Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects
Abstract
:1. Introduction
2. Identification and Selection of Literature
3. Process of Oil Synthesis and Function of Related Genes
4. Key Enzymes Affecting Carbon Source Distribution
4.1. Phosphoenolpyruvate Carboxylase, PEPCase
4.2. Pyruvate Dehydrogenase Complex, PDHC
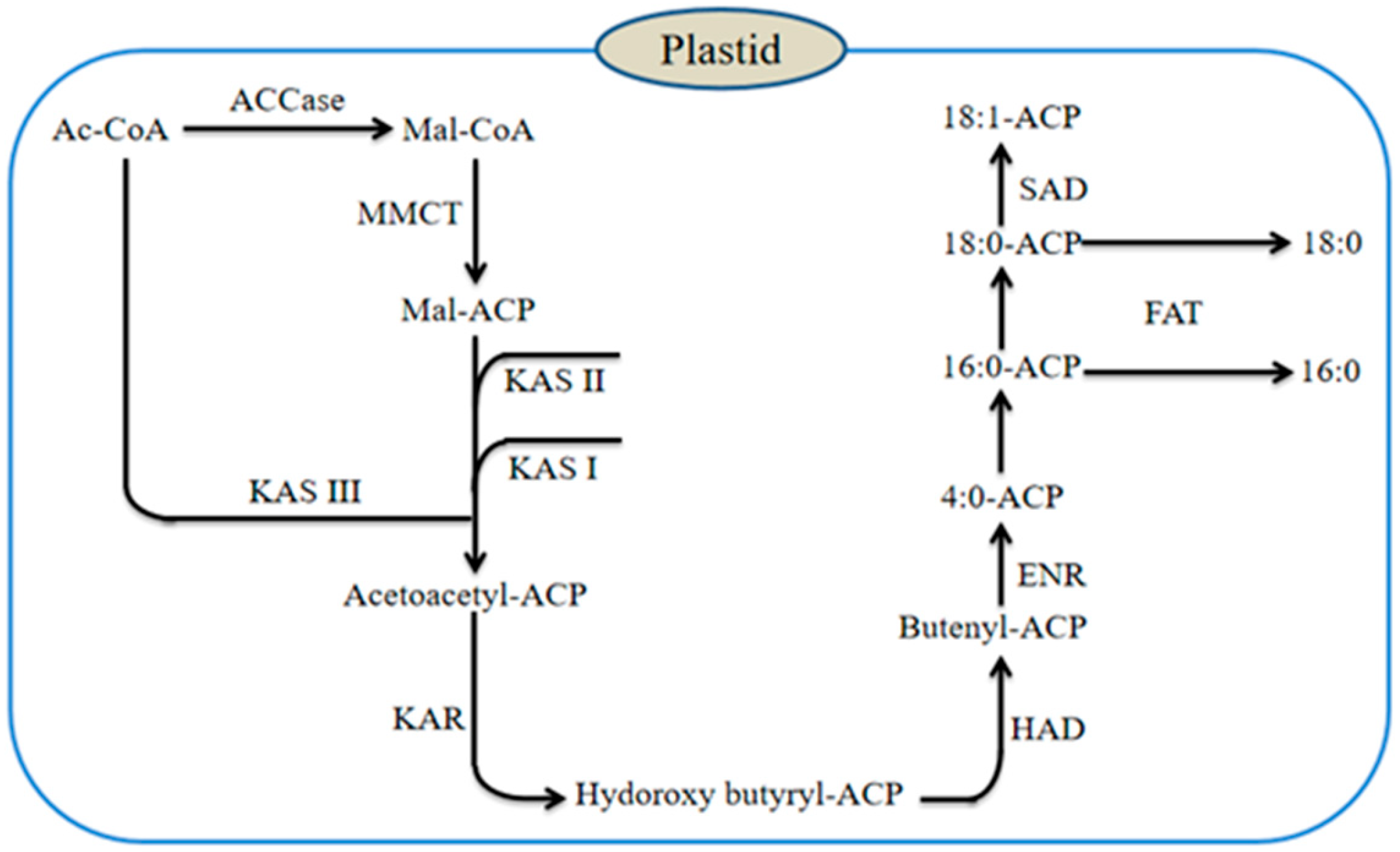
5. Key Enzymes in De Novo Fatty Acid Synthesis
5.1. Acetyl-CoA Carboxylase, ACCase
5.2. Key Enzymes for Carbon Chain Extension
5.2.1. Ketoacyl-ACP Synthase, KAS
5.2.2. Hydroxyacyl-ACP Dehydrase (HAD), Enoyl-ACP Reductase (ENR), β-Ketoacyl-ACP Reductase (KAR)
5.3. Fatty Acid Desaturase
5.4. Diacylglycerol Acyltransferase (DGAT)
5.5. Acyl-ACP Thioesterase, FAT
6. Transcription Factors in the Biosynthesis of Triacylglycerol
6.1. Leaf Cotyledon (LEC)
6.2. WRINKLED1 (WRI)
6.3. DNA Binding with One Finger (Dof)
7. The Strategies for Improving Oil Content through Precursor Transport and Multi-Gene Co-Regulation Involve Several Key Aspects
7.1. Transport of Triacylglycerol Precursors between Organelles
7.2. Multi-Gene Co-Regulation Based on Fatty Acid and Triglyceride Synthesis
8. Discussion
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lin, S.; Zheng, B.; Cheung, P.C.K. Short-chain fatty acids in control of energy metabolism. Crit. Rev. Food Sci. Nutr. 2018, 58, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qin, C.X.; Wang, X.; Ding, N.Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, H.; Wang, Z.; Fu, Y.; Chen, Y.; Wang, X. Enzymatic synthesis of branched chain fatty acid-enriched structured triacylglycerols via esterification with glycerol. Food Chem. 2023, 429, 136943. [Google Scholar] [CrossRef]
- Ziętek, M.; Celewicz, Z.; Szczuko, M. Short-Chain Fatty Acids, Maternal Microbiota and Metabolism in Pregnancy. Nutrients 2021, 13, 1244. [Google Scholar] [CrossRef]
- Duarte Lau, F.; Giugliano, R.P. Adenosine Triphosphate Citrate Lyase and Fatty Acid Synthesis Inhibition: A Narrative Review. JAMA Cardiol. 2023, 8, 879–887. [Google Scholar] [CrossRef]
- Worthmann, A.; Ridder, J.; Piel, S.Y.L.; Evangelakos, I.; Musfeldt, M.; Voß, H.; O’Farrell, M.; Fischer, A.W.; Adak, S.; Sundd, M.; et al. Fatty acid synthesis suppresses dietary polyunsaturated fatty acid use. Nat. Commun. 2024, 15, 45. [Google Scholar] [CrossRef]
- Yu, S.; Song, J.H.; Kim, H.S.; Hong, S.; Park, S.K.; Park, S.H.; Lee, J.; Chae, Y.C.; Park, J.H.; Lee, Y.G. Patulin alleviates hepatic lipid accumulation by regulating lipogenesis and mitochondrial respiration. Life Sci. 2023, 326, 121816. [Google Scholar] [CrossRef]
- Zhukov, A.; Popov, V. Synthesis of C(20-38) Fatty Acids in Plant Tissues. Int. J. Mol. Sci. 2022, 23, 4731. [Google Scholar] [CrossRef]
- Lung, S.C.; Chye, M.L. Arabidopsis acyl-CoA-binding proteins regulate the synthesis of lipid signals. New Phytol. 2019, 223, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Woodfield, H.K.; Cazenave-Gassiot, A.; Haslam, R.P.; Guschina, I.A.; Wenk, M.R.; Harwood, J.L. Using lipidomics to reveal details of lipid accumulation in developing seeds from oilseed rape (Brassica napus L.). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Rahman, A.; Ahmad, N.; Imran, M.; Hafeez, Z. Understanding the Role of Polyunsaturated Fatty Acids in the Development and Prevention of Cancer. Cancer Treat. Res. 2024, 191, 57–93. [Google Scholar] [PubMed]
- Kaur Dhaliwal, L.; Gannaban, R.B.; Shrestha, A.; Shim, J.; Kaur Mangat, P.; Singleton, J.J.; Angeles-Shim, R.B. Integrated morpho-biochemical and transcriptome analyses reveal multidimensional response of upland cotton (Gossypium hirsutum L.) to low temperature stress during seedling establishment. Plant Environ. Interact. 2021, 2, 290–302. [Google Scholar] [CrossRef]
- Comba, A.; Lin, Y.H.; Eynard, A.R.; Valentich, M.A.; Fernandez-Zapico, M.E.; Pasqualini, M.E. Basic aspects of tumor cell fatty acid-regulated signaling and transcription factors. Cancer Metastasis Rev. 2011, 30, 325–342. [Google Scholar] [CrossRef]
- Guyomarc’h, S.; Boutté, Y.; Laplaze, L. AP2/ERF transcription factors orchestrate very long chain fatty acid biosynthesis during Arabidopsis lateral root development. Mol. Plant 2021, 14, 205–207. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Kong, Q.; Lim, A.R.Q.; Lu, S.P.; Zhao, H.; Guo, L.; Yuan, L.; Ma, W. Transcriptional regulation of oil biosynthesis in seed plants: Current understanding, applications, and perspectives. Plant Commun. 2022, 12, 100328. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, W.; Lai, Y.; Zhang, B.; Zhang, D. Edible Plant Oil: Global Status, Health Issues, and Perspectives. Front. Plant Sci. 2020, 11, 1315. [Google Scholar] [CrossRef]
- Valente, F.X.; Cândido, F.G.; Lopes, L.L.; Dias, D.M.; Carvalho, S.D.; Pereira, P.F.; Bressan, J. Effects of coconut oil consumption on energy metabolism, cardiometabolic risk markers, and appetitive responses in women with excess body fat. Eur. J. Nutr. 2018, 57, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Nowinski, S.M.; Solmonson, A.; Rusin, S.F.; Maschek, J.A.; Bensard, C.L.; Fogarty, S.; Jeong, M.Y.; Lettlova, S.; Berg, J.A.; Morgan, J.T.; et al. Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria. eLife 2020, 9, e58041. [Google Scholar] [CrossRef]
- Sagun, J.V.; Yadav, U.P.; Alonso, A.P. Progress in understanding and improving oil content and quality in seeds. Front. Plant Sci. 2023, 14, 1116894. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Phillips, G.N., Jr.; Thorson, J.S. The structural biology of enzymes involved in natural product glycosylation. Nat. Prod. Rep. 2012, 29, 1201–1237. [Google Scholar] [CrossRef]
- Li, X.R.; Wang, L.; Ruan, Y.L. Developmental and molecular physiological evidence for the role of phosphoenolpyruvate carboxylase in rapid cotton fibre elongation. J. Exp. Bot. 2010, 61, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.P.; Li, J.W.; Guo, X.P. Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 2016, 6, 33342–33355. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Lernmark, U.; Gardeström, P. Activity of the mitochondrial pyruvate dehydrogenase complex in plants is stimulated in the presence of malate. Mitochondrion 2014, 19 Pt B, 184–190. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, H.; Cui, X.; Gao, C.; Li, N.; Pu, Y.; Zhang, X.; Zhao, J. Integrated lipidomic and transcriptomic analyses reveal the mechanism of lipid biosynthesis and accumulation during seed development in sesame. Front. Plant Sci. 2023, 14, 1211040. [Google Scholar] [CrossRef]
- Li, D.W.; Xie, W.H.; Hao, T.B.; Cai, J.X.; Zhou, T.B.; Balamurugan, S.; Yang, W.D.; Liu, J.S.; Li, H.Y. Constitutive and Chloroplast Targeted Expression of Acetyl-CoA Carboxylase in Oleaginous Microalgae Elevates Fatty Acid Biosynthesis. Mar. Biotechnol. 2018, 20, 566–572. [Google Scholar] [CrossRef]
- Cui, Y.P.; Liu, Z.J.; Zhao, Y.P. Overexpression of heteromeric GhACCase subunits enhanced oil accumulation in upland cotton. Plant Mol. Biol. Report. 2017, 35, 287–297. [Google Scholar] [CrossRef]
- Anwar, M.; Chen, L.; Xiao, Y.; Wu, J.; Zeng, L.; Li, H.; Wu, Q.; Hu, Z. Recent Advanced Metabolic and Genetic Engineering of Phenylpropanoid Biosynthetic Pathways. Int. J. Mol. Sci. 2021, 22, 9544. [Google Scholar] [CrossRef]
- Deslandes-Hérold, G.; Zanella, M.; Solhaug, E.; Fischer-Stettler, M.; Sharma, M.; Buergy, L.; Herrfurth, C.; Colinas, M.; Feussner, I.; Abt, M.R.; et al. The PRK/Rubisco shunt strongly influences Arabidopsis seed metabolism and oil accumulation, affecting more than carbon recycling. Plant Cell 2023, 35, 808–826. [Google Scholar] [CrossRef]
- Stoll, C.; Lühs, W.; Zarhloul, M.K. Knockout of KASIII regulation changes fatty acid composition in canola (Brassica napus L.). Eur. J. Lipid Sci. Technol. 2006, 108, 277–286. [Google Scholar] [CrossRef]
- Wei, Q.; Li, J.; Zhang, L.; Wu, P.Z.; Chen, Y.P.; Li, M.R.; Jiang, H.W.; Wu, G.J. Cloning and characterization of a β-ketoacyl-acyl carrier protein synthase II from Jatropha curcas. J. Plant Physiol. 2012, 169, 816–824. [Google Scholar] [CrossRef]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [PubMed]
- González-Thuillier, I.; Venegas-Calerón, M.; Sánchez, R.; Garcés, R.; von Wettstein-Knowles, P.; Martínez-Force, E. Sunflower (Helianthus annuus) fatty acid synthase complex: β-hydroxyacyl-[acyl carrier protein] dehydratase genes. Planta 2016, 243, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.M.; Zhao, Y.P. Construction of expression vectors and a preliminarily functional analysis of fatty acid synthetase genes of GhKAR and GhENR in upland cotton. Cotton Sci. 2016, 28, 958–961. [Google Scholar]
- Jako, C.; Kumar, A.; Wei, Y.D.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerel acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef]
- Liu, Z.J.; Zhang, Y.; Wang, Y.M. Construction and transformation of RNAi vector of gene GhDGAT1 in upland cotton. J. Chin. Agric. Univ. 2013, 18, 1–8. [Google Scholar]
- Jing, G.; Tang, D.; Yao, Y.; Su, Y.; Shen, Y.; Bai, Y.; Jing, W.; Zhang, Q.; Lin, F.; Guo, D.; et al. Seed specifically over-expressing DGAT2A enhances oil and linoleic acid contents in soybean seeds. Biochem. Biophys. Res. Commun. 2021, 568, 143–150. [Google Scholar] [CrossRef]
- Oakes, J.; Brackenridge, D.; Colletti, R.; Daley, M.; Hawkins, D.J.; Xiong, H.; Mai, J.; Screen, S.E.; Val, D.; Lardizabal, K.; et al. Expression of fungal diacylglycerol acyltransferase 2 genes to increase kernel oil in maize. Plant Physiol. 2011, 155, 1146–1157. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Angeles-Núnez, J.G.; Tiessen, A. Mutation of the transcription factor LEAFY COTYLEDON2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 2011, 168, 1891–1900. [Google Scholar] [PubMed]
- Shen, B.; Allen, W.B.; Zheng, P. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. Wrinkled1: A novel, low seed oil mutant of Arabidopsis with efficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Zhang, B.; Hao, Y.J.; Huang, J.; Tian, A.G.; Liao, Y.; Zhang, J.S.; Chen, S.Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007, 52, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Ahmad, F. The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene 2019, 690, 11–20. [Google Scholar] [PubMed]
- Campos, F.; Sobrino, T.; Ramos-Cabrer, P.; Castillo, J. Oxaloacetate: A novel neuroprotective for acute ischemic stroke. Int. J. Biochem. Cell Biol. 2012, 44, 262–265. [Google Scholar]
- Chen, J.Q.; Lang, C.X.; Hu, Z.H. Antisense PEP gene regulates to ratio of protein and lipid content in Brassica napus seeds. J. Agric. Biotechnol. 1999, 7, 316–320. [Google Scholar]
- Patel, M.S.; Nemeria, N.S.; Furey, W.; Jordan, F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J. Biol. Chem. 2014, 289, 16615–16623. [Google Scholar] [CrossRef]
- Abo Alrob, O.; Lopaschuk, G.D. Role of CoA and acetyl-CoA in regulating cardiac fatty acid and glucose oxidation. Biochem. Soc. Trans. 2014, 42, 1043–1051. [Google Scholar]
- Caroca, R.; Howell, K.A.; Malinova, I.; Burgos, A.; Tiller, N.; Pellizzer, T.; Annunziata, M.G.; Hasse, C.; Ruf, S.; Karcher, D.; et al. Knockdown of the plastid-encoded acetyl-CoA carboxylase gene uncovers functions in metabolism and development. Plant Physiol. 2021, 185, 1091–1110. [Google Scholar] [CrossRef]
- Zhao, J.R.; Zuo, S.Q.; Xiao, F.; Guo, F.Z.; Chen, L.Y.; Bi, K.; Cheng, D.Y.; Xu, Z.N. Advances in biotin biosynthesis and biotechnological production in microorganisms. World J. Microbiol. Biotechnol. 2024, 40, 163. [Google Scholar] [CrossRef]
- Glasgow, K.; Dillard, M.; Hertenstein, E.; Justin, A.; George, R.; Brady, A. Going Nuclear with Amino Acids and Proteins: Basic Biochemistry and Molecular Biology Primer for the Technologist. J. Nucl. Med. Technol. 2022, 50, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Roessler, P.G.; Ohlrogge, J.B. Cloning and characterization of the gene that encodes in the alga Cyclotella cryptica. J. Biol. Chem. 1993, 268, 19254–19259. [Google Scholar] [CrossRef]
- Kroth, P.G. The biodiversity of carbon assimilation. J. Plant Physiol. 2015, 172, 76–81. [Google Scholar] [CrossRef]
- Sasaki, Y.; Nagano, Y. Plant acetyl-CoA carboxylase: Structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 2004, 68, 1175–1184. [Google Scholar] [CrossRef]
- Roesler, K.R.; Savage, L.J.; Shintani, D.K.; Shorrosh, B.S.; Ohlrogge, J.B. Co-purification, co-immunoprecipitation, and coordinate expression of acetyl-coenzyme A carboxylase activity, biotincarboxylase, and biotin carboxyl carrier protein of higher plants. Planta 1996, 198, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Borgaro, J.G.; Chang, A.; Machutta, C.A.; Zhang, X.; Tonge, P.J. Substrate recognition by β-ketoacyl-ACP synthases. Biochemistry 2011, 50, 10678–10686. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, M.; Zhang, B.; Shrestha, P.; Petrie, J.; Green, A.G.; Singh, S.P. Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding β-ketoacyl-ACP synthase II (KASII). Plant Biotechnol. J. 2017, 15, 132–143. [Google Scholar] [CrossRef]
- Nofiani, R.; Philmus, B.; Nindita, Y. 3-Ketoacyl-ACP synthase (KAS) III homologues and their roles in natural product biosynthesis. Medchemcomm 2019, 10, 1517–1530. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.R.; Sun, Z.J.; Cui, G.Z.; Song, X.J.; Cui, Q. A new strategy for strain improvement of Aurantiochytrium sp. based on heavy-ions mutagenesis and synergistic effects of cold stress and inhibitors of enoyl-ACP reductase. Enzym. Microb. Technol. 2016, 93–94, 182–190. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, C.; Liang, Y.; Sun, R.H.; Gao, L.C.; Liu, T.; Li, D.D. Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. Tree Physiol. 2019, 39, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Ohlrogge, J.B.; Brower, J.A. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar]
- Zhao, L.Q.; Li, R.; Li, W. Cloning of delta-12 oleatedesaturase gene FAD2-1 and construction of its ihpRNA and amiRNA interference vectors from Gossypium hirsutum. Cotton Sci. 2011, 23, 189–192. [Google Scholar]
- Liu, Q.; Surinder, P.S.; Allan, G.G. High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 2002, 129, 1732–1743. [Google Scholar] [CrossRef] [PubMed]
- Craig, W.; Lenzi, P.; Scoth, N.; Palma, M.D.; Saggese, P.; Carbone, V.; Curran, N.M.; Magee, A.M. Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res. 2008, 17, 769–782. [Google Scholar] [CrossRef]
- Cai, M.; Li, W.H.; Wang, J.; Wang, X.; Kong, X.; Yu, Y.; Liu, L. Molecular cloning and expression analysis of stearoyl-ACP desaturase gene (GhSAD2) in upland cotton (Gossypium hirsutum L.). Acta Bot. Boreali-Occident. Sin. 2016, 9, 1713–1720. [Google Scholar]
- Amin, N.B.; Carvajal-Gonzalez, S.; Purkal, J.; Zhu, T.; Crowley, C.; Perez, S.; Chidsey, K.; Kim, A.M.; Goodwin, B. Targeting diacylglycerol acyltransferase 2 for the treatment of nonalcoholic steatohepatitis. Sci. Transl. Med. 2019, 11, eaav9701. [Google Scholar] [CrossRef]
- Cases, S.; Smith, S.J.; Zheng, Y.W.; Myers, H.M.; Lear, S.R.; Sande, E.; Novak, S.; Collins, C.; Welch, C.B.; Lusis, A.J.; et al. Identification of a gene encoding an acyl-CoA: Diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13018–13023. [Google Scholar] [CrossRef]
- Hobbs, D.H.; Lu, C.; Hilis, M.J. Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett. 1999, 452, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Lardizabal, K.D.; Mai, J.T.; Wagner, N.W.; Wyrick, A.; Voelker, T.; Hawkins, D.J. DGAT2 is a new diaclglycerol acyltransferase gene family: Purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyl-transferase activity. J. Biol. Chem. 2001, 276, 38862–38869. [Google Scholar] [CrossRef]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung Tree DGATl and DGA712 have nonredundant ftInctions ifltri-acylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef] [PubMed]
- Wurie, H.R.; Bucketf, L.; Zammit, V.A. Diacylglycerol acyhransferase 2 acts upstream of diacylglycerol acyhransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. FEBS J. 2012, 279, 3033–3047. [Google Scholar] [CrossRef]
- Taylor, D.C.; Zhang, Y.; Kumar, A. Molecular modification of triacylglycerol accumulation by overexpression of DGATl to produce canola with increased seed oil content under field conditions. Botany 2009, 87, 533–543. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Wang, Y.; Liu, J.; Liu, Y.; Cao, X.; Xue, S. Tuning of acyl-ACP thioesterase activity directed for tailored fatty acid synthesis. Appl. Microbiol. Biotechnol. 2018, 102, 3173–3182. [Google Scholar] [CrossRef] [PubMed]
- Aznar-Moreno, J.A.; Sánchez, R.; Gidda, S.K.; Martínez-Force, E.; Moreno-Pérez, A.J.; Venegas Calerón, M.; Garcés, R.; Mullen, R.T.; Salas, J.J. New Insights Into Sunflower (Helianthus annuus L.) FatA and FatB Thioesterases, Their Regulation, Structure and Distribution. Front. Plant Sci. 2018, 9, 1496. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Vega, M.J.; Garcés, R.; Martínez-Force, E. Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds (Helianthus annuus L.). Planta 2015, 221, 868–880. [Google Scholar] [CrossRef]
- Martins-Noguerol, R.; DeAndrés-Gil, C.; Garcés, R.; Salas, J.J.; Martínez-Force, E.; Moreno-Pérez, A.J. Characterization of the acyl-ACP thioesterases from Koelreuteria paniculata reveals a new type of FatB thioesterase. Heliyon 2020, 6, e05237. [Google Scholar] [CrossRef]
- Parcy, F.; Valon, C.; Kohara, A.; Miséra, S.; Giraudat, J. The ABSCISICACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDONl loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 1997, 11, 1265–1277. [Google Scholar]
- To, A.; Valon, C.; Savino, G.; Guilleminot, J.; Devic, M.; Giraudat, J.; Parcy, F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 2006, 18, 1642–1651. [Google Scholar] [CrossRef]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.E.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoët, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Neelakandan, A.K.; Wright, D.A.; Traore, S.M.; Chen, X.; Spalding, M.H.; He, G. CRISPR/Cas9 Based Site-Specific Modification of FAD2 cis-Regulatory Motifs in Peanut (Arachis hypogaea L). Front. Genet. 2022, 13, 849961. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Yang, X.L.; Zhang, F.X.; Zheng, X.; Qu, C.M.; Mu, J.Y.; Fu, F.Y.; Li, J.N.; Guan, R.Z.; Zhang, H.S.; et al. Enhanced seed oil production in canola by conditional expression of brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef]
- Mu, J.Y.; Tan, H.L.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.P.; Liu, F.Z.; Wany, S. In silico Analysis of peanut transcription factor gene WRI1. Mol. Plant Breed. 2012, 3, 363–370. [Google Scholar]
- Liu, J.; Hua, W.; Zhan, G.M. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Pouvreau, B.; Baud, S.; Vernoud, V. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef]
- Li, Q.; Shao, J.H.; Tang, S.H.; Shen, Q.W.; Wang, T.H.; Chen, W.L.; Hong, Y.Y. WRINKLEDl accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front. Plant Sci. 2015, 6, 1015–1029. [Google Scholar] [CrossRef]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewica, A.; Kong, Q.; Ma, W.; Sedbrook, J.; Benning, C. Ectopic expression of WRLNKLED1 affects fatty acid homeostasis in Brachypodiurn distachyon vegetative tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar]
- Zhang, Q.; Zhong, S.; Dong, Q.; Yang, H.; Yang, H.; Tan, F.; Chen, C.; Ren, T.; Shen, J.; Cao, G.; et al. Identification of Photoperiod- and Phytohormone-Responsive DNA-Binding One Zinc Finger (Dof) Transcription Factors in Akebia trifoliata via Genome-Wide Expression Analysis. Int. J. Mol. Sci. 2023, 24, 4973. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Xie, X.; Wu, M.; Lin, Z.; Yin, J.; Lou, S.; Huang, Y.; Hu, Z. Understanding the functions of endogenous DOF transcript factor in Chlamydomonas reinhardtii. Biotechnol. Biofuels 2019, 12, 67. [Google Scholar] [CrossRef]
- Xu, C.; Fan, J.; Shanklin, J. Metabolic and functional connections between cytoplasmic and chloroplast triacylglycerol storage. Prog. Lipid Res. 2020, 80, 101069. [Google Scholar] [CrossRef]
- Sun, J.; Chen, T.; Liu, M.; Zhao, D.; Tao, J. Analysis and Functional Verification of PoWRI1 Gene Associated with Oil Accumulation Process in Paeonia ostii. Int. J. Mol. Sci. 2021, 22, 6996. [Google Scholar] [CrossRef] [PubMed]
- Tjellstrom, H.; Yang, Z.; Allen, D.K.; Ohlrogge, J.B. Rapid kinetic labeling of Arabidopsis cell suspension cultures: Implications for models of lipid export from plastids. Plant Physiol. 2012, 158, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yamaoka, Y.; Ono, H.; Kim, H.; Shim, D.; Maeshima, M.; Martinoia, E.; Cahoon, E.B.; Nishida, I.; Lee, Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2013, 110, 773–778. [Google Scholar] [CrossRef]
- Gräfe, K.; Shanmugarajah, K.; Zobel, T.; Weidtkamp-Peters, S.; Kleinschrodt, D.; Smits, S.H.J.; Schmitt, L. Cloning and expression of selected ABC transporters from the Arabidopsis thaliana ABCG family in Pichia pastoris. PLoS ONE 2019, 14, e0211156. [Google Scholar] [CrossRef]
- El-Kebbaj, R.; Bouchab, H.; Tahri-Joutey, M.; Rabbaa, S.; Limami, Y.; Nasser, B.; Egbujor, M.C.; Tucci, P.; Andreoletti, P.; Saso, L.; et al. The Potential Role of Major Argan Oil Compounds as Nrf2 Regulators and Their Antioxidant Effects. Antioxidants 2024, 13, 344. [Google Scholar] [CrossRef] [PubMed]
- Ting, S.Y.; Lau, N.S.; Sam, K.K.; Janaranjani, M.; Wong, S.C.; Monroig, Ó.; Quah, E.S.H.; Ahmad, A.B.; Him, N.A.I.I.N.; Jaya-Ram, A. Long-chain polyunsaturated fatty acid biosynthesis in a land-crab with advanced terrestrial adaptations: Molecular cloning and functional characterization of two fatty acyl elongases. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2022, 262, 110773. [Google Scholar] [CrossRef]
- Coleman, R.A. It takes a village: Channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J. Lipid Res. 2019, 60, 490–497. [Google Scholar] [CrossRef]
- Xu, Y.; Singer, S.D.; Chen, G. Protein interactomes for plant lipid biosynthesis and their biotechnological applications. Plant Biotechnol. J. 2023, 21, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Ting, N.C.; Sherbina, K.; Khoo, J.S.; Kamaruddin, K.; Chan, P.L.; Chan, K.L.; Halim, M.A.A.; Sritharan, K.; Yaakub, Z.; Mayes, S.; et al. Expression of fatty acid and triacylglycerol synthesis genes in interspecific hybrids of oil palm. Sci. Rep. 2020, 10, 16296. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef]
- Kondakova, T.; Cronan, J.E. Transcriptional regulation of fatty acid cis-trans isomerization in the solvent-tolerant soil bacterium, Pseudomonas putida F1. Environ. Microbiol. 2019, 21, 1659–1676. [Google Scholar] [CrossRef]
- Wang, H.; Luo, J.; Chen, Z.; Cao, W.T.; Xu, H.F.; Gou, D.M.; Zhu, J.J. MicroRNA-24 can control triacylglycerol synthesis in goat mammary epithelial cells by targeting the fatty acid synthase gene. J. Dairy Sci. 2015, 98, 9001–9014. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Huang, D.; Wu, S.; Liu, S.; Wang, C.; Sheng, Y.; Lu, X.; Broxmeyer, H.E.; Wan, J.; Yang, L. Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. Nat. Cell Biol. 2023, 25, 1033–1046. [Google Scholar] [CrossRef]
- Wang, S.; Zha, L.; Cui, X.; Yeh, Y.T.; Liu, R.; Jing, J.; Shi, H.; Chen, W.; Hanover, J.; Yin, J.; et al. Epigenetic Regulation of Hepatic Lipid Metabolism by DNA Methylation. Adv. Sci. 2023, 10, e2206068. [Google Scholar] [CrossRef]
- Violi, F.; Pastori, D.; Pignatelli, P.; Carnevale, R. Nutrition, Thrombosis, and Cardiovascular Disease. Circ. Res. 2020, 126, 1415–1442. [Google Scholar] [CrossRef]
- Durrett, T.P.; McClosky, D.D.; Tumaney, A.W. A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds. Proc. Natl. Acad. Sci. USA 2010, 107, 9464–9469. [Google Scholar] [CrossRef]
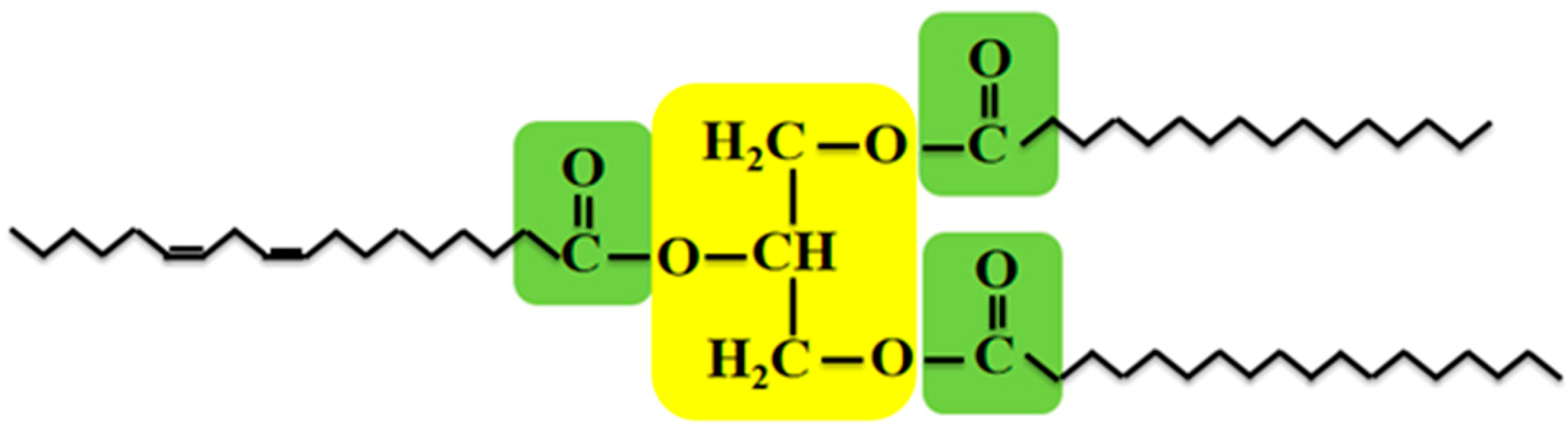

| Genes Encoding the Key Enzymes and TFs | Function Confirmed in Original Species | Function Confirmed in Other Plants |
|---|---|---|
| Phosphoenolpyruvate carboxylase, PEPCase | CsPEPCase, HvPEPCase, RcPEPCase, BnPEPCi, OsPEPCi, AhPEPCi, GhPEPCase [23,24] | StPEPCase |
| Pyruvate dehydrogenase complex, PDHC | SoPDHC, PsPDHC [25], AtPDHC, BvPDHC [26] | |
| Acetyl CoA carboxylase, ACCase | AtACCase, GmACCase [27], RcACCase, AhACCase, GhACCase [28], NtACCase [29] | AtACCase [30] |
| β-ketoacyl-ACP synthase, KAS | AhKASI, BnKASIII [31], AtKASI, JcKASII [32] | |
| β-ketoacyl-ACP reductase, KAR | EgKAR, PdKAR [33], AhKR [34], GhKAR [35] | |
| Hydroxyacyl-ACP dehydrase, HAD | EgHAD, PdHAD [33], AhHD [34], GhHAD [35] | |
| Enoyl-ACP reductase, ENR | EgENR, PdENR [33], AhER, GhENR [35] | |
| Acyl-ACP thioesterase, FAT | AhFATB, AtFAT, RcFAT | LnFAT [29] |
| Glyceral 3-phosphate acyltransferase, GPAT | AtGPAT5, AtGPAT4, AtGPAT6 | MaGPAT |
| Diacylglycerol acyltransferase, DGAT | EgDGAT2 [33], AtDGAT1 [36], OeDGAT2 [37] | UrDGAT [38], NcDGAT2 [39] |
| Leafy cotyledon, LEC | AtLEC1 [40], AhLEC1, AtLEC2 [41], ZmLEC1 [42] | |
| Transcription factor WRINKLED1, WRI | EgWRI1 [33], ZmWRI1 [42], AtWRI1 [43] | BnWRI1 [43] |
| DNA binding with one finger, Dof | ZmDof [44] | GmDof [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Wu, Q.; Yang, Y.; Li, Q.; Li, R.; Ye, J. Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects. Genes 2024, 15, 1125. https://doi.org/10.3390/genes15091125
Zhou L, Wu Q, Yang Y, Li Q, Li R, Ye J. Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects. Genes. 2024; 15(9):1125. https://doi.org/10.3390/genes15091125
Chicago/Turabian StyleZhou, Lixia, Qiufei Wu, Yaodong Yang, Qihong Li, Rui Li, and Jianqiu Ye. 2024. "Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects" Genes 15, no. 9: 1125. https://doi.org/10.3390/genes15091125
APA StyleZhou, L., Wu, Q., Yang, Y., Li, Q., Li, R., & Ye, J. (2024). Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects. Genes, 15(9), 1125. https://doi.org/10.3390/genes15091125






