Identifying Genetic Predisposition to Dozer Lamb Syndrome: A Semi-Lethal Muscle Weakness Disease in Sheep
Abstract
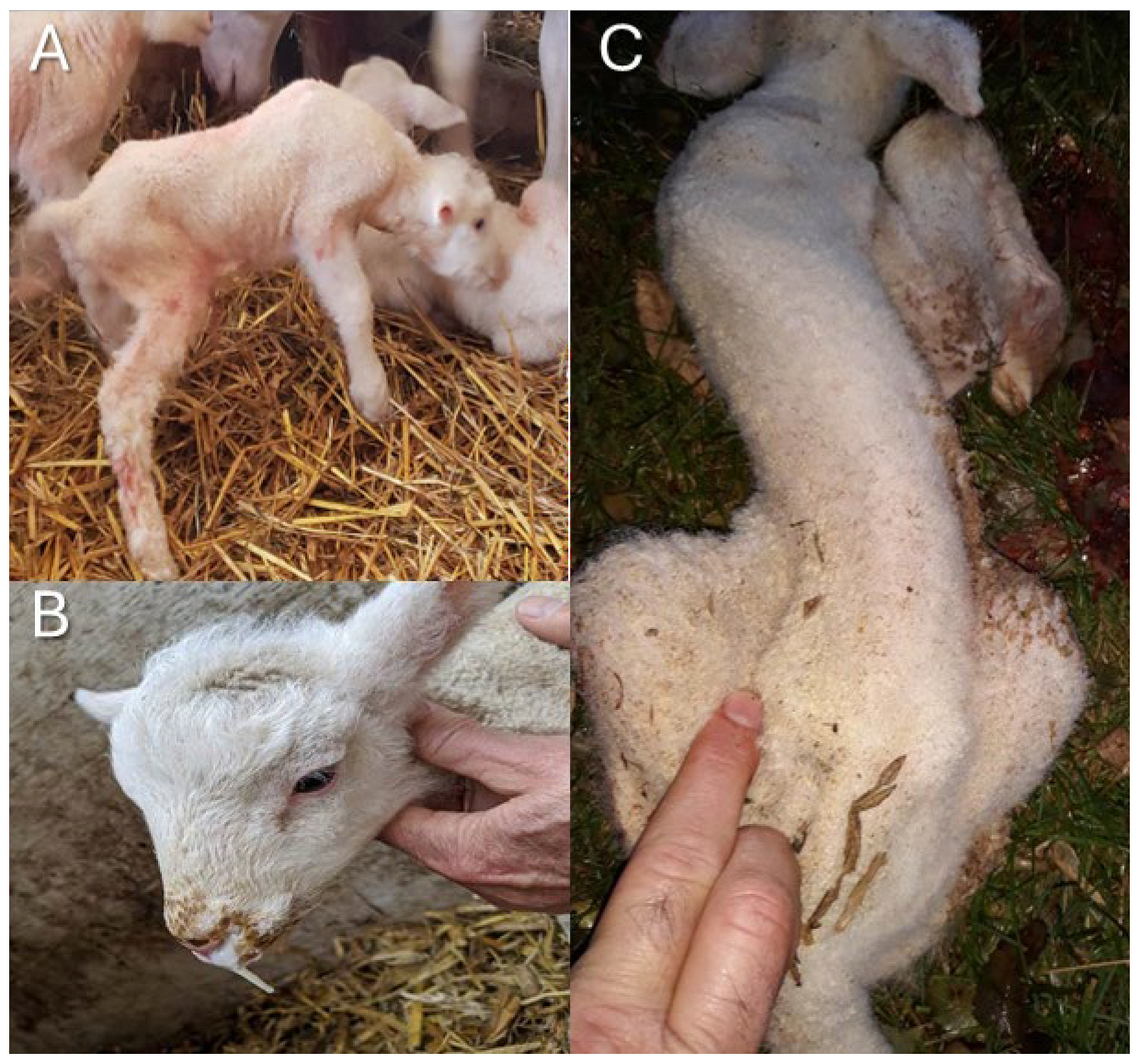
1. Introduction
2. Materials and Methods
2.1. Animals and Samples
2.2. DNA Extraction and Genotyping
2.3. Genome-Wide Association Study
2.4. Runs of Homozygosity
2.5. Sequence Data Used for Fine Mapping
3. Results
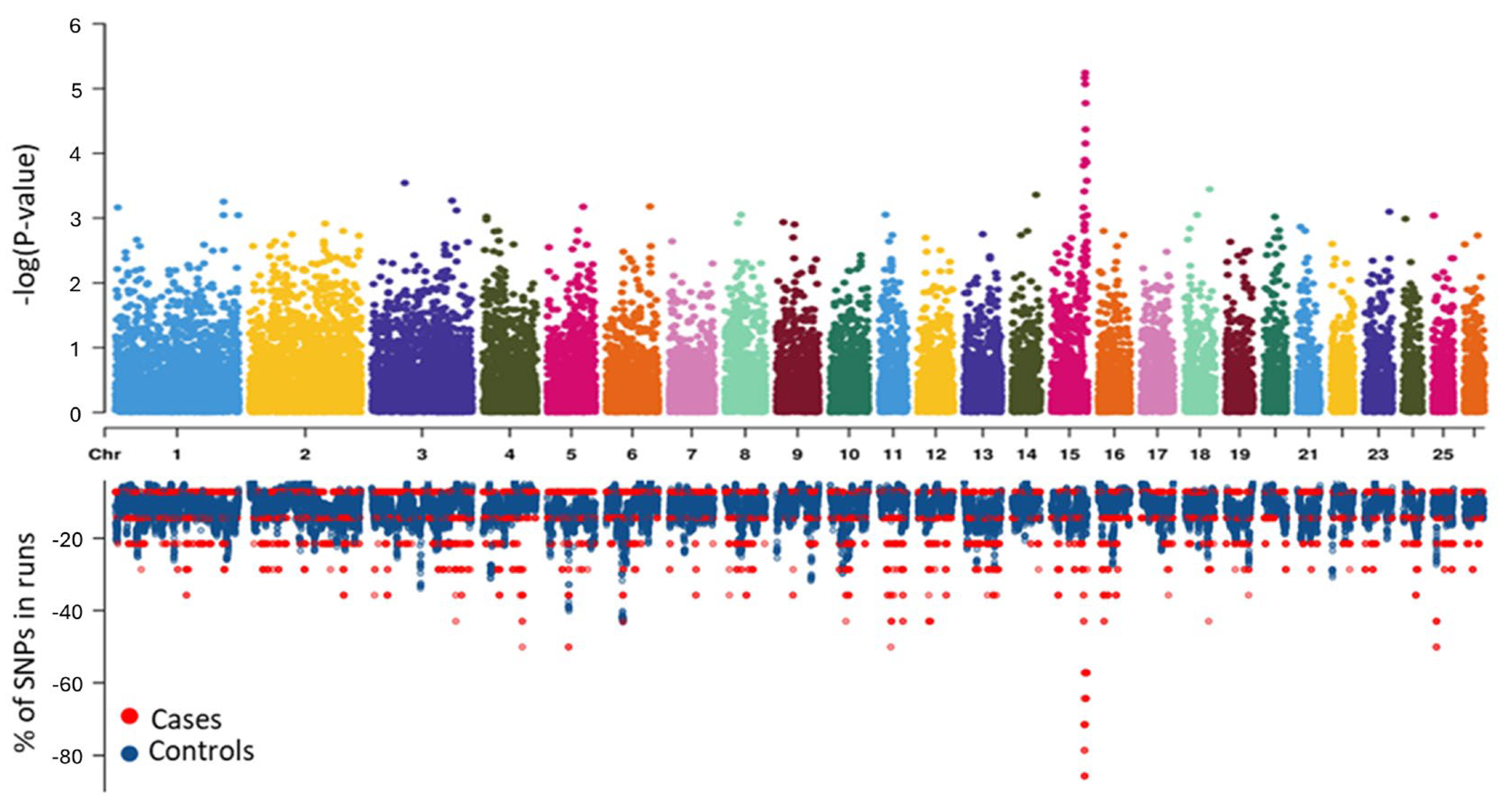
3.1. Genome Wide Association Study
3.2. Runs of Homozygosity
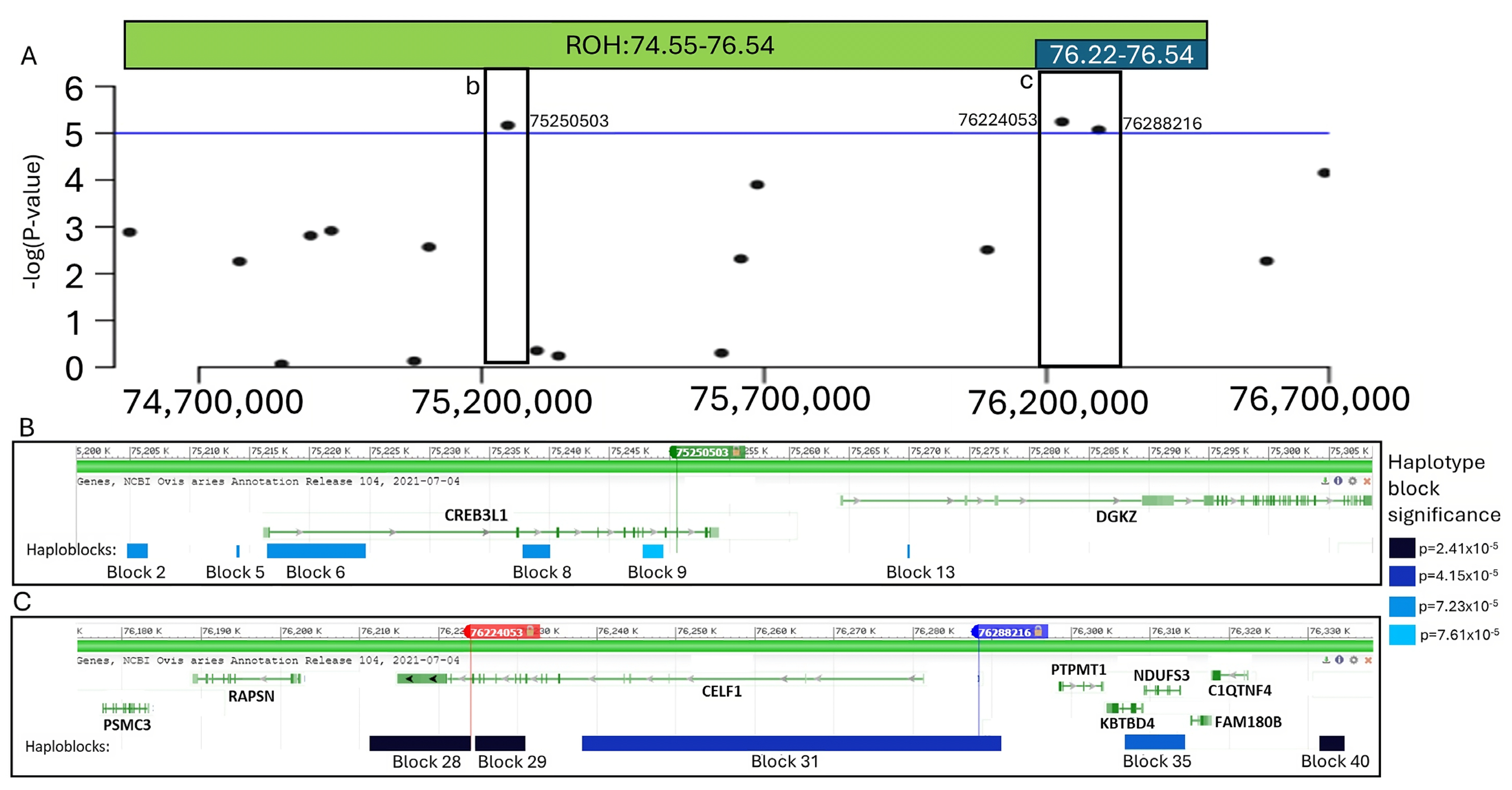
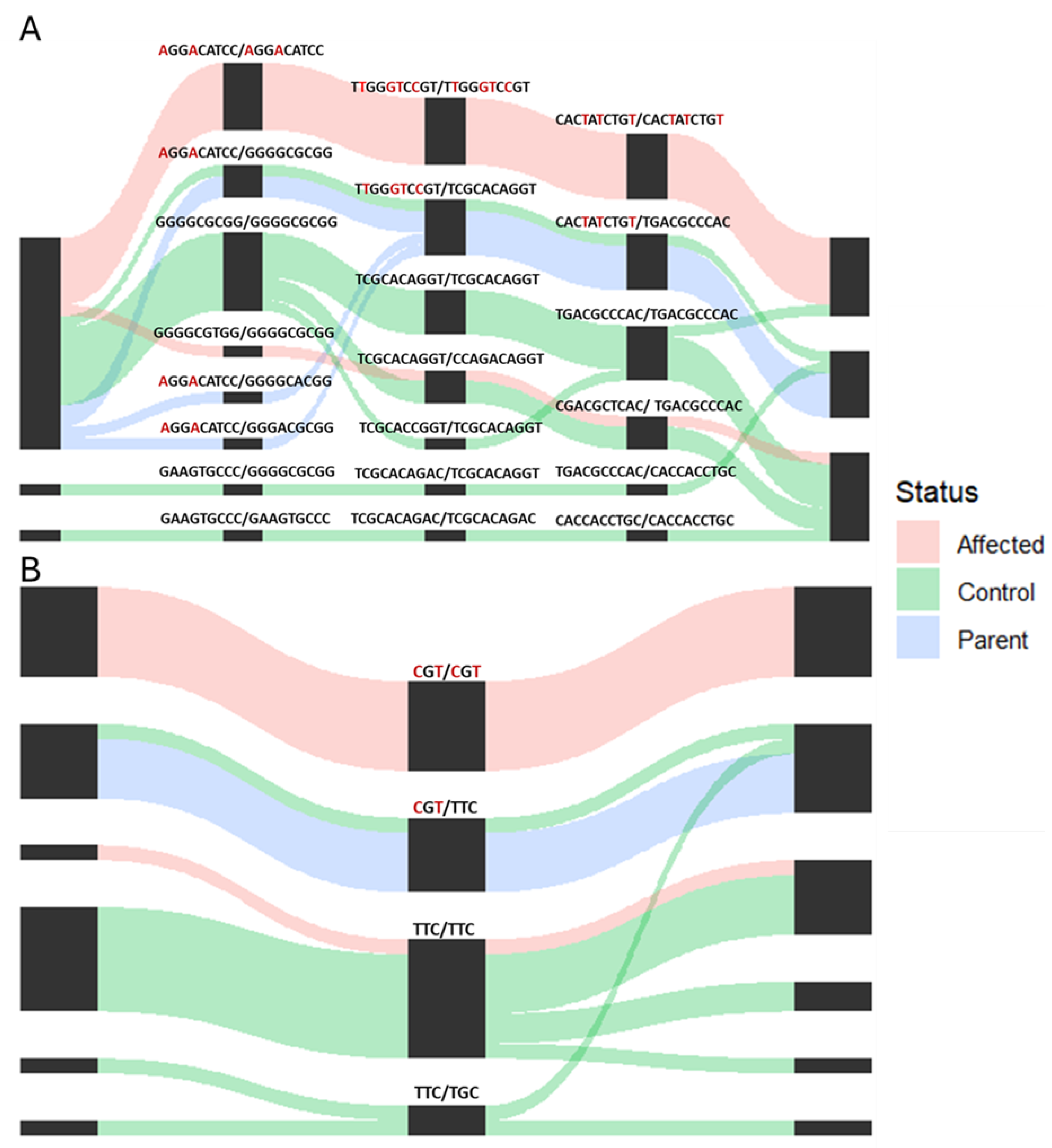
3.3. Whole Genome Fine Mapping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flinn, T.; Kleemann, D.O.; Swinbourne, A.M.; Kelly, J.M.; Weaver, A.C.; Walker, S.K.; Gatford, K.L.; Kind, K.L.; van Wettere, W.H.E.J. Neonatal lamb mortality: Major risk factors and the potential ameliorative role of melatonin. J. Anim. Sci. Biotechnol. 2020, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, C.M. Genetic and physiological determinants of maternal behavior and lamb survival: Implications for low-input sheep management1,2. J. Anim. Sci. 2008, 86 (Suppl. S14), E246–E258. [Google Scholar] [CrossRef] [PubMed]
- Nagy, H.; Veerapaneni, K.D. Myopathy; StatPearls Publishing: St. Petersburg, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK562290/ (accessed on 15 November 2023).
- Shelton, G.D.; Minor, K.M.; Friedenberg, S.G.; Cullen, J.N.; Guo, L.T.; Mickelson, J.R. Current Classification of Canine Muscular Dystrophies and Identification of New Variants. Genes 2023, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Mickelson, J.R.; Valberg, S.J. The Genetics of Skeletal Muscle Disorders in Horses. Annu. Rev. Anim. Biosci. 2015, 3, 197–217. [Google Scholar] [CrossRef]
- Ravenscroft, G.; Bryson-Richardson, R.J.; Nowak, K.J.; Laing, N.G. Recent advances in understanding congenital myopathies. F1000Research 2018, 7, 1921. [Google Scholar] [CrossRef]
- Lehmann-Horn, F.; Jurkat-Rott, K. Voltage-Gated Ion Channels and Hereditary Disease. Physiol. Rev. 1999, 79, 1317–1372. [Google Scholar] [CrossRef]
- Stirm, M.; Fonteyne, L.M.; Shashikadze, B.; Stöckl, J.B.; Kurome, M.; Keßler, B.; Zakhartchenko, V.; Kemter, E.; Blum, H.; Arnold, G.J.; et al. Pig models for Duchenne muscular dystrophy–from disease mechanisms to validation of new diagnostic and therapeutic concepts. Neuromuscul. Disord. 2022, 32, 543–556. [Google Scholar] [CrossRef]
- Huang, K.; Bi, F.-F.; Yang, H. A Systematic Review and Meta-Analysis of the Prevalence of Congenital Myopathy. Front. Neurol. 2021, 12, 761636. [Google Scholar] [CrossRef]
- North, K.N.; Wang, C.H.; Clarke, N.; Jungbluth, H.; Vainzof, M.; Dowling, J.J.; Amburgey, K.; Quijano-Roy, S.; Beggs, A.H.; Sewry, C.; et al. Approach to the diagnosis of congenital myopathies. Neuromuscul. Disord. 2014, 24, 97–116. [Google Scholar] [CrossRef]
- Sewry, C.A.; Wallgren-Pettersson, C. Myopathology in congenital myopathies. Neuropathol. Appl. Neurobiol. 2017, 43, 5–23. [Google Scholar] [CrossRef]
- Ewert, K. Attention Polypay Producers: Have lambs like this ever been born to your Polypay ewes? American Polypay Sheep Association News. 2022, pp. 6–7. Available online: https://www.polypay.org/assets/pdfs/2022-01-Newsletter.pdf (accessed on 15 April 2022).
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Habor Laboratory: Cold Spring Harbor, NY, USA, 1989; Available online: https://www.cabdirect.org/cabdirect/abstract/19901616061 (accessed on 11 January 2025).
- Notter, D.R.; Mousel, M.R.; Lewis, G.S.; Leymaster, K.A.; Taylor, J.B. Evaluation of Rambouillet, Polypay, and Romanov–White Dorper × Rambouillet ewes mated to terminal sires in an extensive rangeland production system: Lamb production. J. Anim. Sci. 2017, 95, 3851–3862. [Google Scholar] [CrossRef] [PubMed]
- Stegemiller, M.R.; Redden, R.R.; Notter, D.R.; Taylor, T.; Taylor, J.B.; Cockett, N.E.; Heaton, M.P.; Kalbfleisch, T.S.; Murdoch, B.M. Using whole genome sequence to compare variant callers and breed differences of US sheep. Front. Genet. 2023, 13, 1060882. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira MA, R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. detectRUNS: Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. CRAN (The Comprehensive R Archive Network), 6 February 2018. Available online: https://cran.r-project.org/web/packages/detectRUNS/index.html (accessed on 11 January 2025).
- Szumilas, M. Explaining Odds Ratios. J. Can. Acad. Child Adolesc. Psychiatry 2010, 19, 227. [Google Scholar] [PubMed]
- Davenport, K.M.; Bickhart, D.M.; Worley, K.; Murali, S.C.; Salavati, M.; Clark, E.L.; Cockett, N.E.; Heaton, M.P.; Smith TP, L.; Murdoch, B.M.; et al. An improved ovine reference genome assembly to facilitate in-depth functional annotation of the sheep genome. GigaScience 2022, 11, giab096. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Barrett, J.C. Haploview: Visualization and Analysis of SNP Genotype Data. Cold Spring Harb. Protoc. 2009, 2009, pdb.ip71. [Google Scholar] [CrossRef]
- Triant, D.A.; Walsh, A.T.; Hartley, G.A.; Petry, B.; Stegemiller, M.R.; Nelson, B.M.; McKendrick, M.M.; Fuller, E.P.; Cockett, N.E.; Koltes, J.E.; et al. AgAnimalGenomes: Browsers for viewing and manually annotating farm animal genomes. Mamm. Genome 2023, 34, 418–436. [Google Scholar] [CrossRef]
- Kingdom, R.; Wright, C.F. Incomplete penetrance and variable expressivity: From clinical studies to population cohorts. Front. Genet. 2022, 13, 920390. [Google Scholar] [CrossRef]
- Blech-Hermoni, Y.; Stillwagon, S.J.; Ladd, A.N. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev. Dyn. 2013, 242, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.N.; Charlet-B, N.; Cooper, T.A. The CELF Family of RNA Binding Proteins Is Implicated in Cell-Specific and Developmentally Regulated Alternative Splicing. Mol. Cell. Biol. 2001, 21, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.N.; Stenberg, M.G.; Swanson, M.S.; Cooper, T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005, 233, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Corbeil-Girard, L.-P.; Klein, A.F.; Sasseville, A.M.-J.; Lavoie, H.; Dicaire, M.-J.; Saint-Denis, A.; Pagé, M.; Duranceau, A.; Codère, F.; Bouchard, J.-P.; et al. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol. Dis. 2005, 18, 551–567. [Google Scholar] [CrossRef] [PubMed]
- Harper, P.S. Congenital myotonic dystrophy in Britain. I. Clinical aspects. Arch. Dis. Child. 1975, 50, 505–513. [Google Scholar] [CrossRef]
- Ho, G.; Carey, K.A.; Cardamone, M.; Farrar, M.A. Myotonic dystrophy type 1: Clinical manifestations in children and adolescents. Arch. Dis. Child. 2019, 104, 48–52. [Google Scholar] [CrossRef]
- Savić Pavićević, D.; Miladinović, J.; Brkušanin, M.; Šviković, S.; Brajušković, G.; Romac, S.; Djurica, S. Molecular Genetics and Genetic Testing in Myotonic Dystrophy Type 1. BioMed Res. Int. 2013, 2013, e391821. [Google Scholar] [CrossRef]
- Thornell, L.-E.; Lindstöm, M.; Renault, V.; Klein, A.; Mouly, V.; Ansved, T.; Butler-Browne, G.; Furling, D. Satellite cell dysfunction contributes to the progressive muscle atrophy in myotonic dystrophy type 1. Neuropathol. Appl. Neurobiol. 2009, 35, 603–613. [Google Scholar] [CrossRef]
- Furling, D.; Coiffier, L.; Mouly, V.; Barbet, J.P.; Lacau St Guily, J.; Taneja, K.; Gourdon, G.; Junien, C.; Butler-Browne, G.S. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001, 10, 2079–2087. [Google Scholar] [CrossRef]
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.-P.; Hudson, T.; et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992, 68, 799–808. [Google Scholar] [CrossRef]
- Kuyumcu-Martinez, N.M.; Wang, G.-S.; Cooper, T.A. Increased Steady-State Levels of CUGBP1 in Myotonic Dystrophy 1 Are Due to PKC-Mediated Hyperphosphorylation. Mol. Cell 2007, 28, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, N.A.; Roma, P.; Polina, I.; Zong-Jin, Q.; Timchennko, L.T. Overexpression f CUG Triplet Repeat-binding Protein, CUGBP1, in Mice Inhibits Myogenesis. J. Biol. Chem. 2004, 279, 13129–13139. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Shen, X.; Chen, X.; Liang, R.; Azares, A.R.; Liu, Y. Celf1 regulates cell cycle and is partially responsible for defective myoblast differentiation in myotonic dystrophy RNA toxicity. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.C.; Guan, X.; Xia, Z.; Cooper, T.A. Increased nuclear but not cytoplasmic activities of CELF1 protein leads to muscle wasting. Hum. Mol. Genet. 2020, 29, 1729–1744. [Google Scholar] [CrossRef]
- Vlasova-St Louis, I.; Dickson, A.M.; Bohjanen, P.R.; Wilusz, C.J. CELFish ways to modulate mRNA decay. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 695–707. [Google Scholar] [CrossRef]
- Itai, T.; Hamanaka, K.; Sasaki, K.; Wagner, M.; Kotzaeridou, U.; Brösse, I.; Ries, M.; Kobayashi, Y.; Tohyama, J.; Kato, M.; et al. De novo variants in CELF2 that disrupt the nuclear localization signal cause developmental and epileptic encephalopathy. Hum. Mutat. 2021, 42, 66–76. [Google Scholar] [CrossRef]
- Kajdasz, A.; Niewiadomska, D.; Sekrecki, M.; Sobczak, K. Distribution of alternative untranslated regions within the mRNA of the CELF1 splicing factor affects its expression. Sci. Rep. 2022, 12, 190. [Google Scholar] [CrossRef]
- MacPherson, M.J.; Erickson, S.L.; Kopp, D.; Wen, P.; Aghanoori, M.-R.; Kedia, S.; Burns KM, L.; Vitobello, A.; Tran Mau-Them, F.; Thomas, Q.; et al. Nucleocytoplasmic transport of the RNA-binding protein CELF2 regulates neural stem cell fates. Cell Rep. 2021, 35, 109226. [Google Scholar] [CrossRef]
- Froehner, S.C.; Luetje, C.W.; Scotland, P.B.; Patrick, J. The postsynaptic 43K protein clusters muscle nicotinic acetylcholine receptors in Xenopus oocytes. Neuron 1990, 5, 403–410. [Google Scholar] [CrossRef]
- Estephan, E.d.P.; Zambon, A.A.; Marchiori, P.E.; da Silva AM, S.; Caldas, V.M.; Moreno CA, M.; Reed, U.C.; Horvath, R.; Töpf, A.; Lochmüller, H.; et al. Clinical variability of early-onset congenital myasthenic syndrome due to biallelic RAPSN mutations in Brazil. Neuromuscul. Disord. 2018, 28, 961–964. [Google Scholar] [CrossRef]
- Burke, G.; Cossins, J.; Maxwell, S.; Owens, G.; Vincent, A.; Robb, S.; Nicolle, M.; Hilton-Jones, D.; Newsom-Davis, J.; Palace, J.; et al. Rapsyn mutations in hereditary myasthenia: Distinct early- and late-onset phenotypes. Neurology 2003, 61, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Leshinsky-Silver, E.; Shapira, D.; Yosovitz, K.; Ginsberg, M.; Lerman-Sagie, T.; Lev, D. A novel mutation in the TPR6 domain of the RAPSN gene associated with congenital myasthenic syndrome. J. Neurol. Sci. 2012, 316, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, J.; Yang, C. The Neuronal Transcription Factor Creb3l1 Potential Upregulates Ntrk2 in the Hypertensive Microenvironment to Promote Vascular Smooth Muscle Cell-Neuron Interaction and Prevent Neurons from Ferroptosis: A Bioinformatic Research of scRNA-seq Data. Dis. Markers 2022, 2022, e8339759. [Google Scholar] [CrossRef]
- You, J.-S.; Lincoln, H.C.; Kim, C.-R.; Frey, J.W.; Goodman, C.A.; Zhong, X.-P.; Hornberger, T.A. The Role of Diacylglycerol Kinase ζ and Phosphatidic Acid in the Mechanical Activation of Mammalian Target of Rapamycin (mTOR) Signaling and Skeletal Muscle Hypertrophy. J. Biol. Chem. 2014, 289, 1551–1563. [Google Scholar] [CrossRef]
- Pereira, C.V.; Peralta, S.; Arguello, T.; Bacman, S.R.; Diaz, F.; Moraes, C.T. Myopathy reversion in mice after restauration of mitochondrial complex I. EMBO Mol. Med. 2020, 12, e10674. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, Q.; Li, S.; Li, Z.; Brotto, M.; Weiss, D.; Prosdocimo, D.; Xu, C.; Reddy, A.; Puchowicz, M.; et al. Loss of PTPMT1 limits mitochondrial utilization of carbohydrates and leads to muscle atrophy and heart failure in tissue-specific knockout mice. eLife 2023, 12, RP86944. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
| Chr | Position (bp) | rs Number | p-Value | Region of Interest | Genes Within the ROI |
|---|---|---|---|---|---|
| 15 | 75,250,503 | rs407879401 | 6.81 × 10−6 | ROI 1: 75,200,503–75,300,503 | CREB3L1, DGKZ |
| 15 | 76,224,053 | rs628219689 | 5.71 × 10−6 | ROI 2: 76,174,053–76,338,216 | PSMC3, RAPSN, CELF1, PTPMT1, KBTBD4, NDUFS3, FAM180B, C1QTNF4 |
| 15 | 76,288,216 | rs608876284 | 8.52 × 10−6 |
| Haplotype | Segment 1 | Segment 2 | Segment 3 | Segment 4 | Segment 5 | Affected Frequency | Control Frequency | Chi-Square p-Value | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | G | T | C | A | C | A | A | G | G | A | C | A | T | C | C | T | T | G | G | G | T | C | C | G | T | C | A | C | T | A | T | C | T | G | T | G | C | A | G | C | G | T | A | 0.857 | 0.179 | 2.41 × 10−5 |
| 2 | G | T | C | A | C | A | G | G | G | G | C | G | C | G | G | T | C | G | C | A | C | A | G | G | T | T | G | A | C | G | C | C | C | A | C | A | T | C | A | A | A | C | T | 0.071 | 0.536 | 0.0035 |
| 3 | G | T | C | A | C | A | G | G | G | G | C | G | T | G | G | C | C | A | G | A | C | A | G | G | T | T | G | A | C | G | C | C | C | A | C | A | T | C | A | A | A | C | T | 0.071 | 0.000 | 0.1523 |
| 4 | A | C | T | G | T | T | G | A | A | G | T | G | C | C | C | T | C | G | C | A | C | A | G | A | C | C | A | C | C | A | C | C | T | G | C | G | C | A | G | C | G | T | A | 0.000 | 0.107 | 0.2037 |
| 5 | G | T | C | A | C | A | G | G | G | G | C | G | C | G | G | C | C | A | G | A | C | A | G | G | T | C | G | A | C | G | C | T | C | A | C | A | T | C | A | A | A | C | T | 0.000 | 0.071 | 0.3055 |
| 6 | G | T | C | A | C | A | G | G | G | A | C | G | C | G | G | T | C | G | C | A | C | A | G | G | T | T | G | A | C | G | C | C | C | A | C | A | T | C | A | A | A | C | T | 0.000 | 0.036 | 0.4742 |
| 7 | G | T | C | A | C | A | G | G | G | G | C | A | C | G | G | T | C | G | C | A | C | A | G | G | T | T | G | A | C | G | C | C | C | A | C | A | T | C | A | A | A | C | T | 0.000 | 0.036 | 0.4742 |
| 8 | G | T | C | A | C | A | G | G | G | G | C | G | C | G | G | T | C | G | C | A | C | C | G | G | T | T | G | A | C | G | C | C | C | A | C | A | T | C | A | A | A | C | T | 0.000 | 0.036 | 0.4742 |
| Haplotype | Segment 1 | Segment 2 | Segment 3 | Frequency of Affected Haplotypes | Frequency of Control Haplotypes | Chi-Square p-Value | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | G | T | A | A | C | C | A | A | A | T | T | T | T | A | T | A | C | G | T | G | C | G | 0.857 | 0.179 | 2.41 × 10−5 |
| 2 | C | C | G | G | T | T | G | G | G | C | C | C | G | G | C | G | T | T | C | T | T | A | 0.071 | 0.571 | 0.0019 |
| 3 | C | C | G | G | T | T | A | G | G | C | C | C | G | G | C | G | T | T | C | T | T | A | 0.071 | 0.000 | 0.1523 |
| 4 | C | C | G | G | T | T | G | G | G | C | C | C | G | G | C | G | T | T | C | G | T | A | 0.000 | 0.143 | 0.1371 |
| 5 | G | T | A | A | C | C | G | A | A | T | T | T | T | A | T | A | T | G | C | T | C | G | 0.000 | 0.036 | 0.4742 |
| 6 | G | T | A | A | C | C | G | A | A | T | T | T | T | A | T | A | T | G | C | G | C | G | 0.000 | 0.036 | 0.4742 |
| 7 | G | C | G | G | T | T | G | G | G | C | C | C | G | G | C | G | T | T | C | T | T | A | 0.000 | 0.036 | 0.4742 |
| Haplotype | Frequency of Affected Haplotypes | Frequency of Control Haplotypes | Chi-Square p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | G | T | G | T | G | 0.857 | 0.179 | 2.41 × 10−5 |
| 2 | G | C | G | C | G | 0.071 | 0.321 | 0.0729 |
| 3 | A | C | A | C | A | 0.071 | 0.250 | 0.1647 |
| 4 | A | C | A | C | G | 0.000 | 0.143 | 0.1371 |
| 5 | G | C | G | T | G | 0.000 | 0.071 | 0.3055 |
| 6 | A | C | G | C | G | 0.000 | 0.036 | 0.4742 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stegemiller, M.R.; Highland, M.A.; Ewert, K.M.; Neaton, H.; Biller, D.S.; Murdoch, B.M. Identifying Genetic Predisposition to Dozer Lamb Syndrome: A Semi-Lethal Muscle Weakness Disease in Sheep. Genes 2025, 16, 83. https://doi.org/10.3390/genes16010083
Stegemiller MR, Highland MA, Ewert KM, Neaton H, Biller DS, Murdoch BM. Identifying Genetic Predisposition to Dozer Lamb Syndrome: A Semi-Lethal Muscle Weakness Disease in Sheep. Genes. 2025; 16(1):83. https://doi.org/10.3390/genes16010083
Chicago/Turabian StyleStegemiller, Morgan R., Margaret A. Highland, Kathleen M. Ewert, Holly Neaton, David S. Biller, and Brenda M. Murdoch. 2025. "Identifying Genetic Predisposition to Dozer Lamb Syndrome: A Semi-Lethal Muscle Weakness Disease in Sheep" Genes 16, no. 1: 83. https://doi.org/10.3390/genes16010083
APA StyleStegemiller, M. R., Highland, M. A., Ewert, K. M., Neaton, H., Biller, D. S., & Murdoch, B. M. (2025). Identifying Genetic Predisposition to Dozer Lamb Syndrome: A Semi-Lethal Muscle Weakness Disease in Sheep. Genes, 16(1), 83. https://doi.org/10.3390/genes16010083





