Key Genes FECH and ALAS2 under Acute High-Altitude Exposure: A Gene Expression and Network Analysis Based on Expression Profile Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preliminary Processing
2.2. Collection and Preliminary Processing of External Datasets
2.3. Identification of Differentially Expressed Genes
2.4. Functional Annotation and Enrichment Analysis
2.5. Construction and Validation of Protein–Protein Interaction Network
3. Results
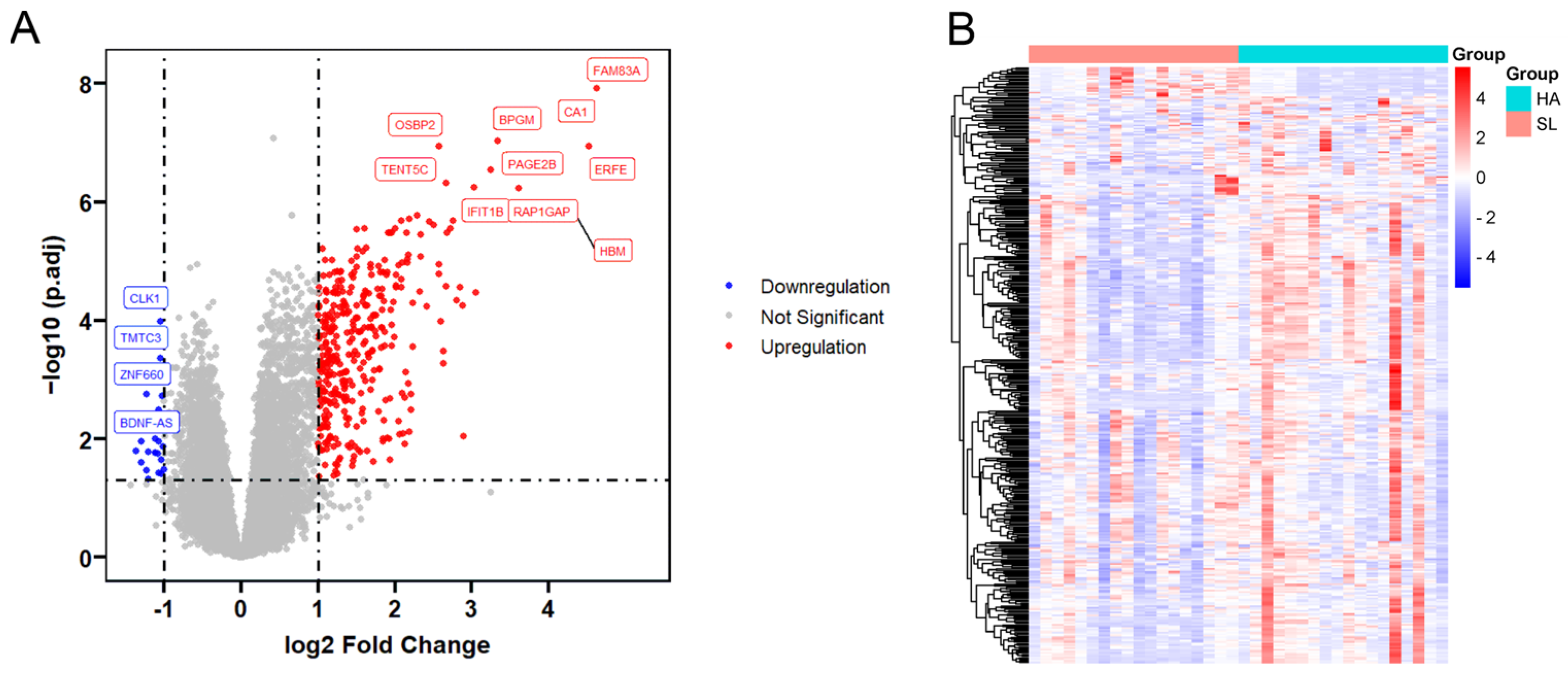
3.1. Identification of Differentially Expressed mRNAs
3.2. GO and KEGG Enrichment Analysis
3.3. Significant Differentially Expressed Genes
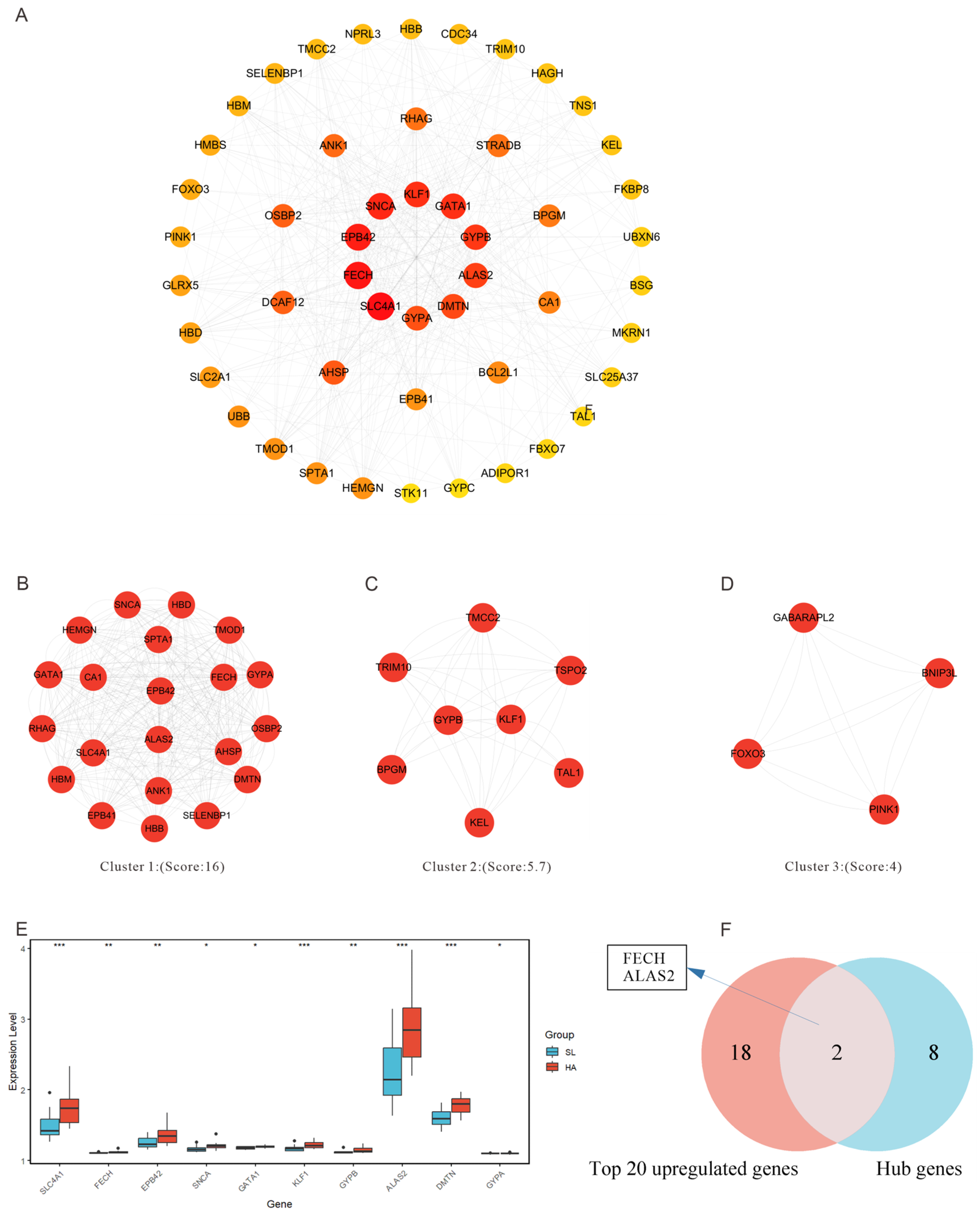
3.4. Analysis of Protein Interaction Networks in Key Pathways
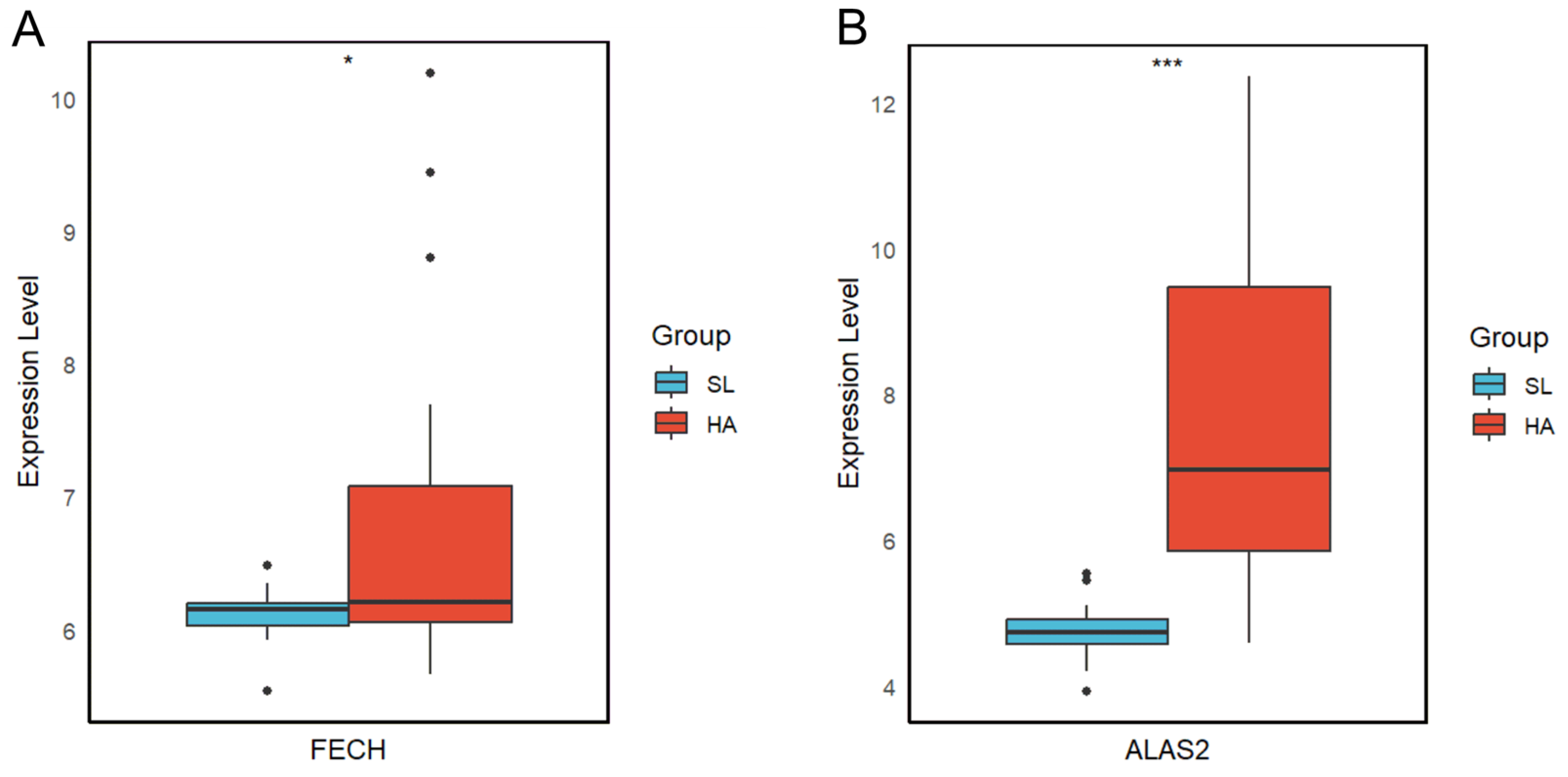
3.5. Candidate Targets for High-Altitude Hypoxia Acclimatization
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bärtsch, P.; Swenson, E.R. Clinical practice: Acute high-altitude illnesses. N. Engl. J. Med. 2013, 368, 2294–2302. [Google Scholar] [CrossRef] [PubMed]
- Savioli, G.; Ceresa, I.F.; Gori, G.; Fumoso, F.; Gri, N.; Floris, V.; Varesi, A.; Martuscelli, E.; Marchisio, S.; Longhitano, Y.; et al. Pathophysiology and Therapy of High-Altitude Sickness: Practical Approach in Emergency and Critical Care. J. Clin. Med. 2022, 11, 3937. [Google Scholar] [CrossRef] [PubMed]
- Luks, A.M.; Swenson, E.R.; Bärtsch, P. Acute high-altitude sickness. Eur. Respir. Rev. 2017, 26, 160096. [Google Scholar] [CrossRef] [PubMed]
- San, T.; Polat, S.; Cingi, C.; Eskiizmir, G.; Oghan, F.; Cakir, B. Effects of high altitude on sleep and respiratory system and theirs adaptations. Sci. World J. 2013, 2013, 241569. [Google Scholar] [CrossRef] [PubMed]
- Naeije, R. Physiological adaptation of the cardiovascular system to high altitude. Prog. Cardiovasc. Dis. 2010, 52, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Richalet, J.P.; Hermand, E.; Lhuissier, F.J. Cardiovascular physiology and pathophysiology at high altitude. Nat. Rev. Cardiol. 2024, 21, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, M.; Mairbäurl, H.; Livshits, L.; Seide, S.; Hackbusch, M.; Malczyk, M.; Kraut, S.; Gassmann, N.N.; Weissmann, N.; Muckenthaler, M.U. The increase in hemoglobin concentration with altitude varies among human populations. Ann. N. Y. Acad. Sci. 2019, 1450, 204–220. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.; Noviana, M.; Hou, M. Nitric oxide in red blood cell adaptation to hypoxia. Acta Biochim. Biophys. Sin. 2018, 50, 621–634. [Google Scholar] [CrossRef]
- Masson, N.; Willam, C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001, 20, 5197–5206. [Google Scholar] [CrossRef]
- Azad, P.; Caldwell, A.B.; Ramachandran, S.; Spann, N.J.; Akbari, A.; Villafuerte, F.C.; Bermudez, D.; Zhao, H.; Poulsen, O.; Zhou, D.; et al. ARID1B, a molecular suppressor of erythropoiesis, is essential for the prevention of Monge’s disease. Exp. Mol. Med. 2022, 54, 777–787. [Google Scholar] [CrossRef]
- Weidemann, A.; Johnson, R.S. Biology of HIF-1alpha. Cell Death Differ. 2008, 15, 621–627. [Google Scholar] [CrossRef]
- Liu, F.; Hu, C.; Ding, J.; Fu, C.; Wang, S.; Li, T. GATA-1 Promotes Erythroid Differentiation Through the Upregulation of miR-451a and miR-210-3p Expressions in CD34(+) Cells in High-Altitude Polycythemia. High. Alt. Med. Biol. 2023, 24, 59–67. [Google Scholar] [CrossRef]
- Taylor, C.T. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem. J. 2008, 409, 19–26. [Google Scholar] [CrossRef]
- Gaur, P.; Saini, S.; Ray, K.; Asanbekovna, K.N.; Akunov, A.; Maripov, A.; Sarybaev, A.; Singh, S.B.; Kumar, B.; Vats, P. Temporal transcriptome analysis suggest modulation of multiple pathways and gene network involved in cell-cell interaction during early phase of high altitude exposure. PLoS ONE 2020, 15, e0238117. [Google Scholar] [CrossRef]
- Yasukochi, Y.; Shin, S.; Wakabayashi, H.; Maeda, T. Transcriptomic Changes in Young Japanese Males After Exposure to Acute Hypobaric Hypoxia. Front. Genet. 2020, 11, 559074. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, J.; Hohenauer, E.; Burtscher, M.; Millet, G.P.; Egg, M. Environmental and behavioral regulation of HIF-mitochondria crosstalk. Free Radic. Biol. Med. 2023, 206, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Varshney, R.; Sethy, N.K. Human adaptation to high altitude: A review of convergence between genomic and proteomic signatures. Hum. Genom. 2022, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhang, H.; Jiang, Y. Methazolamide in high-altitude illnesses. Eur. J. Pharm. Sci. 2020, 148, 105326. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, C.; Zhang, Y.; Dzieciatkowska, M.; Brown, B.C.; Zhang, W.; Xie, T.; Abdulmalik, O.; Song, A.; Tong, C.; et al. Erythrocyte transglutaminase-2 combats hypoxia and chronic kidney disease by promoting oxygen delivery and carnitine homeostasis. Cell Metab. 2022, 34, 299–316.e6. [Google Scholar] [CrossRef]
- Kautz, L.; Jung, G.; Valore, E.V.; Rivella, S.; Nemeth, E.; Ganz, T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat. Genet. 2014, 46, 678–684. [Google Scholar] [CrossRef]
- Kautz, L.; Jung, G.; Du, X.; Gabayan, V.; Chapman, J.; Nasoff, M.; Nemeth, E.; Ganz, T. Erythroferrone contributes to hepcidin suppression and iron overload in a mouse model of β-thalassemia. Blood 2015, 126, 2031–2037. [Google Scholar] [CrossRef] [PubMed]
- Peacock, A.J. ABC of oxygen: Oxygen at high altitude. BMJ 1998, 317, 1063–1066. [Google Scholar] [CrossRef]
- Sarkar, M.; Niranjan, N.; Banyal, P.K. Mechanisms of hypoxemia. Lung India 2017, 34, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.A.; Sun, Y.; Yuan, Z.; Wang, L.; Dong, Q.; Zhou, Y.; Zheng, G.; Aschner, M.; Zou, Y.; Luo, W. Insight into the Effects of High-Altitude Hypoxic Exposure on Learning and Memory. Oxid. Med. Cell Longev. 2022, 2022, 4163188. [Google Scholar] [CrossRef] [PubMed]
- West, J.B. High-altitude medicine. Am. J. Respir. Crit. Care Med. 2012, 186, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Linqing, L.; Yuhan, Q.; Erfei, L.; Yong, Q.; Dong, W.; Chengchun, T.; Gaoliang, Y.; Bo, L. Hypoxia-induced PINK1/Parkin-mediated mitophagy promotes pulmonary vascular remodeling. Biochem. Biophys. Res. Commun. 2021, 534, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Saraji, A.; Sydykov, A.; Schäfer, K.; Garcia-Castro, C.F.; Henneke, I.; Alebrahimdehkordi, N.; Kosanovic, D.; Hadzic, S.; Guenther, A.; Hecker, M.; et al. PINK1-mediated Mitophagy Contributes to Pulmonary Vascular Remodeling in Pulmonary Hypertension. Am. J. Respir. Cell Mol. Biol. 2021, 65, 226–228. [Google Scholar] [CrossRef]
- Greene, A.W.; Grenier, K.; Aguileta, M.A.; Muise, S.; Farazifard, R.; Haque, M.E.; McBride, H.M.; Park, D.S.; Fon, E.A. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012, 13, 378–385. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Zhang, J.; Gao, H.; Wang, S.; Li, S.; Wei, P.; Liang, J.; Yu, G.; Wang, X.; et al. Cul4A-DDB1-mediated monoubiquitination of phosphoglycerate dehydrogenase promotes colorectal cancer metastasis via increased S-adenosylmethionine. J. Clin. Investig. 2021, 131, e146187. [Google Scholar] [CrossRef]
- Pan, S.; Fan, M.; Liu, Z.; Li, X.; Wang, H. Serine, glycine and one-carbon metabolism in cancer (Review). Int. J. Oncol. 2021, 58, 158–170. [Google Scholar] [CrossRef]
- Kimura, H.; Hamasaki, N.; Yamamoto, M.; Tomonaga, M. Circulation of red blood cells having high levels of 2,3-bisphosphoglycerate protects rat brain from ischemic metabolic changes during hemodilution. Stroke 1995, 26, 1431–1436; discussion 1436–1437. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.M.; Klier, M.; Deitmer, J.W. Carbonic anhydrases and their interplay with acid/base-coupled membrane transporters. Subcell. Biochem. 2014, 75, 105–134. [Google Scholar] [CrossRef]
- Seifter, J.L.; Chang, H.Y. Extracellular Acid-Base Balance and Ion Transport Between Body Fluid Compartments. Physiology 2017, 32, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Kalli, A.C.; Reithmeier, R.A.F. Interaction of the human erythrocyte Band 3 anion exchanger 1 (AE1, SLC4A1) with lipids and glycophorin A: Molecular organization of the Wright (Wr) blood group antigen. PLoS Comput. Biol. 2018, 14, e1006284. [Google Scholar] [CrossRef] [PubMed]
- Faggiano, S.; Ronda, L.; Bruno, S.; Abbruzzetti, S.; Viappiani, C.; Bettati, S.; Mozzarelli, A. From hemoglobin allostery to hemoglobin-based oxygen carriers. Mol. Aspects Med. 2022, 84, 101050. [Google Scholar] [CrossRef] [PubMed]
- Dzierzak, E.; Philipsen, S. Erythropoiesis: Development and differentiation. Cold Spring Harb. Perspect. Med. 2013, 3, a011601. [Google Scholar] [CrossRef] [PubMed]
- Piscopo, M.; Notariale, R.; Tortora, F.; Lettieri, G.; Palumbo, G.; Manna, C. Novel Insights into Mercury Effects on Hemoglobin and Membrane Proteins in Human Erythrocytes. Molecules 2020, 25, 3278. [Google Scholar] [CrossRef]
- Ludwig, L.S.; Lareau, C.A.; Bao, E.L.; Nandakumar, S.K.; Muus, C.; Ulirsch, J.C.; Chowdhary, K.; Buenrostro, J.D.; Mohandas, N.; An, X.; et al. Transcriptional States and Chromatin Accessibility Underlying Human Erythropoiesis. Cell Rep. 2019, 27, 3228–3240.e7. [Google Scholar] [CrossRef]
- Vinjamur, D.S.; Alhashem, Y.N.; Mohamad, S.F.; Amin, P.; Williams, D.C., Jr.; Lloyd, J.A. Krüppel-Like Transcription Factor KLF1 Is Required for Optimal γ- and β-Globin Expression in Human Fetal Erythroblasts. PLoS ONE 2016, 11, e0146802. [Google Scholar] [CrossRef]
- Gnanapragasam, M.N.; Planutis, A.; Glassberg, J.A.; Bieker, J.J. Identification of a genomic DNA sequence that quantitatively modulates KLF1 transcription factor expression in differentiating human hematopoietic cells. Sci. Rep. 2023, 13, 7589. [Google Scholar] [CrossRef]
- Stojanovski, B.M.; Hunter, G.A.; Na, I.; Uversky, V.N.; Jiang, R.H.Y.; Ferreira, G.C. 5-Aminolevulinate synthase catalysis: The catcher in heme biosynthesis. Mol. Genet. Metab. 2019, 128, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, Y.; Ren, Z.; Yi, L. Delta-aminolevulinate synthase 2 polymorphism is associated with maximal oxygen uptake after Living-high exercise-high training-low in a male Chinese population. Int. J. Clin. Exp. Med. 2015, 8, 21617–21622. [Google Scholar]
- Tanimura, N.; Miller, E.; Igarashi, K.; Yang, D.; Burstyn, J.N.; Dewey, C.N.; Bresnick, E.H. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2016, 17, 249–265. [Google Scholar] [CrossRef]
- Doty, R.T.; Yan, X.; Lausted, C.; Munday, A.D.; Yang, Z.; Yi, D.; Jabbari, N.; Liu, L.; Keel, S.B.; Tian, Q.; et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood 2019, 133, 457–469. [Google Scholar] [CrossRef]
- Obi, C.D.; Bhuiyan, T.; Dailey, H.A.; Medlock, A.E. Ferrochelatase: Mapping the Intersection of Iron and Porphyrin Metabolism in the Mitochondria. Front. Cell Dev. Biol. 2022, 10, 894591. [Google Scholar] [CrossRef]
- Medlock, A.E.; Shiferaw, M.T.; Marcero, J.R.; Vashisht, A.A.; Wohlschlegel, J.A.; Phillips, J.D.; Dailey, H.A. Identification of the Mitochondrial Heme Metabolism Complex. PLoS ONE 2015, 10, e0135896. [Google Scholar] [CrossRef] [PubMed]
- Medlock, A.E.; Dailey, H.A. New Avenues of Heme Synthesis Regulation. Int. J. Mol. Sci. 2022, 23, 7467. [Google Scholar] [CrossRef] [PubMed]
- Obi, C.D.; Dailey, H.A.; Jami-Alahmadi, Y.; Wohlschlegel, J.A.; Medlock, A.E. Proteomic Analysis of Ferrochelatase Interactome in Erythroid and Non-Erythroid Cells. Life 2023, 13, 577. [Google Scholar] [CrossRef]
- Saunders, P.U.; Garvican-Lewis, L.A.; Schmidt, W.F.; Gore, C.J. Relationship between changes in haemoglobin mass and maximal oxygen uptake after hypoxic exposure. Br. J. Sports Med. 2013, 47 (Suppl. S1), i26–i30. [Google Scholar] [CrossRef]
- Siqués, P.; Brito, J.; León-Velarde, F.; Barrios, L.; De La Cruz, J.J.; López, V.; Herruzo, R. Hematological and lipid profile changes in sea-level natives after exposure to 3550-m altitude for 8 months. High Alt. Med. Biol. 2007, 8, 286–295. [Google Scholar] [CrossRef]
- Akunov, A.; Sydykov, A.; Toktash, T.; Doolotova, A.; Sarybaev, A. Hemoglobin Changes After Long-Term Intermittent Work at High Altitude. Front. Physiol. 2018, 9, 1552. [Google Scholar] [CrossRef] [PubMed]
- Keel, S.B.; Doty, R.T.; Yang, Z.; Quigley, J.G.; Chen, J.; Knoblaugh, S.; Kingsley, P.D.; De Domenico, I.; Vaughn, M.B.; Kaplan, J.; et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 2008, 319, 825–828. [Google Scholar] [CrossRef] [PubMed]



| Dataset | Age * | n | Height * | Weight * | SBP * | DBP * | HR * | SpO2 * | Smoker/Drinker |
|---|---|---|---|---|---|---|---|---|---|
| GSE133702 | 19–23 | 30 | — | — | — | — | — | — | No |
| GSE103940 | 21.6 ± 2.0 | 53 | 172 ± 1.0 | 64.9 ± 2.3 | — | — | — | — | — |
| GSE75665 | 20–23 | 10 | — | — | 116 ± 15.0 | 58 ± 5.7 | 65 ± 10.1 | 97 ± 1.0 | — |
| total number of participants | — | 93 | — | — | — | — | — | — |
| Gene Name | Description | Expression Level * | Degree |
|---|---|---|---|
| SLC4A1 | solute carrier family 4 member 1 (Diego blood group) | Up | 94 |
| FECH | ferrochelatase | Up | 90 |
| EPB42 | erythrocyte membrane protein band 4.2 | Up | 86 |
| SNCA | synuclein α | Up | 84 |
| GATA1 | GATA binding protein 1 | Up | 76 |
| KLF1 | KLF transcription factor 1 | Up | 76 |
| GYPB | glycophorin B (MNS blood group) | Up | 74 |
| ALAS2 | 5′-aminolevulinate synthase 2 | Up | 72 |
| DMTN | dematin actin binding protein | Up | 70 |
| GYPA | glycophorin A (MNS blood group) | Up | 68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zhu, L.; Shi, D.; Gao, J.; Fan, M. Key Genes FECH and ALAS2 under Acute High-Altitude Exposure: A Gene Expression and Network Analysis Based on Expression Profile Data. Genes 2024, 15, 1075. https://doi.org/10.3390/genes15081075
Zhao Y, Zhu L, Shi D, Gao J, Fan M. Key Genes FECH and ALAS2 under Acute High-Altitude Exposure: A Gene Expression and Network Analysis Based on Expression Profile Data. Genes. 2024; 15(8):1075. https://doi.org/10.3390/genes15081075
Chicago/Turabian StyleZhao, Yifan, Lingling Zhu, Dawei Shi, Jiayue Gao, and Ming Fan. 2024. "Key Genes FECH and ALAS2 under Acute High-Altitude Exposure: A Gene Expression and Network Analysis Based on Expression Profile Data" Genes 15, no. 8: 1075. https://doi.org/10.3390/genes15081075
APA StyleZhao, Y., Zhu, L., Shi, D., Gao, J., & Fan, M. (2024). Key Genes FECH and ALAS2 under Acute High-Altitude Exposure: A Gene Expression and Network Analysis Based on Expression Profile Data. Genes, 15(8), 1075. https://doi.org/10.3390/genes15081075






