Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Management
2.2. Blood Sample Collection and Measurement
2.3. Liver Tissue Sample Collection
2.4. Transcriptomics Analysis and Data Processing
2.5. Construction of Gene Co-Expression Network
2.6. Identification and Functional Enrichment Analysis of Intersection Genes between WGCNA Modules and DEGs
2.7. Screening the Hub Genes Highly Associated with Milk Yield Using Linear Mixed-Effects Models
2.8. Random Forest Machine Learning Model Validated the Importance of Hub Genes for Milk Production
2.9. Statistical Analysis
3. Results
3.1. Milk Yield Positively Correlates with Feed Conversion Efficiency
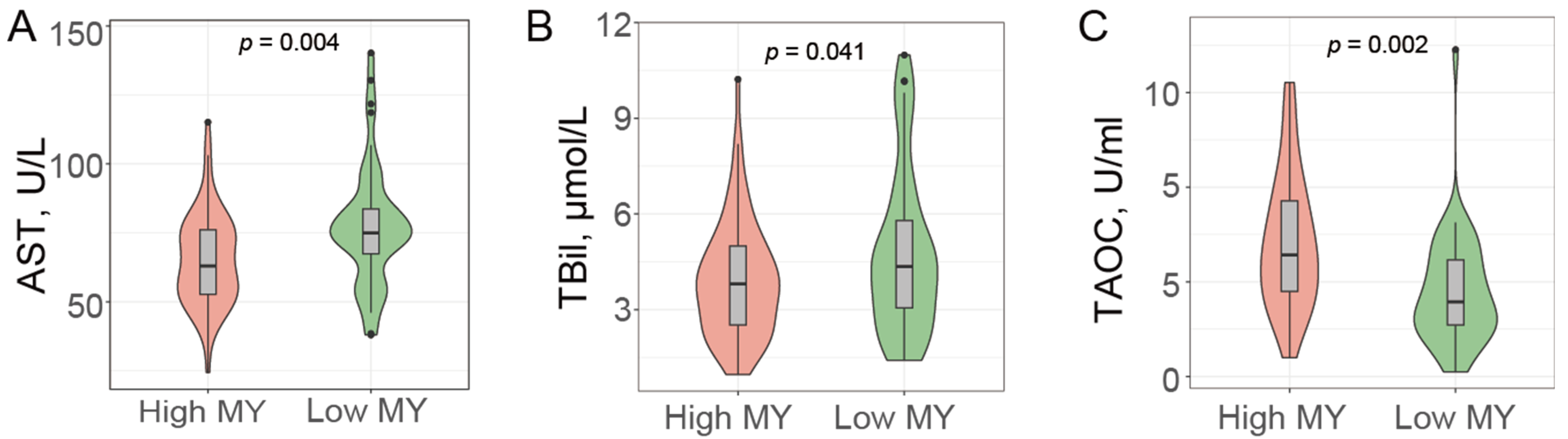
3.2. Liver Health Status and Cows’ Milk Production
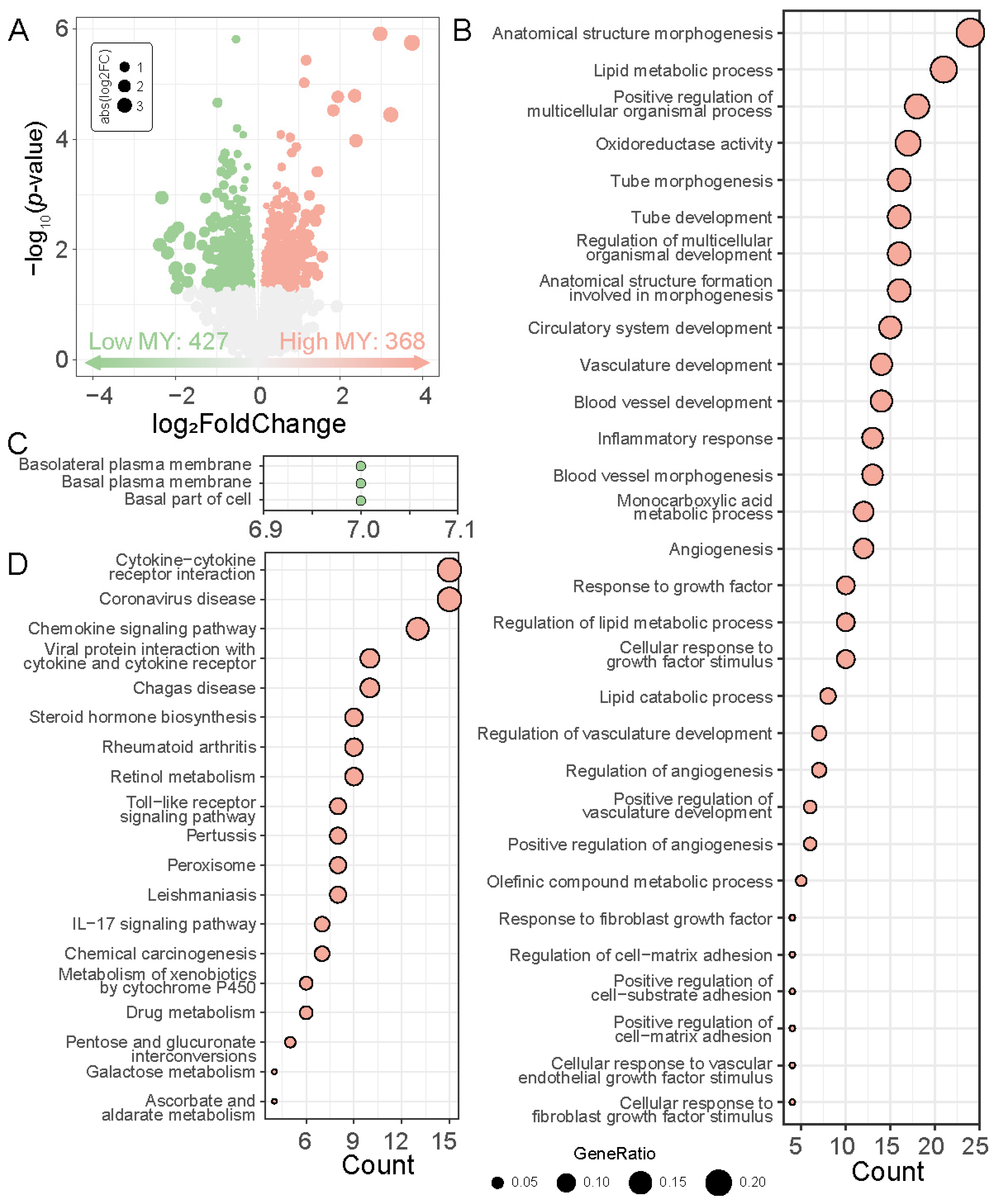
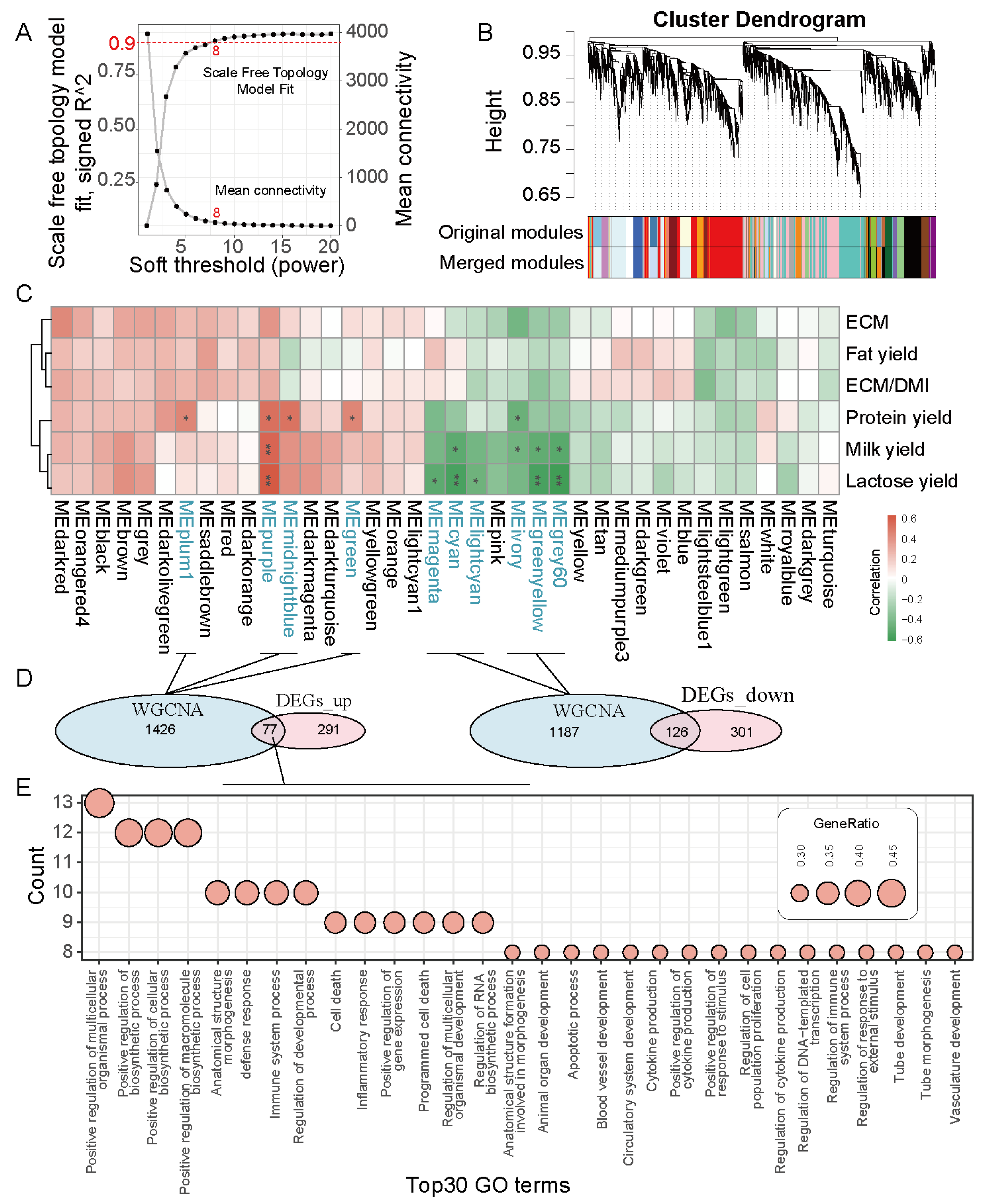
3.3. WGCNA Construction and Key Module Genes Identification
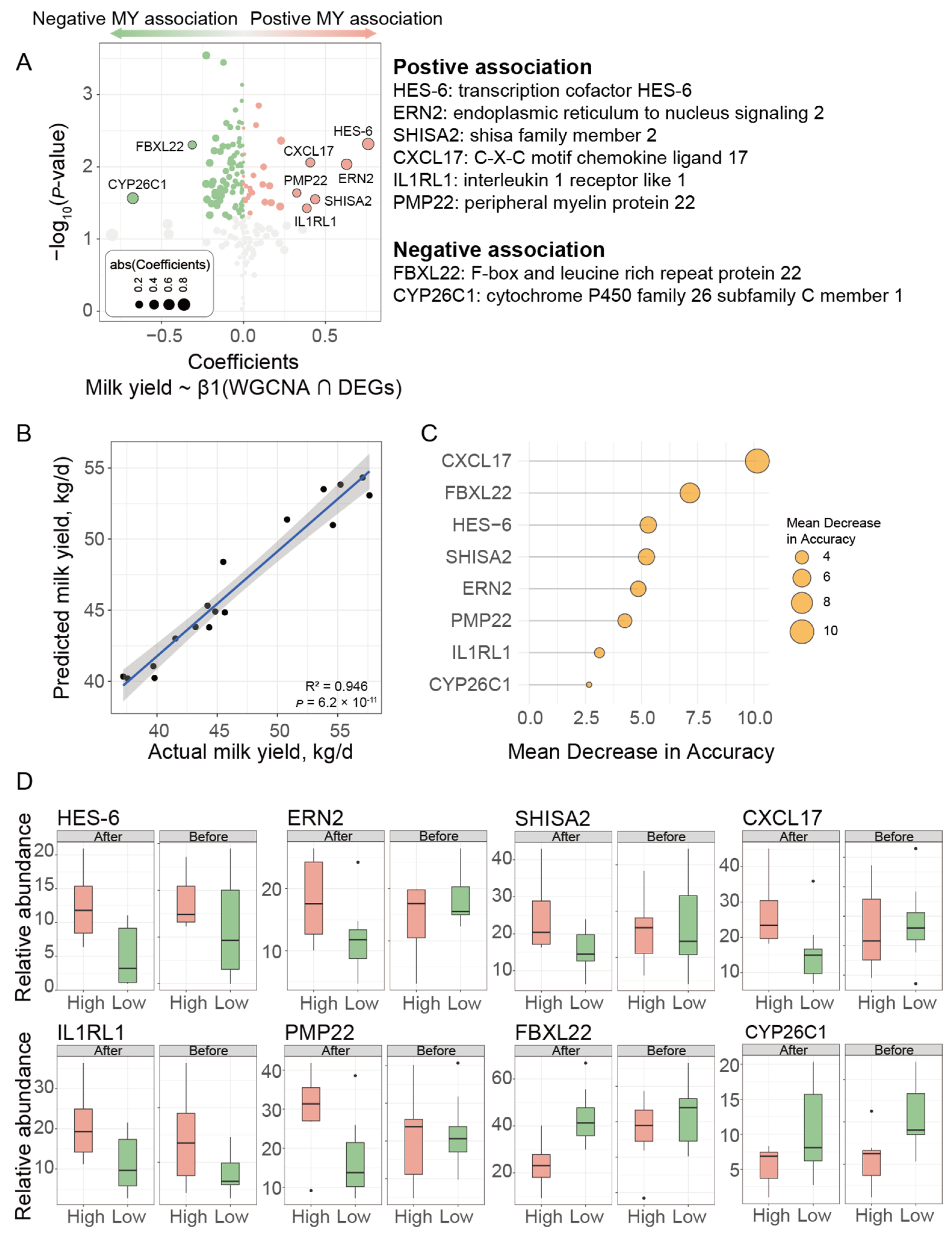
3.4. Hub Genes Further Filtered with Linear Mixed-Effects Model and Verification Using Machine Learning
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holter, J.B.; West, J.W.; McGilliard, M.L. Predicting ad libitum dry matter intake and yield of holstein cows. J. Dairy Sci. 1997, 80, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- John, A.J.; Freeman, M.J.; Kerrisk, K.F.; Garcia, S.C.; Clark, C.E.F. Robot utilisation of pasture-based dairy cows with varying levels of milking frequency. Animal 2019, 13, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- McBride, B.W.; Kelly, J.M. Energy cost of absorption and metabolism in the ruminant gastrointestinal tract and liver: A review. J. Anim. Sci. 1990, 68, 2997–3010. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Gao, S.T.; Girma, D.D.; Bionaz, M.; Ma, L.; Bu, D.P. Hepatic transcriptomic adaptation from prepartum to postpartum in dairy cows. J. Dairy Sci. 2021, 104, 1053–1072. [Google Scholar] [CrossRef]
- Drackley, J.K.; Dann, H.M.; Douglas, N.; Guretzky, N.A.J.; Litherland, N.B.; Underwood, J.P.; Loor, J.J. Physiological and pathological adaptations in dairy cows that may increase susceptibility to periparturient diseases and disorders. Ital. J. Anim. Sci. 2005, 4, 323–344. [Google Scholar] [CrossRef]
- Graber, M.; Kohler, S.; Kaufmann, T.; Doherr, M.G.; Bruckmaier, R.M.; van Dorland, H.A. A field study on characteristics and diversity of gene expression in the liver of dairy cows during the transition period. J. Dairy Sci. 2010, 93, 5200–5215. [Google Scholar] [CrossRef]
- Schlegel, G.; Ringseis, R.; Keller, J.; Schwarz, F.J.; Eder, K. Changes in the expression of hepatic genes involved in cholesterol homeostasis in dairy cows in the transition period and at different stages of lactation. J. Dairy Sci. 2012, 95, 3826–3836. [Google Scholar] [CrossRef]
- Ha, N.T.; Drögemüller, C.; Reimer, C.; Schmitz-Hsu, F.; Bruckmaier, R.M.; Simianer, H.; Gross, J.J. Liver transcriptome analysis reveals important factors involved in the metabolic adaptation of the transition cow. J. Dairy Sci. 2017, 100, 9311–9323. [Google Scholar] [CrossRef]
- Li, Q.; Liang, R.; Li, Y.; Gao, Y.; Li, Q.; Sun, D.; Li, J. Identification of candidate genes for milk production traits by rna sequencing on bovine liver at different lactation stages. BMC Genet. 2020, 21, 72. [Google Scholar] [CrossRef]
- Council, N.R. Nutrient Requirements of Dairy Cattle: Seventh Revised Edition, 2001; The National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Chen, L.; Thorup, V.M.; Kudahl, A.B.; Østergaard, S. Effects of heat stress on feed intake, milk yield, milk composition, and feed efficiency in dairy cows: A meta-analysis. J. Dairy Sci. 2024, 107, 3207–3218. [Google Scholar] [CrossRef] [PubMed]
- Sjaunja, L.O.; Baevre, L.; Junkkarinen, L.; Pedersen, J.; Setala, J. A Nordic Proposal for an Energy Corrected Milk (ecm) Formula. Available online: https://www.researchgate.net/publication/284193091_A_Nordic_proposal_for_an_energy_corrected_milk_ECM_formula (accessed on 16 September 2024).
- Girma, D.D.; Ma, L.; Wang, F.; Jiang, Q.R.; Callaway, T.R.; Drackley, J.K.; Bu, D.P. Effects of close-up dietary energy level and supplementing rumen-protected lysine on energy metabolites and milk production in transition cows. J. Dairy Sci. 2019, 102, 7059–7072. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Bionaz, M.; Wang, M.; Nan, X.; Ma, L.; Wang, J. Transcriptome difference and potential crosstalk between liver and mammary tissue in mid-lactation primiparous dairy cows. PLoS ONE 2017, 12, e0173082. [Google Scholar] [CrossRef] [PubMed]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed fastqc emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read rna-seq alignments with stringtie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An r package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Wgcna: An r package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Fu, S.; Wu, D.; Jiang, W.; Li, J.; Long, J.; Jia, C.; Zhou, T. Molecular biomarkers in drug-induced liver injury: Challenges and future perspectives. Front. Pharmacol. 2019, 10, 1667. [Google Scholar] [CrossRef] [PubMed]
- Bayat, A.R.; Rinne, M.; Kuoppala, K.; Ahvenjärvi, S.; Huhtanen, P. Ruminal large and small particle kinetics in dairy cows fed primary growth and regrowth grass silages harvested at two stages of growth. Anim. Feed Sci. Technol. 2011, 165, 51–60. [Google Scholar] [CrossRef]
- Craig, A.L.; Gordon, A.W.; Hamill, G.; Ferris, C.P. Milk composition and production efficiency within feed-to-yield systems on commercial dairy farms in northern ireland. Animals 2022, 12, 1771. [Google Scholar] [CrossRef] [PubMed]
- Huntington, G.B. Energy metabolism in the digestive tract and liver of cattle: Influence of physiological state and nutrition. Reprod. Nutr. Dev. 1990, 30, 35–47. [Google Scholar] [CrossRef]
- Reynolds, C.K.; Harmon, D.L.; Cecava, M.J. Absorption and delivery of nutrients for milk protein synthesis by portal-drained viscera. J. Dairy Sci. 1994, 77, 2787–2808. [Google Scholar] [CrossRef]
- Ning, M.; Zhao, Y.; Dai, D.M.; Yao, C.; Liu, H.T.; Fang, L.Z.; Wang, B.; Zhang, Y.; Cao, J. Gene co-expression network and differential expression analyses of subcutaneous white adipose tissue reveal novel insights into the pathological mechanisms underlying ketosis in dairy cows. J. Dairy Sci. 2023, 106, 5018–5028. [Google Scholar] [CrossRef]
- Stojević, Z.; Pirsljin, J.; Milinković-Tur, S.; Zdelar-Tuk, M.; Beer Ljubic, B. Activities of ast, alt and ggt in clinically healthy dairy cows during lactation and in the dry period. Vet. Arh. 2005, 75, 67–73. [Google Scholar]
- Walter, L.L.; Gärtner, T.; Gernand, E.; Wehrend, A.; Donat, K. Effects of parity and stage of lactation on trend and variability of metabolic markers in dairy cows. Animals 2022, 12, 1008. [Google Scholar] [CrossRef]
- Schären, M.; Riefke, B.; Slopianka, M.; Keck, M.; Gruendemann, S.; Wichard, J.; Brunner, N.; Klein, S.; Snedec, T.; Theinert, K.B.; et al. Aspects of transition cow metabolomics—Part III: Alterations in the metabolome of liver and blood throughout the transition period in cows with different liver metabotypes. J. Dairy Sci. 2021, 104, 9245–9262. [Google Scholar] [CrossRef]
- Nemati, M.; Menatian, S.; Joz Ghasemi, S.; Hooshmandfar, R.; Taheri, M.; Saifi, T. Effect of protected-glutamine supplementation on performance, milk composition and some blood metabolites in fresh holstein cows. Iran. J. Vet. Res. 2018, 19, 225–228. [Google Scholar]
- Garcia, A.B.; Angeli, N.; Machado, L.; de Cardoso, F.C.; Gonzalez, F. Relationships between heat stress and metabolic and milk parameters in dairy cows in southern brazil. Trop. Anim. Health Prod. 2015, 47, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, C.; Bernabucci, U.; Basiricò, L. Effect of antioxidant supplementation on milk yield and quality in italian mediterranean lactating buffaloes. Animals 2022, 12, 1903. [Google Scholar] [CrossRef] [PubMed]
- Matra, M.; Wanapat, M. Phytonutrient pellet supplementation enhanced rumen fermentation efficiency and milk production of lactating holstein-friesian crossbred cows. Anim. Nutr. 2022, 9, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Koch, C.; Romberg, F.J.; Winkler, A.; Dusel, G.; Herzog, E.; Most, E.; Eder, K. The effect of grape seed and grape marc meal extract on milk performance and the expression of genes of endoplasmic reticulum stress and inflammation in the liver of dairy cows in early lactation. J. Dairy Sci. 2015, 98, 8856–8868. [Google Scholar] [CrossRef]
- Gessner, D.K.; Winkler, A.; Koch, C.; Dusel, G.; Liebisch, G.; Ringseis, R.; Eder, K. Analysis of hepatic transcript profile and plasma lipid profile in early lactating dairy cows fed grape seed and grape marc meal extract. BMC Genom. 2017, 18, 253. [Google Scholar] [CrossRef]
- Abu Hafsa, S.H.; Hassan, A.A. Grape seed alleviates lindane-induced oxidative stress and improves growth performance, caecal fermentation and antioxidant capacity in growing rabbits. J. Anim. Physiol. Anim. Nutr. 2022, 106, 899–909. [Google Scholar] [CrossRef]
- Ao, X.; Kim, I.H. Effects of grape seed extract on performance, immunity, antioxidant capacity, and meat quality in pekin ducks. Poult. Sci. 2020, 99, 2078–2086. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, X.; Chen, D.; He, J.; Zheng, P.; Luo, Y.; Yu, B.; Huang, Z. Dietary grape seed proanthocyanidin extract supplementation improves antioxidant capacity and lipid metabolism in finishing pigs. Anim. Biotechnol. 2023, 34, 4021–4031. [Google Scholar] [CrossRef]
- Mabrouk, M.; El Ayed, M.; Démosthènes, A.; Aissouni, Y.; Aouani, E.; Daulhac-Terrail, L.; Mokni, M.; Bégou, M. Antioxidant effect of grape seed extract corrects experimental autoimmune encephalomyelitis behavioral dysfunctions, demyelination, and glial activation. Front. Immunol. 2022, 13, 960355. [Google Scholar] [CrossRef]
- White, H.M. Adsa foundation scholar award: Influencing hepatic metabolism: Can nutrient partitioning be modulated to optimize metabolic health in the transition dairy cow? J. Dairy Sci. 2020, 103, 6741–6750. [Google Scholar] [CrossRef]
- McCabe, C.J.; Boerman, J.P. Invited review: Quantifying protein mobilization in dairy cows during the transition period. Appl. Anim. Behav. Sci. 2020, 36, 389–396. [Google Scholar] [CrossRef]
- Robinson, M.W.; Harmon, C.; O’Farrelly, C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016, 13, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Choreño-Parra, J.A.; Thirunavukkarasu, S.; Zúñiga, J.; Khader, S.A. The protective and pathogenic roles of cxcl17 in human health and disease: Potential in respiratory medicine. Cytokine Growth Factor Rev. 2020, 53, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Ma, X.; Wang, F.; Chen, D.; Lin, Y.; Wang, Y.; Liu, W.; Li, Y. Effect of cxcl17 on subcutaneous preadipocytes proliferation in goats. Animals 2023, 13, 1757. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Zhen, Z.; Ma, X.; Yu, W.; Zeng, H.; Li, L. Cxcl17 promotes cell metastasis and inhibits autophagy via the lkb1-ampk pathway in hepatocellular carcinoma. Gene 2019, 690, 129–136. [Google Scholar] [CrossRef]
- Li, L.; Yan, J.; Xu, J.; Liu, C.Q.; Zhen, Z.J.; Chen, H.W.; Ji, Y.; Wu, Z.P.; Hu, J.Y.; Zheng, L.; et al. Cxcl17 expression predicts poor prognosis and correlates with adverse immune infiltration in hepatocellular carcinoma. PLoS ONE 2014, 9, e110064. [Google Scholar] [CrossRef]
- Krossa, I.; Strub, T.; Martel, A.; Nahon-Esteve, S.; Lassalle, S.; Hofman, P.; Baillif, S.; Ballotti, R.; Bertolotto, C. Recent advances in understanding the role of hes6 in cancers. Theranostics 2022, 12, 4374–4385. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, P.; Zhang, J.; Gong, H.; Zhang, X.; Song, J.; Nie, L.; Peng, Y.; Li, Y.; Peng, H.; et al. The novel gata1-interacting protein hes6 is an essential transcriptional cofactor for human erythropoiesis. Nucleic Acids Res. 2023, 51, 4774–4790. [Google Scholar] [CrossRef]
- Tamura, K.; Furihata, M.; Satake, H.; Hashida, H.; Kawada, C.; Osakabe, H.; Fukuhara, H.; Kumagai, N.; Iiyama, T.; Shuin, T.; et al. Shisa2 enhances the aggressive phenotype in prostate cancer through the regulation of wnt5a expression. Oncol. Lett. 2017, 14, 6650–6658. [Google Scholar] [CrossRef]
- Li, J.; Parker, B.; Martyn, C.; Natarajan, C.; Guo, J. The pmp22 gene and its related diseases. Mol. Neurobiol. 2013, 47, 673–698. [Google Scholar] [CrossRef]
- Akhabir, L.; Sandford, A. Genetics of interleukin 1 receptor-like 1 in immune and inflammatory diseases. Curr. Genom. 2010, 11, 591–606. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, C.; Zhu La, A.L.T.; Gao, S.; Gao, W.; Ma, L.; Bu, D.; Zhang, W. Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production. Genes 2024, 15, 1229. https://doi.org/10.3390/genes15091229
Du C, Zhu La ALT, Gao S, Gao W, Ma L, Bu D, Zhang W. Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production. Genes. 2024; 15(9):1229. https://doi.org/10.3390/genes15091229
Chicago/Turabian StyleDu, Chao, A La Teng Zhu La, Shengtao Gao, Wenshuo Gao, Lu Ma, Dengpan Bu, and Wenju Zhang. 2024. "Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production" Genes 15, no. 9: 1229. https://doi.org/10.3390/genes15091229
APA StyleDu, C., Zhu La, A. L. T., Gao, S., Gao, W., Ma, L., Bu, D., & Zhang, W. (2024). Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production. Genes, 15(9), 1229. https://doi.org/10.3390/genes15091229




