RNA-Seq and WGCNA Analyses Reveal Key Regulatory Modules and Genes for Salt Tolerance in Cotton
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Planting and Sample Preparation
2.2. Library Construction and Sequencing
2.3. Sequencing Data Quality Control and Analysis
2.4. Differentially Expressed Gene Screening and Analysis
2.5. Weighted Gene Co-Expression Network Construction
2.6. qRT-PCR Validation
3. Results
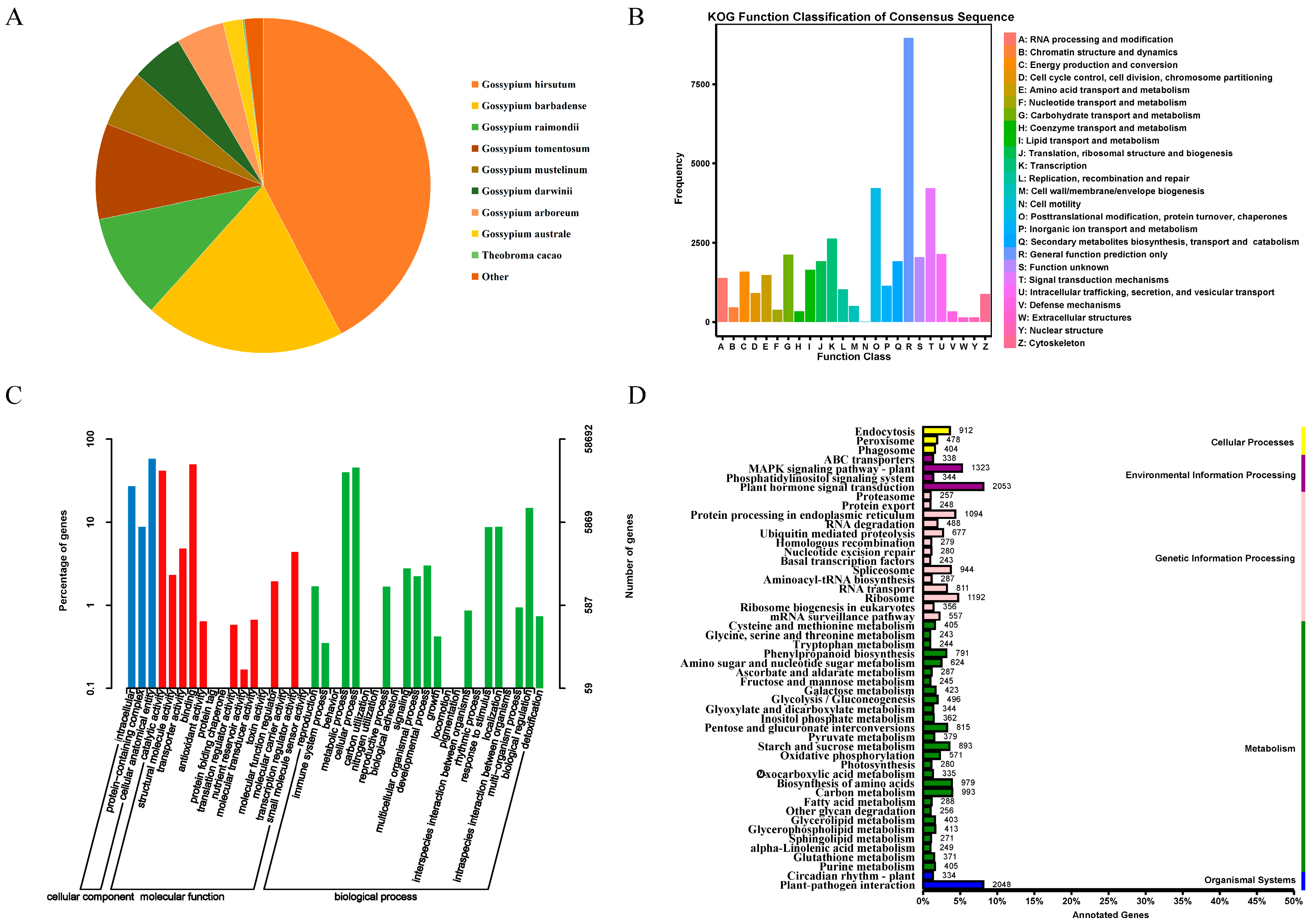
3.1. Transcriptome Sequencing and Assembly
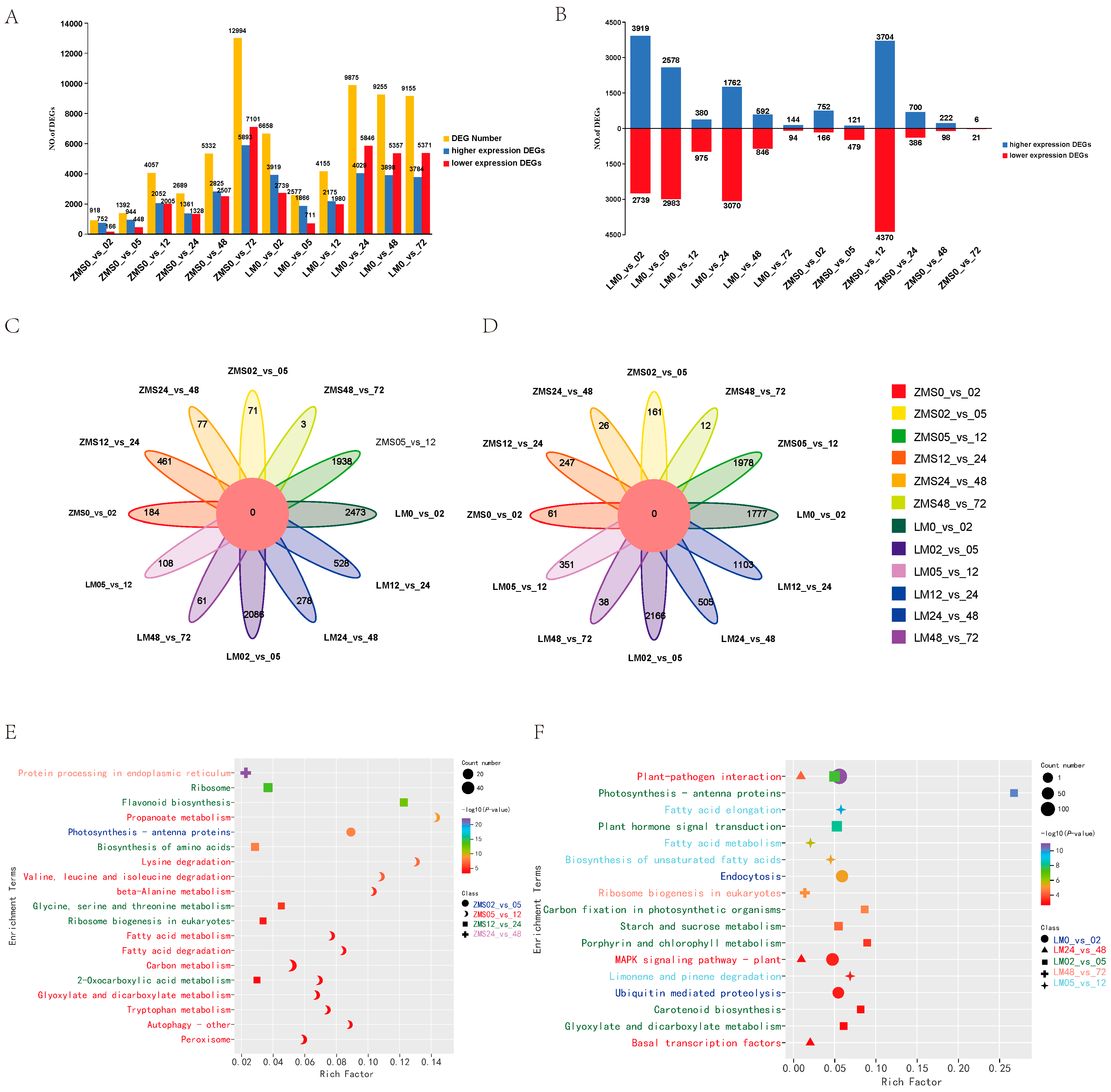
3.2. Differentially Expressed Gene Analysis
3.3. WGCNA Analysis
3.4. Module Visualization and Screening of Hub Genes
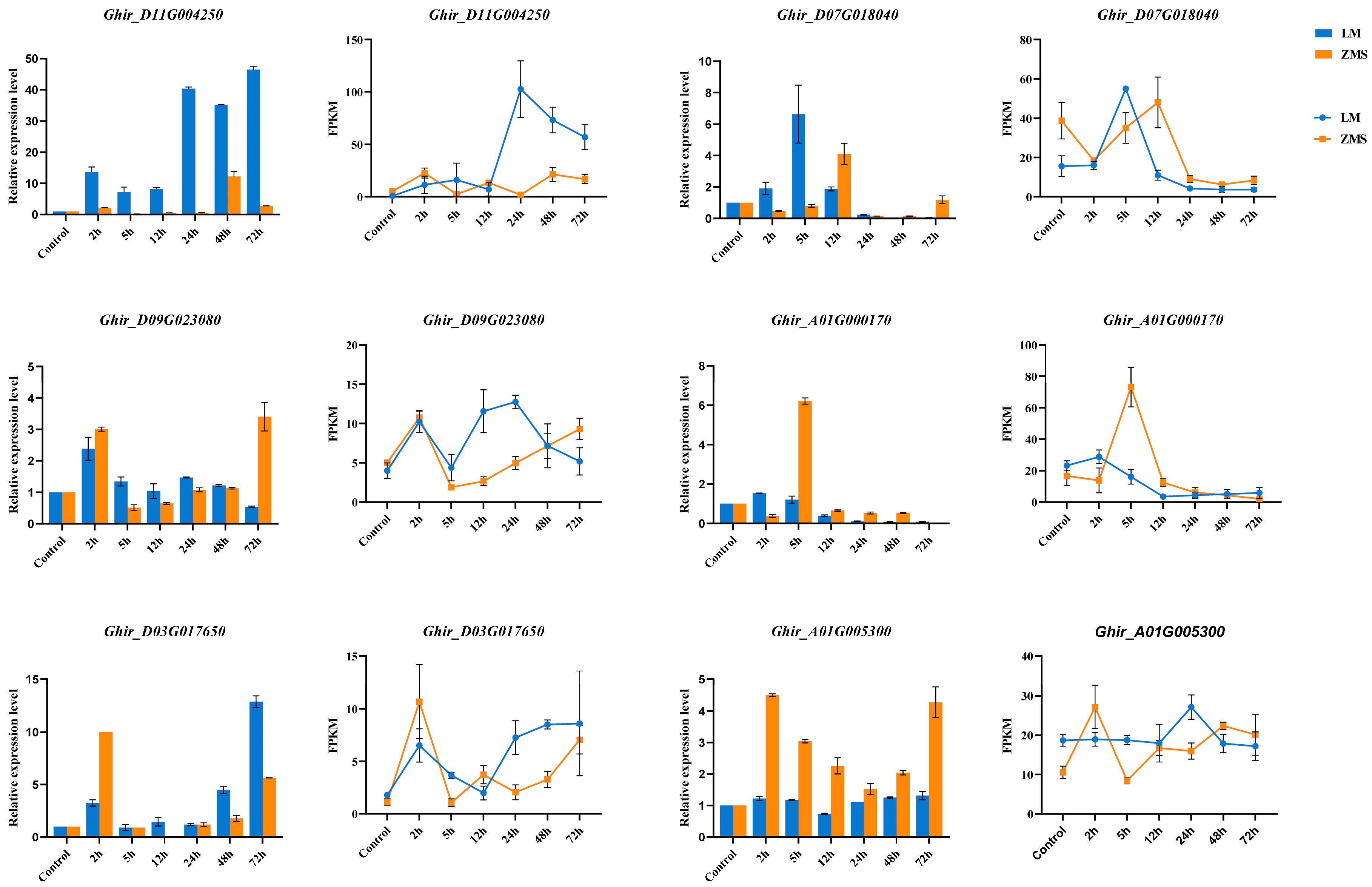
3.5. DEG Validation Using qRT-PCR
4. Discussion
4.1. RNA-Seq Mining of Salt Tolerance Genes
4.2. Metabolic Networks Regulate Cotton’s Resistance to Salt Stress
4.3. TFs Reveal Salt Tolerance Strategies in Cotton
4.4. Salt Stress Affects Hubgene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2022, 38, 39–67. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef] [PubMed]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef]
- Chen, J.; Mueller, V. Coastal climate change, soil salinity and human migration in Bangladesh. Nat. Clim. Chang. 2018, 8, 981–985. [Google Scholar] [CrossRef]
- Gong, Z. Plant abiotic stress: New insights into the factors that activate and modulate plant responses. J. Integr. Plant Biol. 2021, 63, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2022, 15, 1030890. [Google Scholar] [CrossRef]
- Rastogi, S.; Shah, S.; Kumar, R.; Vashisth, D.; Akhtar, M.Q.; Kumar, A.; Dwivedi, U.N.; Shasany, A.K. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought and salinity. PLoS ONE 2019, 14, e0210903. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; García, A.; Benítez, Á.; Martínez, C.; Jamilena, M. Comparative RNA-Seq Analysis between Monoecious and Androecious Plants Reveals Regulatory Mechanisms Controlling Female Flowering in Cucurbita pepo. Int. J. Mol. Sci. 2023, 24, 17195. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, X.; Xie, W.; Lin, C.; Wang, Q.; Tao, Z. ETHYLENE INSENSITIVE3/EIN3-LIKE1 modulate FLOWERING LOCUS C expression via histone demethylase interaction. Plant Physiol. 2023, 192, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Aleem, M.; Raza, M.M.; Haider, M.S.; Atif, R.M.; Ali, Z.; Bhat, J.A.; Zhao, T. Comprehensive RNA-seq analysis revealed molecular pathways and genes associated with drought tolerance in wild soybean (Glycine soja Sieb. and Zucc.). Physiol. Plant. 2021, 172, 707–732. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, P.; Cheng, S.; Zhao, Z.; Liu, Y.; Wei, Y.; Lu, Q.; Han, J.; Cai, X.; Zhou, Z.; et al. Protoplast Dissociation and Transcriptome Analysis Provides Insights to Salt Stress Response in Cotton. Int. J. Mol. Sci. 2022, 23, 2845. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Mehari, T.G.; Fang, H.; Han, J.; Huo, X.; Zhang, J.; Chen, Y.; Wang, D.; Zhuang, Z.; Ditta, A.; et al. Transcriptome, proteome and functional characterization reveals salt stress tolerance mechanisms in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 2023, 16, 1092616. [Google Scholar] [CrossRef]
- Wang, D.; Lu, X.; Chen, X.; Wang, S.; Wang, J.; Guo, L.; Yin, Z.; Chen, Q.; Ye, W. Temporal salt stress-induced transcriptome alterations and regulatory mechanisms revealed by PacBio long-reads RNA sequencing in Gossypium hirsutum. BMC Genom. 2020, 21, 838. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Guo, Q.; Ji, W.; Wang, H.; Tao, J.; Xu, P.; Chen, X.; Ali, W.; Wu, X.; Shen, X.; et al. Transcriptome Expression Profiling Reveals the Molecular Response to Salt Stress in Gossypium anomalum Seedlings. Plants 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Guo, Q.; Meng, S.; Zhang, X.; Xu, Z.; Guo, W.; Shen, X. Genome-wide association analysis reveals genetic variations and candidate genes associated with salt tolerance related traits in Gossypium hirsutum. BMC Genom. 2021, 22, 26. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Julien, P.; Kuhn, M.; Mering, C.; Muller, J.; Doerks, T.; Bork, P. eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2007, 36, D250–D254. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Yao, W.; Ge, W.; Jiang, T.; Zhou, B. Transcriptome Sequencing and WGCNA Reveal Key Genes in Response to Leaf Blight in Poplar. Int. J. Mol. Sci. 2023, 24, 10047. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Rao, A.; Barkley, D.; França, G.S.; Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; You, L.; Hardillo, J.A.U.; Chien, M.P. Spatial Transcriptomic Technologies. Cells 2023, 12, 2042. [Google Scholar] [CrossRef] [PubMed]
- Szádeczky-Kardoss, I.; Szaker, H.M.; Verma, R.; Darkó, É.; Pettkó-Szandtner, A.; Silhavy, D.; Csorba, T. Elongation factor TFIIS is essential for heat stress adaptation in plants. Nucleic Acids Res. 2022, 50, 1927–1950. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, Q.; Zhang, D.; Wang, J.; Cao, T.; Bozorov, T.A.; Cheng, L.; Zhang, D. Transcriptome Reveals the Molecular Mechanism of the ScALDH21 Gene from the Desert Moss Syntrichia caninervis Conferring Resistance to Salt Stress in Cotton. Int. J. Mol. Sci. 2023, 24, 5822. [Google Scholar] [CrossRef] [PubMed]
- Gul, H.S.; Ulfat, M.; Zafar, Z.U.; Haider, W.; Ali, Z.; Manzoor, H.; Afzal, S.; Ashraf, M.; Athar, H.U. Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes 2022, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Zahra, N.; Al Hinai, M.S.; Hafeez, M.B.; Rehman, A.; Wahid, A.; Siddique, K.H.M.; Farooq, M. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 2022, 178, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Liu, Y.; Zhang, T.Y.; Zhou, Z.F.; Huang, J.Y.; Zhou, T.; Hua, Y.P. Integrated physiological and transcriptional dissection reveals the core genes involving nutrient transport and osmoregulatory substance biosynthesis in allohexaploid wheat seedlings under salt stress. BMC Plant Biol. 2022, 22, 502. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Liu, Z.; Ding, Z.; Yisilam, G.; Wang, Q.; Tian, X. Metabolomic analysis reveals key metabolites and metabolic pathways in Suaeda salsa under salt and drought stress. Funct. Plant Biol. 2023, 50, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hua, Y.; Wang, S.; Liu, X.; Zou, L.; Chen, C.; Zhao, H.; Yan, Y. Analysis of the Prunellae Spica transcriptome under salt stress. Plant Physiol. Biochem. 2020, 156, 314–322. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M. Synthesis versus degradation: Directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef]
- Bohn, M.; Lüthje, S.; Sperling, P.; Heinz, E.; Dörffling, K. Plasma membrane lipid alterations induced by cold acclimation and abscisic acid treatment of winter wheat seedlings differing in frost resistance. J. Plant Physiol. 2007, 164, 146–156. [Google Scholar] [CrossRef]
- Lin, H.; Wu, L. Effects of salt stress on root plasma membrane characteristics of salt-tolerant and salt-sensitive buffalograss clones. Environ. Exp. Bot. 1996, 36, 239–254. [Google Scholar] [CrossRef]
- Wu, J.; Seliskar, D.M.; Gallagher, J.L. Stress tolerance in the marsh plant Spartina patens: Impact of NaCl on growth and root plasma membrane lipid composition. Physiol. Plant. 1998, 102, 307–317. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Rupasinghe, T.W.T.; Roessner, U.; Barkla, B.J. Salt stress alters membrane lipid content and lipid biosynthesis pathways in the plasma membrane and tonoplast. Plant Physiol. 2022, 189, 805–826. [Google Scholar] [CrossRef]
- Duarte, B.; Matos, A.R.; Marques, J.C.; Caçador, I. Leaf fatty acid remodeling in the salt-excreting halophytic grass Spartina patens along a salinity gradient. Plant Physiol. Biochem. 2018, 124, 112–116. [Google Scholar] [CrossRef]
- Rehman, H.M.; Chen, S.; Zhang, S.; Khalid, M.; Uzair, M.; Wilmarth, P.A.; Ahmad, S.; Lam, H.M. Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins. Int. J. Mol. Sci. 2022, 23, 13270. [Google Scholar] [CrossRef]
- Khomutov, G.; Fry, I.V.; Huflejt, M.E.; Packer, L. Membrane lipid composition, fluidity, and surface charge changes in response to growth of the fresh water cyanobacterium Synechococcus 6311 under high salinity. Arch. Biochem. Biophys. 1990, 277, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, W.; Li, M.; Wan, S.; Sui, N. Antioxidants and unsaturated fatty acids are involved in salt tolerance in peanut. Acta Physiol. Plant. 2017, 207, 39. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wen, W.; Shi, Z.; Gu, Q.; Ahammed, G.J.; Cao, K.; Jahan, S.M.; Shu, S.; Wang, J.; et al. Hydrogen peroxide mediates spermidine-induced autophagy to alleviate salt stress in cucumber. Autophagy 2021, 17, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Bao, Y.; Lu, Y.; He, F.; Wang, S.; Wang, D.; Yu, X.; Yin, W.; Xia, X.; Liu, C. Poplar Autophagy Receptor NBR1 Enhances Salt Stress Tolerance by Regulating Selective Autophagy and Antioxidant System. Front. Plant Sci. 2021, 11, 568411. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef]
- He, A.; Dean, J.M.; Lodhi, I.J. Peroxisomes as cellular adaptors to metabolic and environmental stress. Trends Cell Biol. 2021, 31, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Jiao, B.; Wang, J.; Yang, Y.; Yang, F.; Geng, Z.; Zhao, G.; Liu, Y.; Dong, F.; Wang, Y.; et al. Comparative Analysis of miRNA Expression Profiles under Salt Stress in Wheat. Genes 2023, 14, 1586. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Liu, H.; Islam, F.; Li, L.; Farooq, M.A.; Ruan, S.; Zhou, W. OsPEX11, a Peroxisomal Biogenesis Factor 11, Contributes to Salt Stress Tolerance in Oryza sativa. Front. Plant Sci. 2016, 15, 1357. [Google Scholar] [CrossRef]
- Qiu, T.; Du, K.; Jing, Y.; Zeng, Q.; Liu, Z.; Li, Y.; Ren, Y.; Yang, J.; Kang, X. Integrated transcriptome and miRNA sequencing approaches provide insights into salt tolerance in allotriploid Populus cathayana. Planta 2021, 254, 25. [Google Scholar] [CrossRef]
- Deslandes, L.; Rivas, S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012, 17, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wei, L.; Liu, T.; Ma, J.; Huang, K.; Guo, H.; Huang, Y.; Zhang, L.; Zhao, J.; Tsuda, K.; et al. Suppression of ETI by PTI priming to balance plant growth and defense through an MPK3/MPK6-WRKYs-PP2Cs module. Mol. Plant 2023, 16, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Zarattini, M.; Farjadm, M.; Launay, A.; Cannella, D.; Soulié, M.C.; Bernacchia, G.; Fagard, M. Every cloud has a silver lining: How abiotic stresses affect gene expression in plant-pathogen interactions. J. Exp. Bot. 2021, 72, 1020–1033. [Google Scholar] [CrossRef]
- Wu, B.; Munkhtuya, Y.; Li, J.; Hu, Y.; Zhang, Q.; Zhang, Z. Comparative Transcriptional Profiling and Physiological Responses of Two Contrasting Oat Genotypes under Salt Stress. Sci. Rep. 2018, 8, 16248. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234–235, 80–93. [Google Scholar] [CrossRef]
- Li, H.; Duijts, K.; Pasini, C.; van Santen, J.E.; Lamers, J.; de Zeeuw, T.; Verstappen, F.; Wang, N.; Zeeman, S.C.; Santelia, D.; et al. Effective root responses to salinity stress include maintained cell expansion and carbon allocation. New Phytol. 2023, 238, 1942–1956. [Google Scholar] [CrossRef]
- Hu, T.; Hu, L.; Zhang, X.; Zhang, P.; Zhao, Z.; Fu, J. Differential responses of CO2 assimilation.; carbohydrate allocation and gene expression to NaCl stress in perennial ryegrass with different salt tolerance. PLoS ONE 2013, 8, e66090. [Google Scholar] [CrossRef]
- Li, C.; Ji, J.; Wang, G.; Li, Z.; Wang, Y.; Fan, Y. Over-Expression of LcPDS, LcZDS, and LcCRTISO, Genes from Wolfberry for Carotenoid Biosynthesis, Enhanced Carotenoid Accumulation, and Salt Tolerance in Tobacco. Front. Plant Sci. 2020, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Jia, L.; Zhang, J. ABA signal in rice under stress conditions. Rice 2012, 5, 1. [Google Scholar] [CrossRef]
- Benech Arnold, R.L.; Fenner, M.; Edwards, P.J. Changes in germinability, ABA content and ABA embryonic sensitivity in developing seeds of Sorghum bicolor (L.) Moench. induced by water stress during grain filling. New Phytol. 1991, 118, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Chen, G.; Wang, Y.; Huang, Y.; Marchant, D.B.; Wang, Y.; Yang, Q.; Dai, F.; Hills, A.; Franks, P.J.; et al. Evolutionary Conservation of ABA Signaling for Stomatal Closure. Plant Physiol. 2017, 174, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 2, 1979. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Sun, H.; Hakim; Yang, X.; Zhang, X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 2018, 162, 439–454. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A Novel Sweetpotato WRKY Transcription Factor, IbWRKY2, Positively Regulates Drought and Salt Tolerance in Transgenic Arabidopsis. Biomolecules 2020, 10, 506. [Google Scholar] [CrossRef]
- Kang, G.; Yan, D.; Chen, X.; Li, Y.; Yang, L.; Zeng, R. Molecular characterization and functional analysis of a novel WRKY transcription factor HbWRKY83 possibly involved in rubber production of Hevea brasiliensis. Plant Physiol. Biochem. 2020, 155, 483–493. [Google Scholar] [CrossRef]
- Yu, L.; Liu, W.; Guo, Z.; Li, Z.; Jiang, H.; Zou, Q.; Mao, Z.; Fang, H.; Zhang, Z.; Wang, N.; et al. Interaction between MdMYB63 and MdERF106 enhances salt tolerance in apple by mediating Na+/H+ transport. Plant Physiol. Biochem. 2020, 155, 464–471. [Google Scholar] [CrossRef]
- Wang, D.; Sun, Z.; Hu, X.; Xiong, J.; Hu, L.; Xu, Y.; Tang, Y.; Wu, Y. The key regulator LcERF056 enhances salt tolerance by modulating reactive oxygen species-related genes in Lotus corniculatus. BMC Plant Biol. 2021, 21, 605. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Z.; Wang, P.; Qin, C.; He, L.; Kong, L.; Ren, W.; Liu, X.; Ma, W. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int. J. Biol. Macromol. 2023, 240, 124436. [Google Scholar] [CrossRef] [PubMed]
- Rui, Z.; Pan, W.; Zhao, Q.; Hu, H.; Li, X.; Xing, L.; Jia, H.; She, K.; Nie, X. Genome-wide identification.; evolution and expression analysis of NAC gene family under salt stress in wild emmer wheat (Triticum dicoccoides L.). Int. J. Biol. 2023, 230, 123376. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Y.; Wang, S.; Tao, Y.; Wang, Z.; Shu, Y.; Peng, H.; Mijiti, A.; Wang, Z.; Zhang, H.; et al. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 2016, 35, 613–627. [Google Scholar] [CrossRef]
- Waseem, M.; Rong, X.; Li, Z. Dissecting the Role of a Basic Helix-Loop-Helix Transcription Factor, SlbHLH22, Under Salt and Drought Stresses in Transgenic Solanum lycopersicum L. Front. Plant Sci. 2019, 10, 734. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, F.; Ma, Y.; Chong, K.; Xu, Y. Overexpression of OrbHLH001, a putative helix-loop-helix transcription factor causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. J. Plant Physiol. 2013, 17, 93–100. [Google Scholar] [CrossRef]
- Verma, D.; Jalmi, S.K.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef]
- Han, H.; Wang, C.; Yang, X.; Wang, L.; Ye, J.; Xu, F.; Liao, Y.; Zhang, W. Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta 2023, 258, 13. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.C.; Kim, H.S.; Kim, S. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef]
- Pratyusha, D.S.; Sarada, D.V.L. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Lee, W.J.; Le, Q.T.; Hong, S.W.; Lee, H. Growth Performance Can Be Increased Under High Nitrate and High Salt Stress Through Enhanced Nitrate Reductase Activity in Arabidopsis Anthocyanin Over-Producing Mutant Plants. Front. Plant Sci. 2021, 12, 644455. [Google Scholar] [CrossRef]
- Gao, Q.; Li, X.; Xiang, C.; Li, R.; Xie, H.; Liu, J.; Li, X.; Zhang, G.; Yang, S.; Liang, Y.; et al. EbbHLH80 Enhances Salt Responses by Up-Regulating Flavonoid Accumulation and Modulating ROS Levels. Int. J. Mol. Sci. 2023, 24, 11080. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yao, Y.T.; Wang, B.B.; Li, Y.M.; Li, L.; Bao, A.K. Flavonoids are involved in salt tolerance through ROS scavenging in the halophyte Atriplex canescens. Plant Cell Rep. 2023, 43, 5. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.J.; Hong, Y.S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, D.D.; Min, D.H.; Cao, T.; Ning, L.; Jiang, Q.Y.; Sun, X.J.; Zhang, H.; Tang, W.S.; Gao, S.Q.; et al. Foxtail millet MYB-like transcription factor SiMYB16 confers salt tolerance in transgenic rice by regulating phenylpropane pathway. Plant Physiol. Biochem. 2023, 195, 310–321. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Numan, M.; Lubna; Kim, K.M. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 2021, 11, 968. [Google Scholar] [CrossRef]
- Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J.G.; Sato, F. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007, 48, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Li, D.H.; Guo, J.; Bin, W.; Zhao, N.; Wang, K.B.; Li, J.Y.; Li, Z.L.; Hua, H.M. Two new benzylisoquinoline alkaloids from Thalictrum foliolosum and their antioxidant and in vitro antiproliferative properties. Arch. Pharm. Res. 2016, 39, 871–877. [Google Scholar] [CrossRef]
- Madani, H.; Hosseini, B.; Karimzadeh, G.; Rahimi, A. Enhanced thebaine and noscapine production and modulated gene expression of tyrosine/dopa decarboxylase and salutaridinol 7-O-acetyltransferase genes in induced autotetraploid seedlings of Papaver bracteatum Lindl. Acta Physiol. Plant. 2019, 41, 194. [Google Scholar] [CrossRef]
- Gómez-Sagasti, M.T.; Barrutia, O.; Ribas, G.; Garbisu, C.; Becerril, J.M. Early transcriptomic response of Arabidopsis thaliana to polymetallic contamination: Implications for the identification of potential biomarkers of metal exposure. Metallomics 2016, 8, 518–531. [Google Scholar] [CrossRef]
- Moran Lauter, A.N.; Peiffer, G.A.; Yin, T.; Whitham, S.A.; Cook, D.; Shoemaker, R.C.; Graham, M.A. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genom. 2014, 15, 702. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Clemente, R.M.; Vives, V.; Zandalinas, S.I.; López-Climent, M.F.; Muñoz, V.; Gómez-Cadenas, A. Biotechnological approaches to study plant responses to stress. Biomed. Res. Int. 2013, 2013, 654120. [Google Scholar] [CrossRef]
- Hou, S.; Liu, D.; Huang, S.; Luo, D.; Liu, Z.; Xiang, Q.; Wang, P.; Mu, R.; Han, Z.; Chen, S.; et al. The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes. Nat. Commun. 2021, 12, 5494. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J. Characterisation of the Arabidopsis thaliana Leucine-Rich Repeat Receptor Kinase Subfamily XII in Immune Signalling. Ph.D. Dissertation, University of East Anglia, Norwich, UK, 2019. [Google Scholar]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing.; root growth and response to abiotic and, biotic, stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Mueller-Roeber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, J.; Zhang, Y.; Yin, L.; Lu, J. Isolation of a WRKY30 gene from Muscadinia rotundifolia (Michx) and validation of its function under biotic and abiotic stresses. Protoplasma 2015, 252, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Duanmu, H.; Yu, Y.; Chen, C.; Sun, X.; Zhu, P.; Chan, R.; Duan, X.; Li, H.; Cao, L.; et al. GsJ11, identified by genome-wide analysis.; facilitates alkaline tolerance in transgenic plants. Plant Cell Tissue Organ. Cult. 2017, 129, 411–430. [Google Scholar] [CrossRef]
- Chen, K.M.; Holmström, M.; Raksajit, W.; Suorsa, M.; Piippo, M.; Aro, E.M. Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Elkelish, A.A.; Soliman, M.H.; Alhaithloul, H.A.; El-Esawi, M.A. Selenium protects wheat seedlings against salt stress-mediated oxidative damage by up-regulating antioxidants and osmolytes metabolism. Plant Physiol. Biochem. 2019, 137, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, C.; Wuriyanghan, H.; Lei, Z.; Tang, Y.; Zhang, H.; Zhao, X. Exogenous selenium endows salt-tolerant and salt-sensitive soybeans with salt tolerance through plant-microbial coactions. Agronomy 2023, 13, 2271. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Zhao, G.; Geng, Z.; Qi, H.; Dou, H.; Zhang, H. Dynamic transcriptome and co-expression network analysis of the cotton (Gossypium hirsutum) root response to salinity stress at the seedling stage. Acta Physiol. Plant. 2020, 42, 143. [Google Scholar] [CrossRef]
- Siwinska, J.; Siatkowska, K.; Olry, A.; Grosjean, J.; Hehn, A.; Bourgaud, F.; Meharg, A.A.; Carey, M.; Lojkowska, E.; Ihnatowicz, A. Scopoletin 8-hydroxylase: A novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J. Exp. Bot. 2018, 69, 1735–1748. [Google Scholar] [CrossRef]
- Duan, C.; Mao, T.; Sun, S.; Guo, X.; Guo, L.; Huang, L.; Wang, Z.; Zhang, Y.; Li, M.; Sheng, Y.; et al. Constitutive expression of GmF6′H1 from soybean improves salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2019, 141, 446–455. [Google Scholar] [CrossRef]


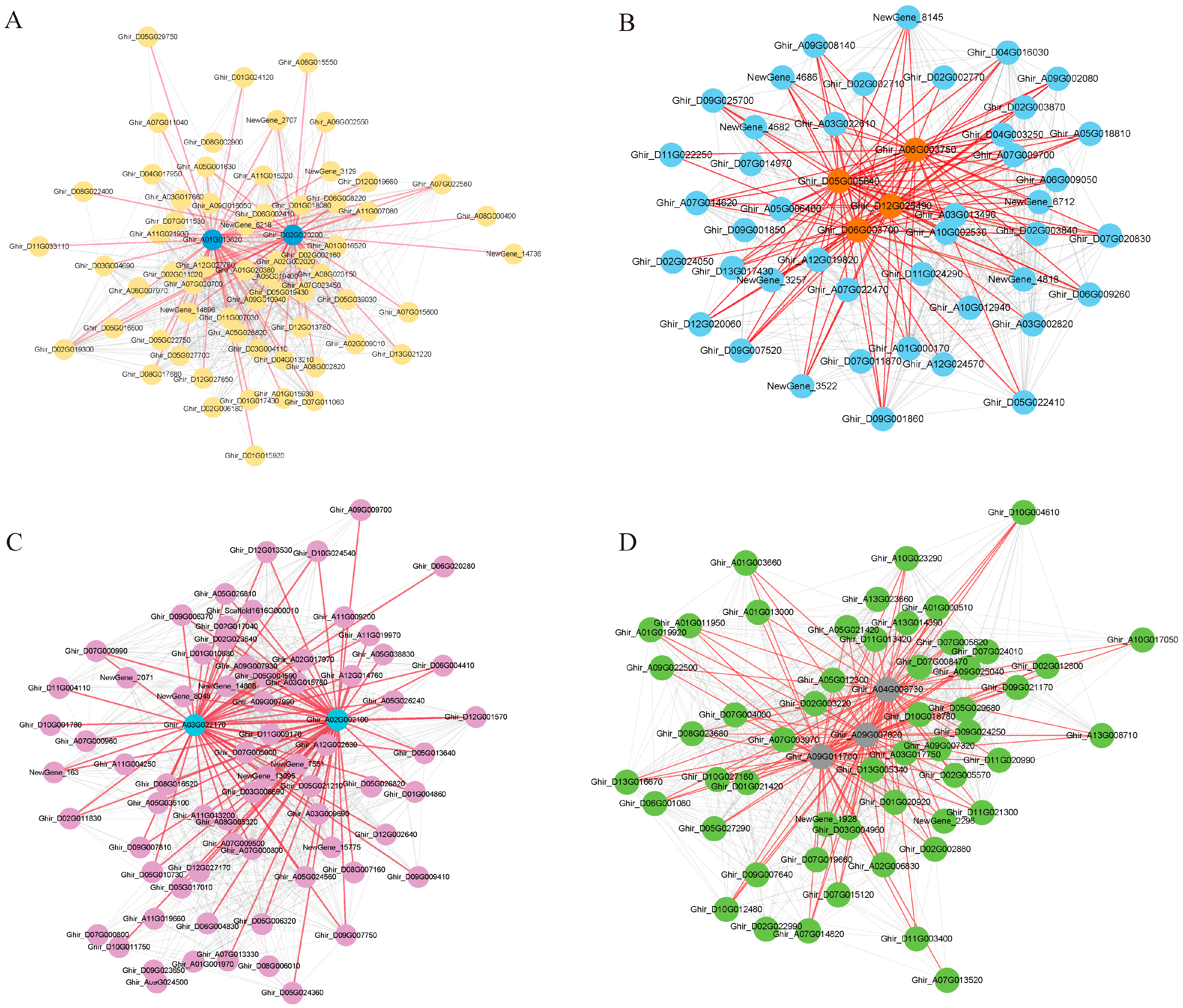
| Modules | Gene ID | Gene Name | Description |
|---|---|---|---|
| Black | Ghir_A01G013620 | COR2 | Non-functional NADPH-dependent codeinone reductase 2 |
| Ghir_D02G020200 | EXL5 | Protein EXORDIUM-like 5 | |
| Blue | Ghir_D05G005640 | MIK2 | MDIS1-interacting receptor-like kinase 2 |
| Ghir_A06G003750 | MIK2 | MDIS1-interacting receptor-like kinase 2 | |
| Ghir_D06G003700 | MIK2 | MDIS1-interacting receptor-like kinase 2 | |
| Ghir_D12G026490 | WRKY30 | Probable WRKY transcription factor 30 | |
| Cyan | Ghir_A03G022170 | ATJ11 | Chaperone protein dnaJ 11, chloroplastic |
| Ghir_A02G002100 | SBP2 | Selenium-binding protein 2 | |
| Grey60 | Ghir_A04G008730 | ASL | Argininosuccinate lyase |
| Ghir_A09G007820 | SKIP27 | F-box protein SKIP27 | |
| Ghir_A09G011700 | F6′H1 | Feruloyl CoA ortho-hydroxylase 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, B.; Li, J.; Zhang, R.; Luo, P.; Wang, Z.; Shi, S.; Gao, W.; Li, S. RNA-Seq and WGCNA Analyses Reveal Key Regulatory Modules and Genes for Salt Tolerance in Cotton. Genes 2024, 15, 1176. https://doi.org/10.3390/genes15091176
Pang B, Li J, Zhang R, Luo P, Wang Z, Shi S, Gao W, Li S. RNA-Seq and WGCNA Analyses Reveal Key Regulatory Modules and Genes for Salt Tolerance in Cotton. Genes. 2024; 15(9):1176. https://doi.org/10.3390/genes15091176
Chicago/Turabian StylePang, Bo, Jing Li, Ru Zhang, Ping Luo, Zhengrui Wang, Shunyu Shi, Wenwei Gao, and Shengmei Li. 2024. "RNA-Seq and WGCNA Analyses Reveal Key Regulatory Modules and Genes for Salt Tolerance in Cotton" Genes 15, no. 9: 1176. https://doi.org/10.3390/genes15091176
APA StylePang, B., Li, J., Zhang, R., Luo, P., Wang, Z., Shi, S., Gao, W., & Li, S. (2024). RNA-Seq and WGCNA Analyses Reveal Key Regulatory Modules and Genes for Salt Tolerance in Cotton. Genes, 15(9), 1176. https://doi.org/10.3390/genes15091176








