Agouti-Signaling Protein and Melanocortin-1-Receptor Mutations Associated with Coat Color Phenotypes in Fallow Deer (Dama dama)
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. DNA Isolation
2.3. Primer Design
2.4. Sequencing
2.5. Genotyping
3. Results and Discussion
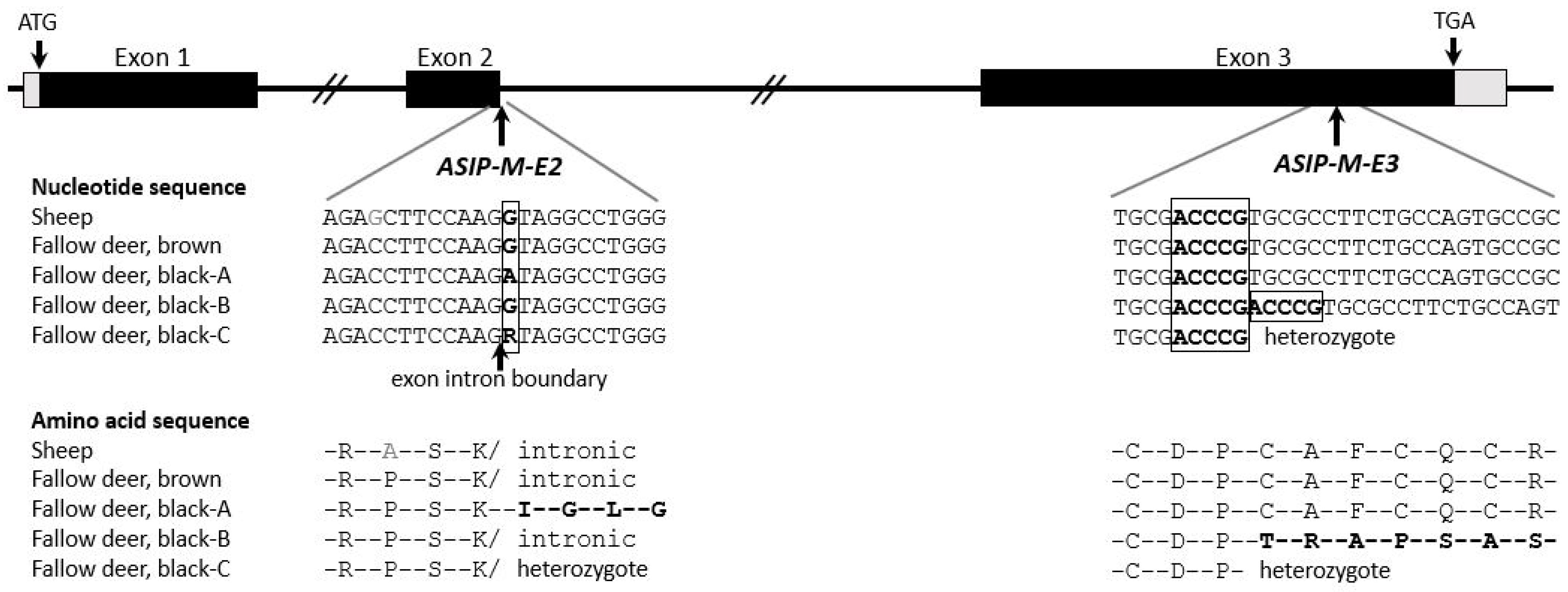
3.1. Agouti-Signaling Protein (ASIP)
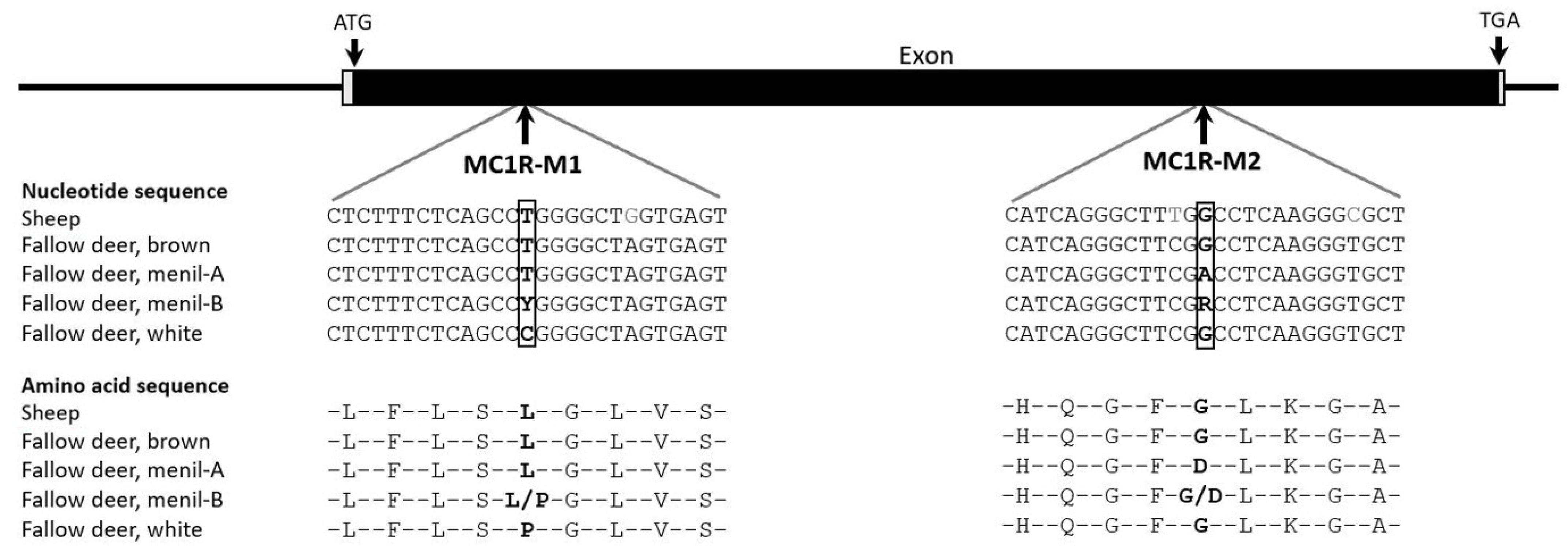
3.2. Melanocortin-1-Receptor (MC1R), Tyrosinase (TYR), and Solute Carrier Family 45 Member 2 (SLC45A2)
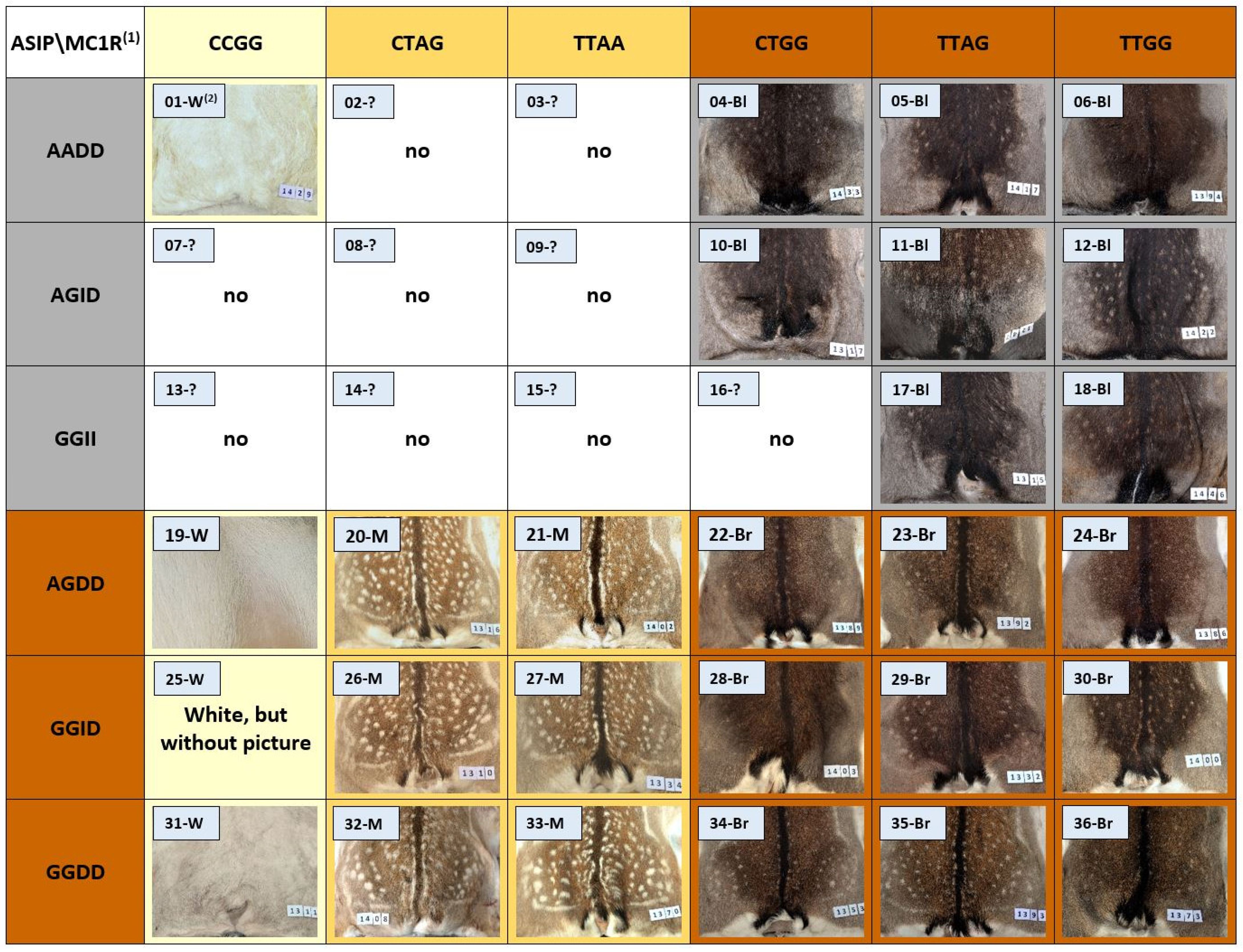
3.3. Combined Effect of ASIP and MC1R Mutations on Coat Coloration
4. Conclusions
- -
- The four mutations found in this study explain the four main colors of fallow deer. There are certainly other mutations in other genes that lead to further variations.
- -
- White fallow deer are neither albinos (pure white with unpigmented eyes and pink skin) nor leucists (pure white, pink skin, but mostly pigmented eyes and occasionally small pigment spots) since they can produce pigments and possess melanocytes.
- -
- Our results demonstrate that diverse MC1R alleles can lead to very different coat colorations, which go far beyond a simple switch from eumelanin to pheomelanin. Previously, it was already concluded that MC1R mutations, depending on their position, can result in a non-functional receptor, a constitutively activated receptor, or a hormonally activated receptor [53].
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klungland, H.; Roed, K.H.; Nesbo, C.L.; Jakobsen, K.S.; Vage, D.I. The melanocyte-stimulating hormone receptor (MC1-R) gene as a tool in evolutionary studies of artiodactyles. Hereditas 1999, 131, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Mundy, N.I.; Kelly, J.; Theron, E.; Hawkins, K. Evolutionary genetics of the melanocortin-1 receptor in vertebrates. Ann. N. Y. Acad. Sci. 2003, 994, 307–312. [Google Scholar] [CrossRef]
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Ludwig, A. Colours of domestication. Biol. Rev. Camb. Philos. Soc. 2011, 86, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, E.; Trindade, F.J. Genetics and Evolution of Mammalian Coat Pigmentation. Annu. Rev. Anim. Biosci. 2021, 9, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Klungland, H.; Vage, D.I. Pigmentary switches in domestic animal species. Ann. N. Y. Acad. Sci. 2003, 994, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Larson, G.; Ribeiro, H.S.; Li, N.; Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009, 5, e1000341. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; Pruvost, M.; Reissmann, M.; Benecke, N.; Brockmann, G.A.; Castanos, P.; Cieslak, M.; Lippold, S.; Llorente, L.; Malaspinas, A.S.; et al. Coat color variation at the beginning of horse domestication. Science 2009, 324, 485. [Google Scholar] [CrossRef] [PubMed]
- Linderholm, A.; Larson, G. The role of humans in facilitating and sustaining coat colour variation in domestic animals. Semin. Cell Dev. Biol. 2013, 24, 587–593. [Google Scholar] [CrossRef]
- Reissmann, M.; Ludwig, A. Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Semin. Cell Dev. Biol. 2013, 24, 576–586. [Google Scholar] [CrossRef]
- Ludwig, A.; Vernesi, C.; Lieckfeldt, D.; Lattenkamp, E.Z.; Wiethölter, A.; Lutz, W. Origin and patterns of genetic diversity of German fallow deer as inferred from mitochondrial DNA. Eur. J. Wildl. Res. 2012, 58, 495–501. [Google Scholar] [CrossRef]
- The British Deer Society. Fallow Deer. Available online: https://bds.org.uk/information-advice/about-deer/deer-species/fallow-deer/ (accessed on 5 May 2024).
- New Zeeland Deerstalkers Association. Fallow Deer. Available online: https://www.deerstalkers.org.nz/resources-2/animals/new-zealands-big-game-animals/fallow-deer/ (accessed on 5 May 2024).
- Reiner, G.; Weber, T.; Nietfeld, F.; Fischer, D.; Wurmser, C.; Fries, R.; Willems, H. A genome-wide scan study identifies a single nucleotide substitution in MC1R gene associated with white coat colour in fallow deer (Dama dama). BMC Genet. 2020, 21, 126. [Google Scholar] [CrossRef] [PubMed]
- Fossil Rim Wildlife Center. Fallow Deer. Available online: https://fossilrim.org/animals/fallow-deer/ (accessed on 5 May 2024).
- Rue, L.L., III. The Deer of North America; Crown Publishers Inc.: New York, NY, USA, 1978; ISBN 978-1592284658. [Google Scholar]
- Rieder, S.; Taourit, S.; Mariat, D.; Langlois, B.; Guerin, G. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm. Genome 2001, 12, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Forestier, L.; Allain, D.; Scotti, E.; Beretti, F.; Deretz-Picoulet, S.; Pecchioli, E.; Vernesi, C.; Robinson, T.J.; Malaney, J.L.; et al. Characterization of the rabbit agouti signaling protein (ASIP) gene: Transcripts and phylogenetic analyses and identification of the causative mutation of the nonagouti black coat colour. Genomics 2010, 95, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Norris, B.J.; Whan, V.A. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, M.; Lutz, W.; Lieckfeldt, D.; Sandoval-Castellanos, E.; Ludwig, A. An Agouti-Signaling-Protein Mutation is Strongly Associated with Melanism in European Roe Deer (Capreolus capreolus). Genes 2020, 11, 647. [Google Scholar] [CrossRef]
- Mariat, D.; Taourit, S.; Guerin, G. A mutation in the MATP gene causes the cream coat colour in the horse. Genet. Sel. Evol. 2003, 35, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Holl, H.M.; Pflug, K.M.; Yates, K.M.; Hoefs-Martin, K.; Shepard, C.; Cook, D.G.; Lafayette, C.; Brooks, S.A. A candidate gene approach identifies variants in SLC45A2 that explain dilute phenotypes, pearl and sunshine, in compound heterozygote horses. Anim. Genet. 2019, 50, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Sevane, N.; Sanz, C.R.; Dunner, S. Explicit evidence for a missense mutation in exon 4 of SLC45A2 gene causing the pearl coat dilution in horses. Anim. Genet. 2019, 50, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Bisbee, D.; Carpenter, M.L.; Hoefs-Martin, K.; Brooks, S.A.; Lafayette, C. Identification of a novel missense variant in SLC45A2 associated with dilute snowdrop phenotype in Gypsy horses. Anim. Genet. 2020, 51, 342–343. [Google Scholar] [CrossRef]
- Xu, X.; Dong, G.X.; Hu, X.S.; Miao, L.; Zhang, X.L.; Zhang, D.L.; Yang, H.D.; Zhang, T.Y.; Zou, Z.T.; Zhang, T.T.; et al. The genetic basis of white tigers. Curr. Biol. 2013, 23, 1031–1035. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Primer3. Version 4.1.0. Available online: https://primer3.ut.ee/ (accessed on 19 April 2022).
- Kreuzer, S.; Reissmann, M.; Brockmann, G.A. Gene test to elucidate the ETEC F4ab/F4ac receptor status in pigs. Vet. Microbiol. 2013, 162, 293–295. [Google Scholar] [CrossRef]
- Miltenberger, R.J.; Wakamatsu, K.; Ito, S.; Woychik, R.P.; Russell, L.B.; Michaud, E.J. Molecular and phenotypic analysis of 25 recessive, homozygous-viable alleles at the mouse agouti locus. Genetics 2002, 160, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Eberle, A.N.; Bodi, J.; Orosz, G.; Suli-Vargha, H.; Jaggin, V.; Zumsteg, U. Antagonist and agonist activities of the mouse agouti protein fragment (91–131) at the melanocortin-1 receptor. J. Recept. Signal Transduct. Res. 2001, 21, 25–45. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Dall’Olio, S.; Gonzalez, E.G.; Finocchiaro, R.; Davoli, R.; Russo, V.; Portolano, B. Missense and nonsense mutations in melanocortin 1 receptor (MC1R) gene of different goat breeds: Association with red and black coat colour phenotypes but with unexpected evidences. BMC Genet. 2009, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Alshanbari, F.; Castaneda, C.; Juras, R.; Hillhouse, A.; Mendoza, M.N.; Gutierrez, G.A.; Ponce de Leon, F.A.; Raudsepp, T. Comparative FISH-Mapping of MC1R, ASIP, and TYRP1 in New and Old World Camelids and Association Analysis With Coat Color Phenotypes in the Dromedary (Camelus dromedarius). Front. Genet. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Hu, M.; Zhu, W.; Yu, F.; Bai, C.; Zhang, J.; Yan, S. A 4-bp deletion in the ASIP gene is associated with the recessive black coat colour in domestic guinea pigs (Cavia porcellus). Anim. Genet. 2019, 50, 190–191. [Google Scholar] [CrossRef]
- Sooriyabandara, M.G.C.; Bandaranayake, A.U.; Hathurusinghe, H.; Jayasundara, S.M.; Marasinghe, M.; Prasad, G.A.T.; Abeywardana, V.; Pinidiya, M.A.; Nilanthi, R.M.R.; Bandaranayake, P.C.G. A unique single nucleotide polymorphism in Agouti Signalling Protein (ASIP) gene changes coat colour of Sri Lankan leopard (Panthera pardus kotiya) to dark black. PLoS ONE 2023, 18, e0269967. [Google Scholar] [CrossRef]
- Trigo, B.B.; Alves, N.F.; Milanesi, M.; Garcia, J.F.; Terefe, E.; Hanotte, O.; Tijjani, A.; Utsunomiya, Y.T. A structural variant at ASIP associated with the darkness of hair coat is found in multiple zebu cattle populations. Anim. Genet. 2023, 54, 544–548. [Google Scholar] [CrossRef]
- Kingsley, E.P.; Manceau, M.; Wiley, C.D.; Hoekstra, H.E. Melanism in peromyscus is caused by independent mutations in agouti. PLoS ONE 2009, 4, e6435. [Google Scholar] [CrossRef]
- Anello, M.; Daverio, M.S.; Rodriguez, S.S.; Romero, S.R.; Renieri, C.; Vidal Rioja, L.; Di Rocco, F. The ASIP gene in the llama (Lama glama): Alternative transcripts, expression and relation with color phenotypes. Gene 2022, 809, 146018. [Google Scholar] [CrossRef] [PubMed]
- Trigo, B.B.; Utsunomiya, A.T.H.; Fortunato, A.; Milanesi, M.; Torrecilha, R.B.P.; Lamb, H.; Nguyen, L.; Ross, E.M.; Hayes, B.; Padula, R.C.M.; et al. Variants at the ASIP locus contribute to coat color darkening in Nellore cattle. Genet. Sel. Evol. 2021, 53, 40. [Google Scholar] [CrossRef]
- Klungland, H.; Vage, D.I.; Gomez-Raya, L.; Adalsteinsson, S.; Lien, S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm. Genome 1995, 6, 636–639. [Google Scholar] [CrossRef]
- Joerg, H.; Fries, H.R.; Meijerink, E.; Stranzinger, G.F. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm. Genome 1996, 7, 317–318. [Google Scholar] [CrossRef]
- Marklund, L.; Moller, M.J.; Sandberg, K.; Andersson, L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome 1996, 7, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Vage, D.I.; Lu, D.; Klungland, H.; Lien, S.; Adalsteinsson, S.; Cone, R.D. A non-epistatic interaction of agouti and extension in the fox, Vulpes vulpes. Nat. Genet. 1997, 15, 311–315. [Google Scholar] [CrossRef]
- Everts, R.E.; Rothuizen, J.; van Oost, B.A. Identification of a premature stop codon in the melanocyte-stimulating hormone receptor gene (MC1R) in Labrador and Golden retrievers with yellow coat colour. Anim. Genet. 2000, 31, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Schioth, H.B.; Phillips, S.R.; Rudzish, R.; Birch-Machin, M.A.; Wikberg, J.E.; Rees, J.L. Loss of function mutations of the human melanocortin 1 receptor are common and are associated with red hair. Biochem. Biophys. Res. Commun. 1999, 260, 488–491. [Google Scholar] [CrossRef]
- Albinism Database. Available online: http://www.ifpcs.org/albinism/index.html (accessed on 29 May 2024).
- Baxter, L.L.; Watkins-Chow, D.E.; Pavan, W.J.; Loftus, S.K. A curated gene list for expanding the horizons of pigmentation biology. Pigment. Cell Melanoma Res. 2019, 32, 348–358. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.; Wu, S.; Ma, T.; Jiang, H.; Zhang, Q. Differential expression of MC1R gene in Liaoning Cashmere goats with different coat colors. Anim. Biotechnol. 2019, 30, 273–278. [Google Scholar] [CrossRef]
- Almathen, F.; Elbir, H.; Bahbahani, H.; Mwacharo, J.; Hanotte, O. Polymorphisms in MC1R and ASIP Genes are Associated with Coat Color Variation in the Arabian Camel. J. Hered. 2018, 109, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Feeley, N.L.; Munyard, K.A. Characterisation of the melanocortin-1 receptor gene in alpaca and identification of possible markers associ-ated with phenotypic variations in colour. Anim. Prod. Sci. 2009, 49, 675–681. [Google Scholar] [CrossRef]
- Gebreselassie, G.; Liang, B.; Berihulay, H.; Islam, R.; Abied, A.; Jiang, L.; Zhao, Z.; Ma, Y. Genomic mapping identifies two genetic variants in the MC1R gene for coat colour variation in Chinese Tan sheep. PLoS ONE 2020, 15, e0235426. [Google Scholar] [CrossRef] [PubMed]
- Hepp, D.; Goncalves, G.L.; Moreira, G.R.; de Freitas, T.R. Epistatic Interaction of the Melanocortin 1 Receptor and Agouti Signaling Protein Genes Modulates Wool Color in the Brazilian Creole Sheep. J. Hered. 2016, 107, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Rochus, C.M.; Westberg Sunesson, K.; Jonas, E.; Mikko, S.; Johansson, A.M. Mutations in ASIP and MC1R: Dominant black and recessive black alleles segregate in native Swedish sheep populations. Anim. Genet. 2019, 50, 712–717. [Google Scholar] [CrossRef]
- Poronui. Fallow Deer. Available online: https://www.poronuihunting.com/fallow-multicolored-camouflage-experts/ (accessed on 5 May 2024).
- Graphodatskaya, D.; Joerg, H.; Stranzinger, G. Molecular and pharmacological characterisation of the MSH-R alleles in Swiss cattle breeds. J. Recept. Signal Transduct. Res. 2002, 22, 421–430. [Google Scholar] [CrossRef]

| Color | Nöbeln | Hunted | Hirschaue | Sum | |||
|---|---|---|---|---|---|---|---|
| Bucks | Old Does | Fawns | Purchase | ||||
| Black | 1 | – | 42 | 8 | – | 11 | 62 |
| Brown | 13 | 14 | 502 | 75 | 17 | 151 | 772 |
| Menil | 3 | – | 90 | 4 | – | 29 | 126 |
| White | – | – | 36 | – | – | 2 | 38 |
| Sum | 17 | 14 | 670 | 87 | 17 | 193 | 998 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reissmann, M.; Ullrich, E.; Bergfeld, U.; Ludwig, A. Agouti-Signaling Protein and Melanocortin-1-Receptor Mutations Associated with Coat Color Phenotypes in Fallow Deer (Dama dama). Genes 2024, 15, 1055. https://doi.org/10.3390/genes15081055
Reissmann M, Ullrich E, Bergfeld U, Ludwig A. Agouti-Signaling Protein and Melanocortin-1-Receptor Mutations Associated with Coat Color Phenotypes in Fallow Deer (Dama dama). Genes. 2024; 15(8):1055. https://doi.org/10.3390/genes15081055
Chicago/Turabian StyleReissmann, Monika, Evelin Ullrich, Uwe Bergfeld, and Arne Ludwig. 2024. "Agouti-Signaling Protein and Melanocortin-1-Receptor Mutations Associated with Coat Color Phenotypes in Fallow Deer (Dama dama)" Genes 15, no. 8: 1055. https://doi.org/10.3390/genes15081055
APA StyleReissmann, M., Ullrich, E., Bergfeld, U., & Ludwig, A. (2024). Agouti-Signaling Protein and Melanocortin-1-Receptor Mutations Associated with Coat Color Phenotypes in Fallow Deer (Dama dama). Genes, 15(8), 1055. https://doi.org/10.3390/genes15081055







