.; Gao, L.; Wei, S.; Xue, Y.; Shang, X. The role of LINC00094/miR-224-5p (miR-497-5p)/Endophilin-1 axis in Memantine mediated protective effects on blood-brain barrier in AD microenvironment. J. Cell. Mol. Med. 2019, 23, 3280–3292. [Google Scholar] [CrossRef] [PubMed]
- Wicik, Z.; Czajka, P.; Eyileten, C.; Fitas, A.; Wolska, M.; Jakubik, D.; von Lewinski, D.; Sourij, H.; Siller-Matula, J.M.; Postula, M. The role of miRNAs in regulation of platelet activity and related diseases—A bioinformatic analysis. Platelets 2022, 33, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Jia, N.; Li, R.; Zhang, Z.; Zhong, Y.; Han, K. miR-143-3p inhibition promotes neuronal survival in an Alzheimer’s disease cell model by targeting neuregulin-1. Folia Neuropathol. 2020, 58, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [Google Scholar] [CrossRef] [PubMed]
- Ułamek-Kozioł, M.; Furmaga-Jabłońska, W.; Januszewski, S.; Brzozowska, J.; Sciślewska, M.; Jabłoński, M.; Pluta, R. Neuronal autophagy: Self-eating or self-cannibalism in Alzheimer’s disease. Neurochem. Res. 2013, 38, 1769–1773. [Google Scholar] [CrossRef] [PubMed]
- Ułamek-Kozioł, M.; Kocki, J.; Bogucka-Kocka, A.; Petniak, A.; Gil-Kulik, P.; Januszewski, S.; Bogucki, J.; Jabłoński, M.; Furmaga-Jabłońska, W.; Brzozowska, J.; et al. Dysregulation of Autophagy, Mitophagy, and Apoptotic Genes in the Medial Temporal Lobe Cortex in an Ischemic Model of Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2016, 54, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, W.; Zhang, S.; Iyaswamy, A.; Sun, J.; Wang, J.; Yang, C. Novel Insight into Functions of Transcription Factor EB (TFEB) in Alzheimer’s Disease and Parkinson’s Disease. Aging Dis. 2023, 14, 652–669. [Google Scholar] [CrossRef]
- Tammineni, P.; Cai, Q. Defective retrograde transport impairs autophagic clearance in Alzheimer disease neurons. Autophagy 2017, 13, 982–984. [Google Scholar] [CrossRef]
- Qiao, L.; Hu, J.; Qiu, X.; Wang, C.; Peng, J.; Zhang, C.; Zhang, M.; Lu, H.; Chen, W. LAMP2A, LAMP2B and LAMP2C: Similar structures, divergent roles. Autophagy 2023, 19, 2837–2852. [Google Scholar] [CrossRef] [PubMed]
- Bourdenx, M.; Martín-Segura, A.; Scrivo, A.; Rodriguez-Navarro, J.A.; Kaushik, S.; Tasset, I.; Diaz, A.; Storm, N.J.; Xin, Q.; Juste, Y.R.; et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 2021, 184, 2696–2714.e2625. [Google Scholar] [CrossRef] [PubMed]
- Gammoh, N. The multifaceted functions of ATG16L1 in autophagy and related processes. J. Cell Sci. 2020, 133, jcs249227. [Google Scholar] [CrossRef] [PubMed]
- Stürner, E.; Behl, C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Minoia, M.; Boncoraglio, A.; Vinet, J.; Morelli, F.F.; Brunsting, J.F.; Poletti, A.; Krom, S.; Reits, E.; Kampinga, H.H.; Carra, S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 2014, 10, 1603–1621. [Google Scholar] [CrossRef] [PubMed]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wells, C.D.; Roach, P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011, 413, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Marzetti, E. Cell Death and Inflammation: The Role of Mitochondria in Health and Disease. Cells 2021, 10, 537. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Shteinfer-Kuzmine, A.; Verma, A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020, 10, 1485. [Google Scholar] [CrossRef]
- Araiso, Y.; Imai, K.; Endo, T. Role of the TOM Complex in Protein Import into Mitochondria: Structural Views. Annu. Rev. Biochem. 2022, 91, 679–703. [Google Scholar] [CrossRef] [PubMed]
- Ellenrieder, L.; Mårtensson, C.U.; Becker, T. Biogenesis of mitochondrial outer membrane proteins, problems and diseases. Biol. Chem. 2015, 396, 1199–1213. [Google Scholar] [CrossRef] [PubMed]
- Chhimpa, N.; Singh, N.; Puri, N.; Kayath, H.P. The Novel Role of Mitochondrial Citrate Synthase and Citrate in the Pathophysiology of Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2023, 94, S453–S472. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, H.; Liu, X.; Tan, L.; Qiao, X.; Ni, J.; Sun, Y.; Liang, J.; Hou, Y.; Dou, H. Pyruvate kinase isoform M2 impairs cognition in systemic lupus erythematosus by promoting microglial synaptic pruning via the β-catenin signaling pathway. J. Neuroinflamm. 2021, 18, 229. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Xu, J.; Zhang, K.; Gu, W.; Nie, L.; Wang, G.; Luo, Y. Refining Cancer Management Using Integrated Liquid Biopsy. Theranostics 2020, 10, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Nous, A.; Engelborghs, S.; Smolders, I. Melatonin levels in the Alzheimer’s disease continuum: A systematic review. Alzheimer’s Res. Ther. 2021, 13, 52. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L. Screening and Identification of Potential Peripheral Blood Biomarkers for Alzheimer’s Disease Based on Bioinformatics Analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e924263. [Google Scholar] [CrossRef]

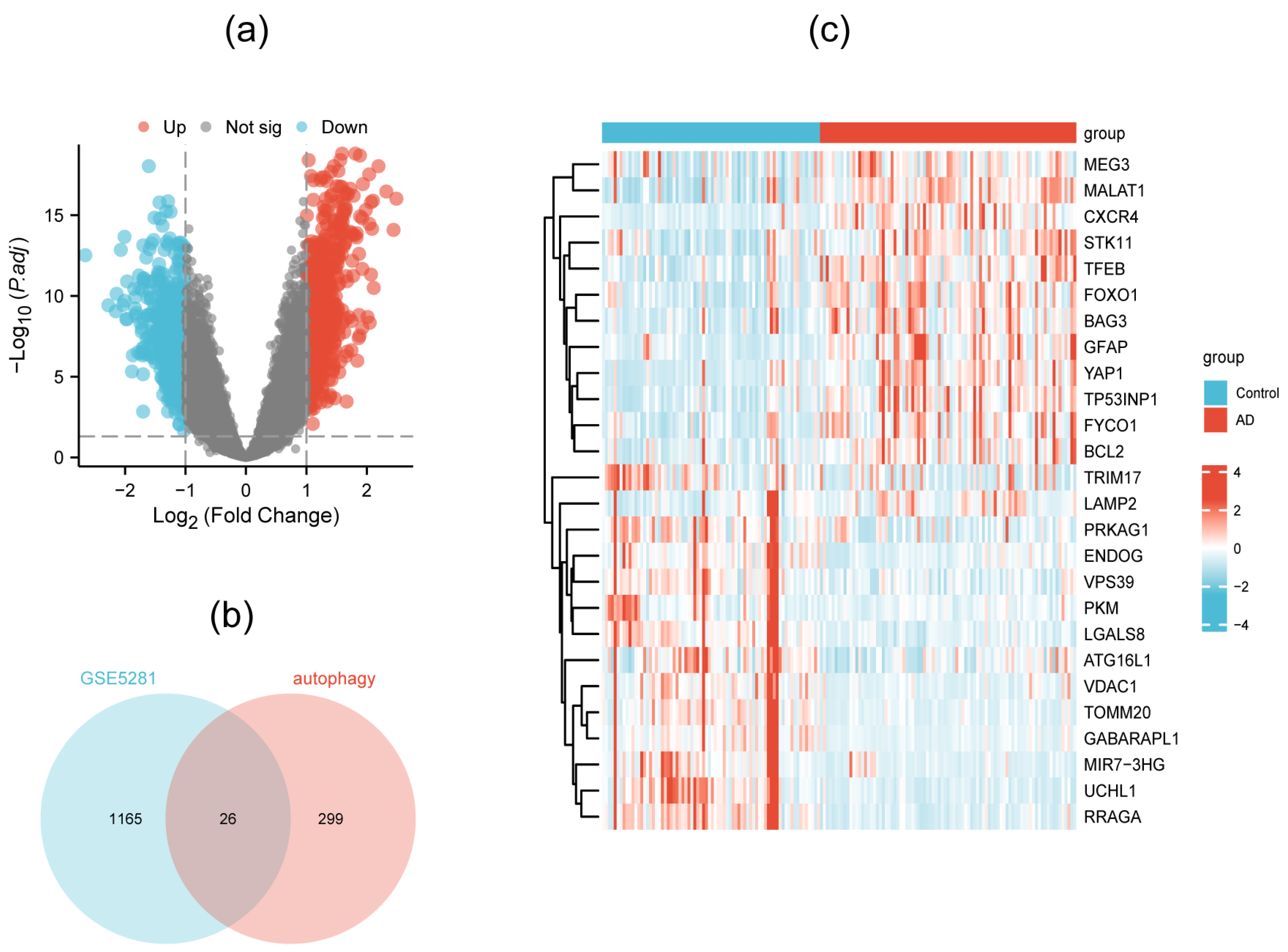
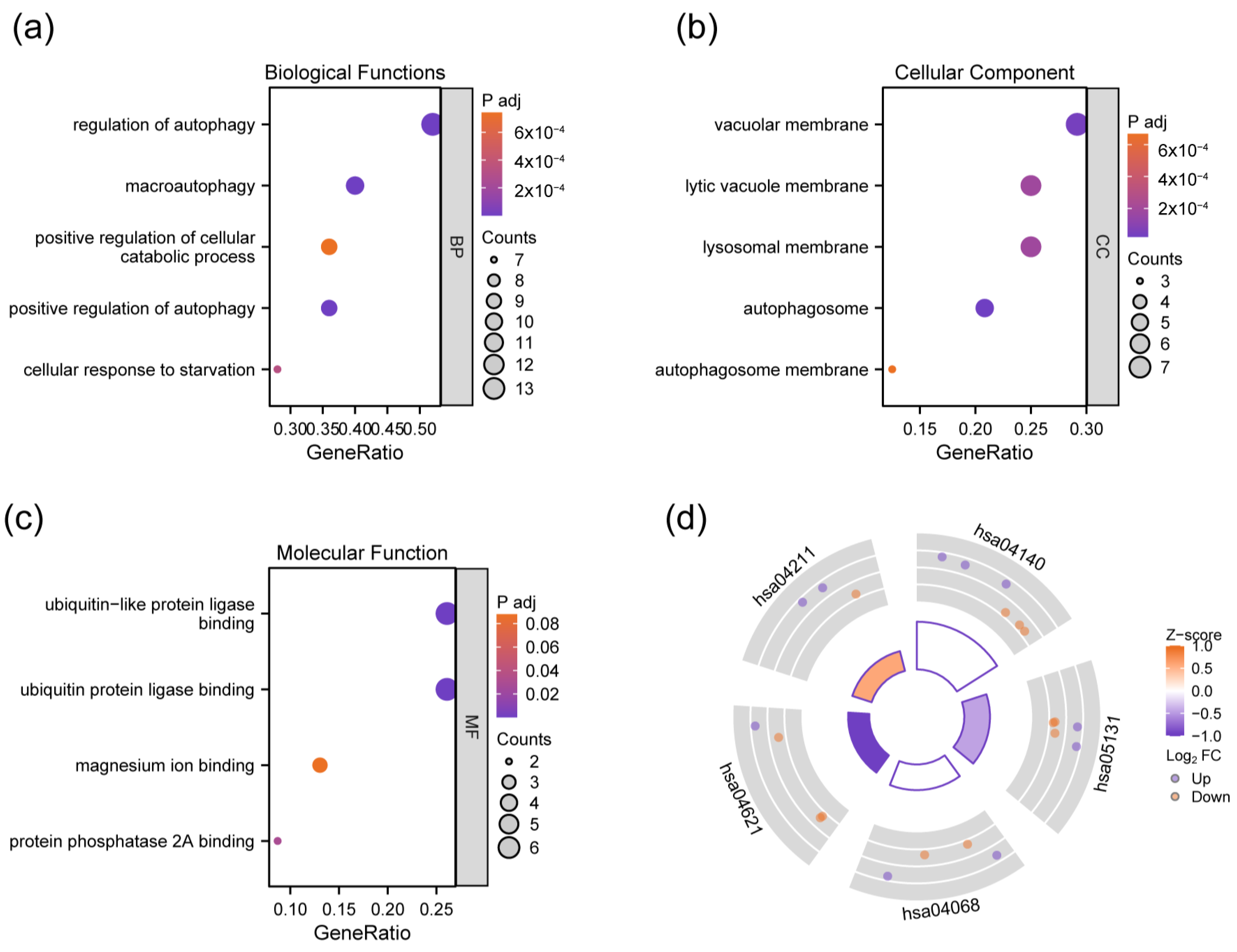
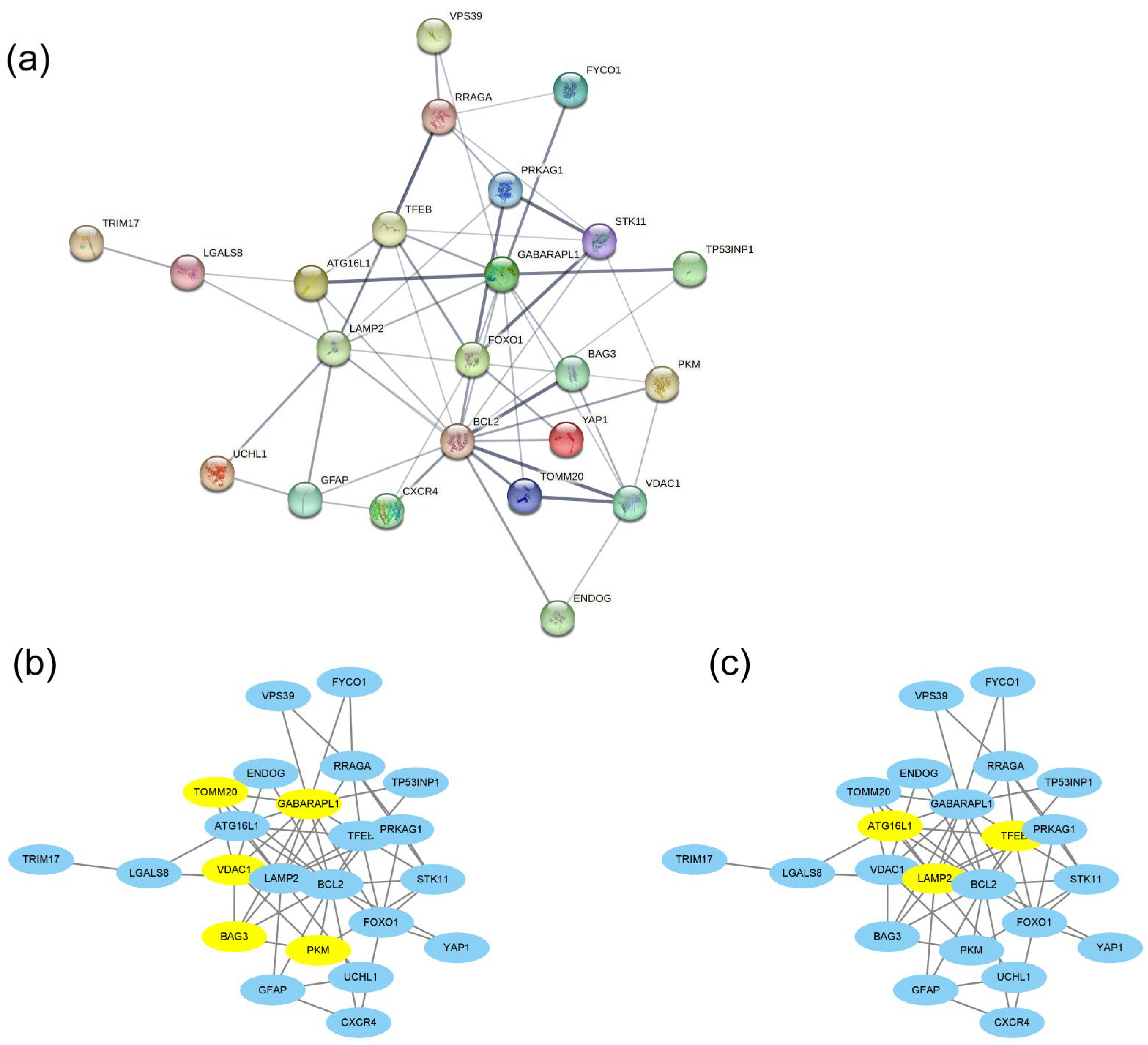


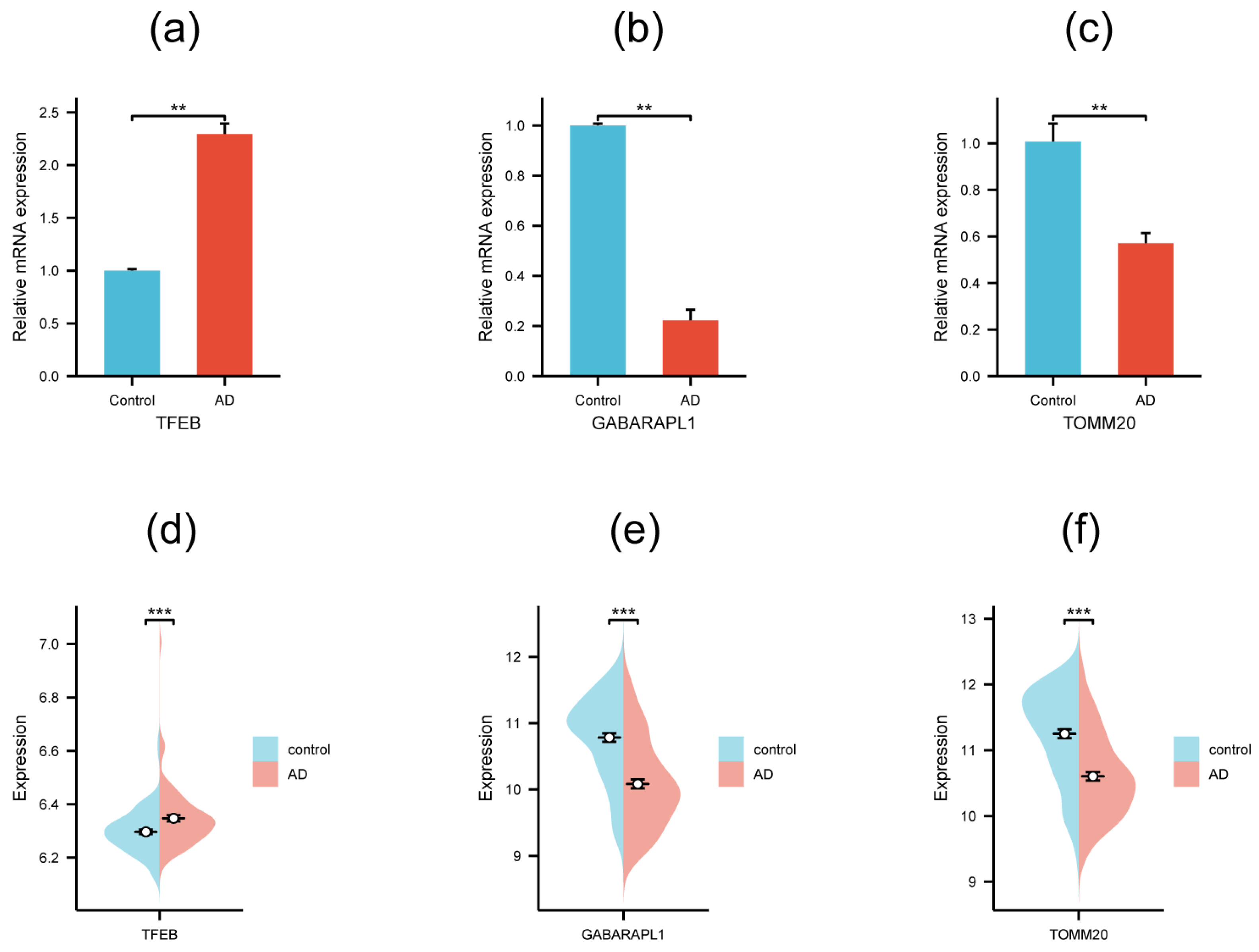
| Gene Symbol | logFC | p.adj | Description |
|---|---|---|---|
| TFEB | 1.80 | 1.75 × 10−14 | Transcription factor EB |
| BAG3 | 1.39 | 2.25 × 10−11 | BAG cochaperone 3 |
| LAMP2 | 1.24 | 5.57 × 10−13 | Lysosomal-associated membrane protein 2 |
| VDAC1 | −1.26 | 3.96 × 10−7 | Voltage-dependent anion channel 1 |
| PKM | −1.06 | 3.05 × 10−5 | Pyruvate kinase M1/2 |
| ATG16L1 | −1.02 | 1.60 × 10−4 | Autophagy-related 16 like 1 |
| TOMM20 | −1.01 | 1.77 × 10−6 | Translocase of outer mitochondrial membrane 20 |
| GABARAPL1 | −1.01 | 1.57 × 10−9 | GABA type A receptor-associated protein-like 1 |
| Gene Symbol | Primer Sequence | |
|---|---|---|
| Forward | Reverse | |
| TFEB | 5′-ACCTGTCCGAGACCTATGGG-3′ | 5′-CGTCCAGACGCATAATGTTGTC-3′ |
| TOMM20 | 5′-GGTACTGCATCTACTTCGACCG-3′ | 5′-TGGTCTACGCCCTTCTCATATTC-3′ |
| GABARAPL1 | 5′-ATGAAGTTCCAGTACAAGGAGGA-3′ | 5′-GCTTTTGGAGCCTTCTCTACAAT-3′ |
| GAPDH | 5′-CCAGCCCAGCAAGGATACTG-3′ | 5′-GGTATTCGAGAGAAGGGAGGGC-3′ |
| Gene Symbol | miRNA Name | Description |
|---|---|---|
| TFEB | hsa-miR-29a-3p | Slightly dysregulated in plasma; potential biomarkers of AD [34]. |
| TFEB | hsa-miR-124-3p | Colocalization with microglia in AD patient hippocampi; reshapes microglia plasticity; relevant with inflammation in AD-associated neurodegeneration [35]. |
| TFEB | hsa-miR-29b-3p | Has diagnostic potential as minimally invasive AD biomarker [36]. |
| TFEB | hsa-miR-29c-3p | Inhibits BACE1 expression; activates the Wnt/β-catenin pathway; plays a therapeutic role in AD [37]. |
| TOMM20 | hsa-let-7a-2-3p | Related to cognitive impairment [38]. |
| TOMM20 | hsa-miR-204-3p | Controls the timing of the dopaminergic differentiation [39]. |
| GABARAPL1 | hsa-miR-195-5p | Upregulated in the AD process [40]. |
| GABARAPL1 | hsa-miR-16-5p | Consistently downregulated in late-stage AD by meta-analysis across the literature [41]. |
| GABARAPL1 | hsa-miR-155-5p | Regulate inflammatory responses in the pathogenesis of AD [42]. |
| GABARAPL1 | hsa-miR-497-5p | The MEM/LINC00094/miR-224-5p (miR-497-5p)/endophilin-1 axis exerts a key role in regulating the BBB permeability in the AD microenvironment [43]. |
| GABARAPL1 | hsa-miR-15b-5p | Involved in platelet reactivity in AD [44]. |
| GABARAPL1 | hsa-miR-143-3p | Upregulated in the serum of AD patients; miR-143-3p inhibition promotes neuronal survival; the miR-143-3p/NRG1 axis is a potential therapeutic target and candidate biomarker for AD [45]. |
| GABARAPL1 | hsa-miR-133b | A novel promising diagnostic biomarker for AD; may have a neuroprotective role in AD and targets EGFR [46]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Su, X.; Qin, J.; Jin, Y.; Zhang, N.; Huang, S. Identification of Autophagy-Related Biomarkers and Diagnostic Model in Alzheimer’s Disease. Genes 2024, 15, 1027. https://doi.org/10.3390/genes15081027
Xu W, Su X, Qin J, Jin Y, Zhang N, Huang S. Identification of Autophagy-Related Biomarkers and Diagnostic Model in Alzheimer’s Disease. Genes. 2024; 15(8):1027. https://doi.org/10.3390/genes15081027
Chicago/Turabian StyleXu, Wei, Xi Su, Jing Qin, Ye Jin, Ning Zhang, and Shasha Huang. 2024. "Identification of Autophagy-Related Biomarkers and Diagnostic Model in Alzheimer’s Disease" Genes 15, no. 8: 1027. https://doi.org/10.3390/genes15081027





