Attention-Guided Probabilistic Diffusion Model for Generating Cell-Type-Specific Gene Regulatory Networks from Gene Expression Profiles
Abstract
1. Introduction
- Hybrid multi-level attention for complex regulation modeling. Planet integrates graph attention for structure-aware local aggregation, cross-attention for preserving global regulatory architecture across diffusion steps, and multi-head self-attention for capturing long-range dependencies. Together, these preserve high-confidence edges, enable structure-guided feature aggregation, and dynamically modulate information across diffusion steps, allowing stable modeling of complex network under high noise and sparsity.
- Dynamic exploitation of time-step information. By means of time embeddings, it maps gene-expression and regulatory features into a shared temporal space: global structure is retained at high-noise steps, local details are emphasized at low-noise steps, and node- and edge-specific signals are fed back into the network structure, improving robustness and biological consistency.
- Efficient diffusion with accelerated sampling. With the training objective unchanged, accelerated sampling achieves generation quality comparable to full-step sampling while using fewer reverse steps, reducing computation and latency and offering a practical path for large-scale network and multi-condition scenarios.
2. Materials and Methods
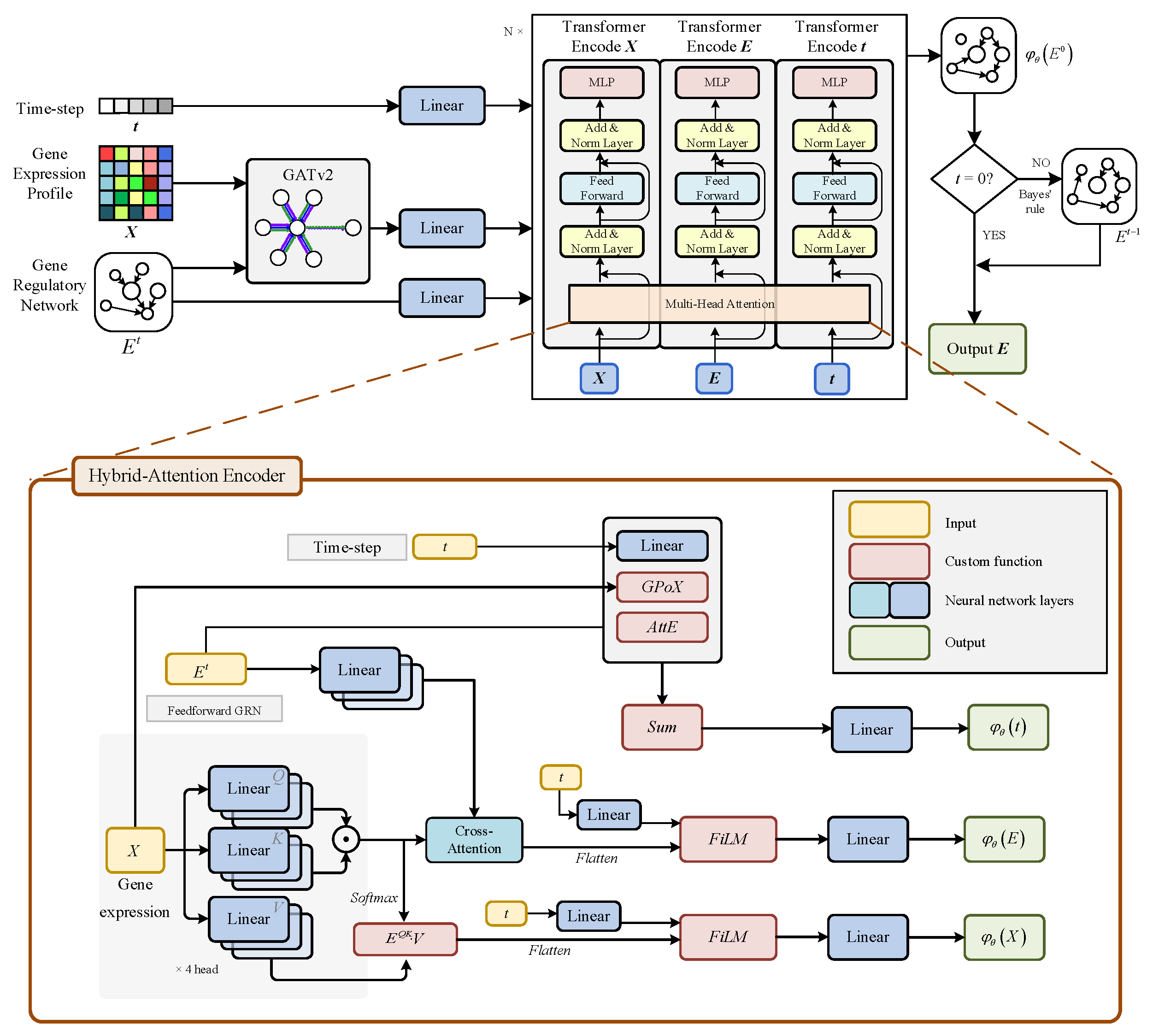
2.1. Planet Framework
2.2. Diffusion Model Framework
2.2.1. Framework Overview
2.2.2. Forward Process
2.2.3. Reverse Process
2.3. Triple Hybrid-Attention Transformer for Generative GRN
2.3.1. Graph Attention Mechanism
2.3.2. Graph Transformer Encoder
2.4. Benchmark Datasets and Preprocessing
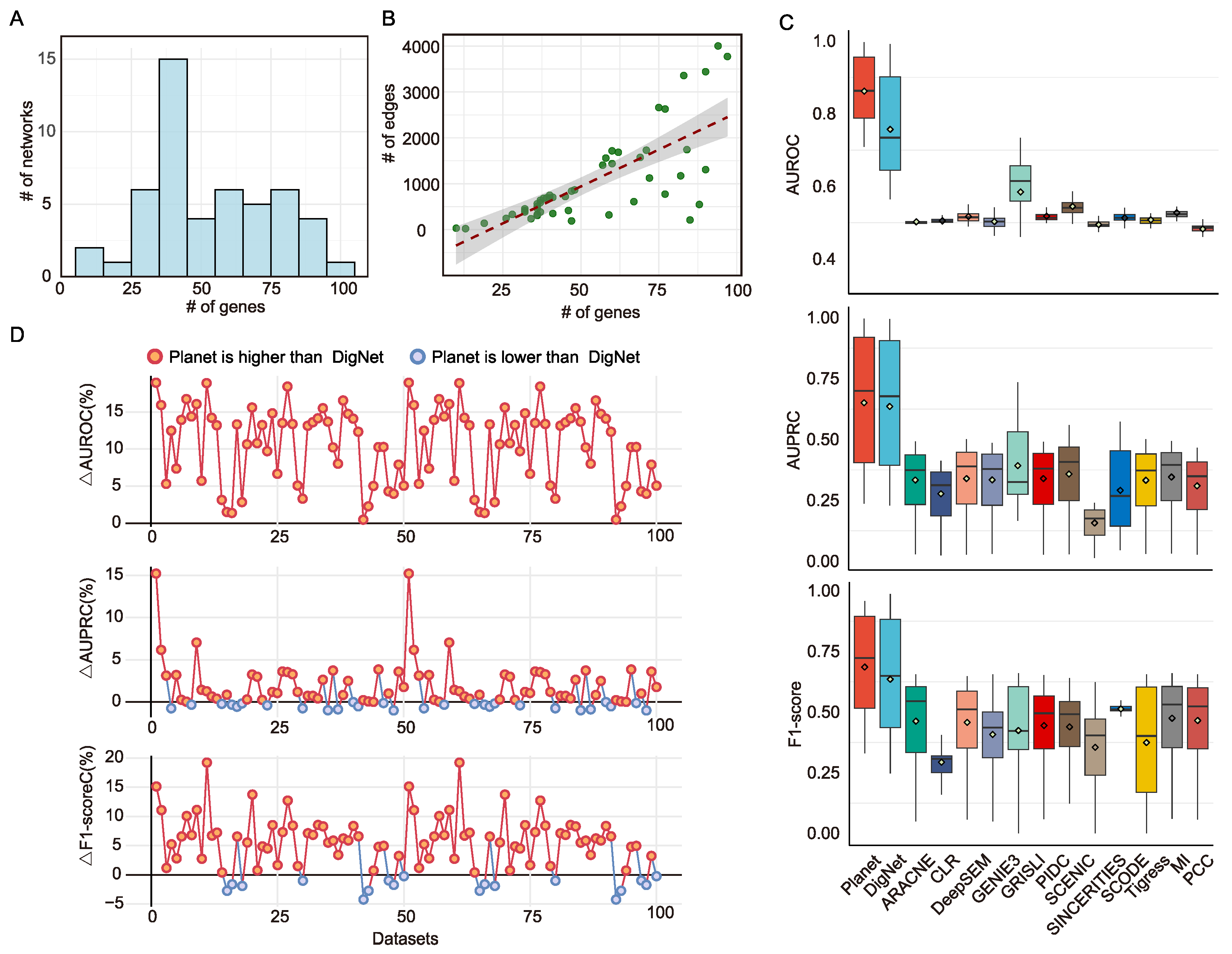
2.4.1. Simulated Datasets
2.4.2. Breast Cancer Dataset
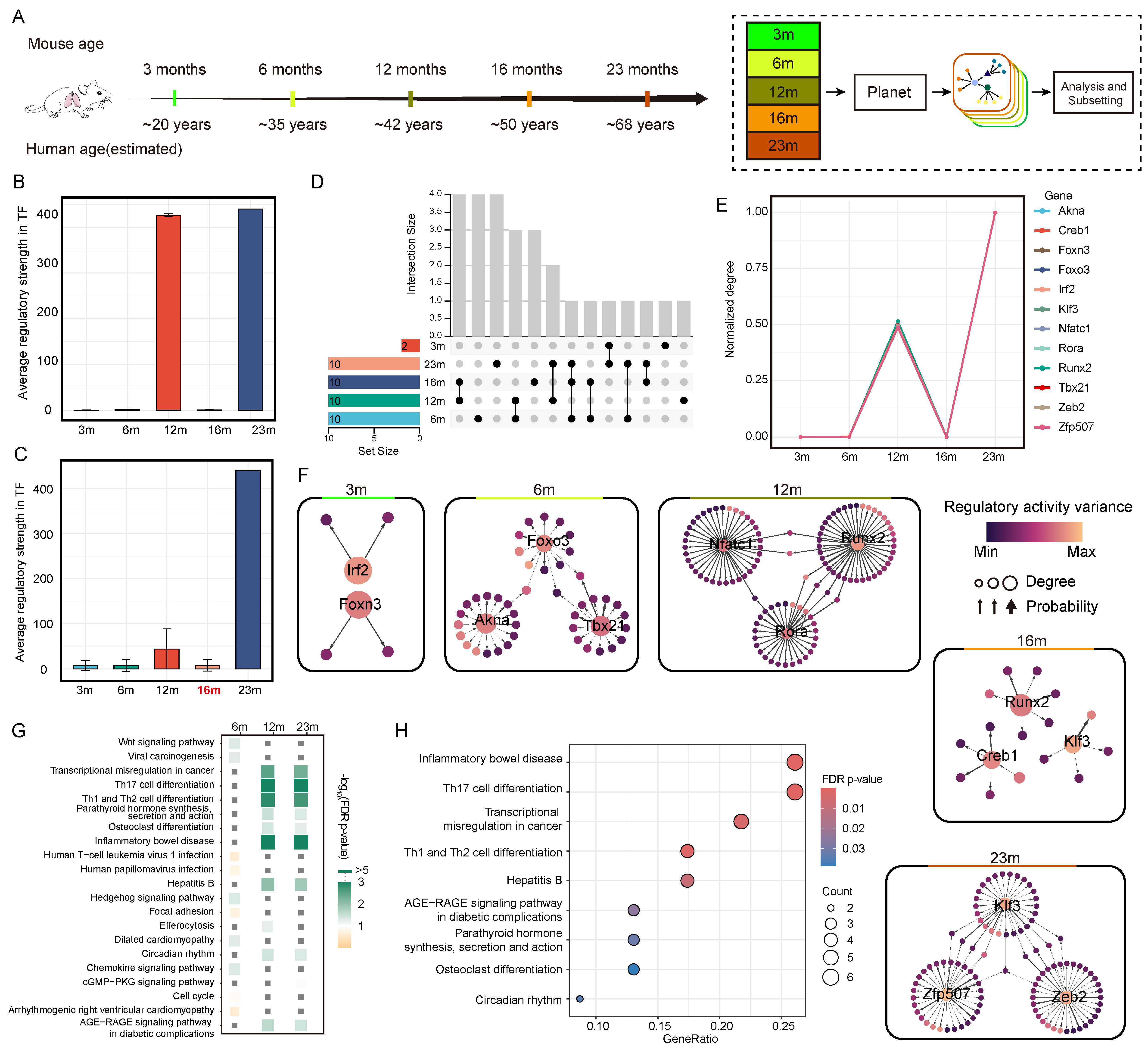
2.4.3. Mouse Aging Dataset
3. Results and Discussion
3.1. Synthetic Benchmarking Confirms Planet Efficacy
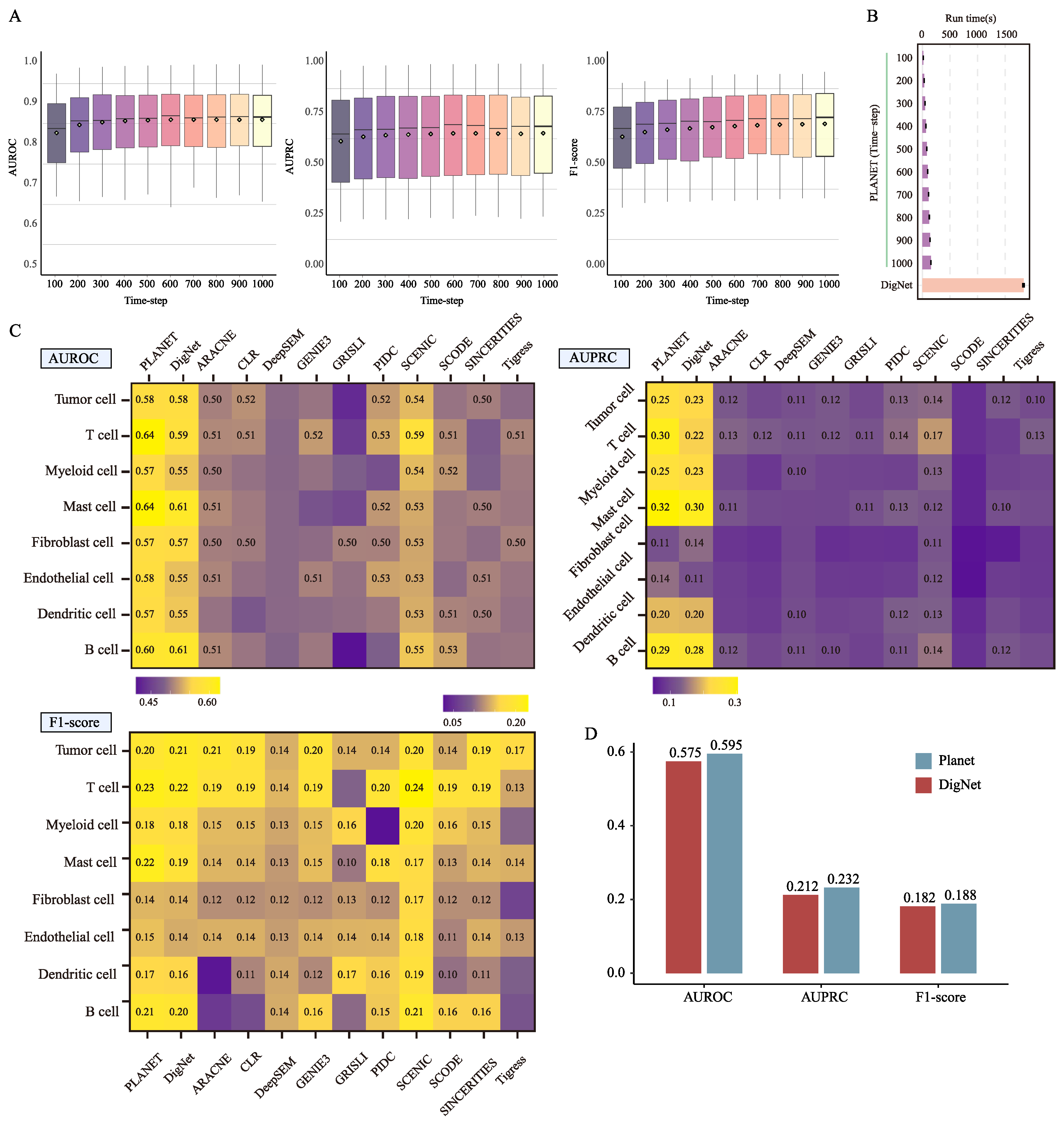
3.2. Accelerated Sampling Strategy and Module Contributions
3.3. Planet Generates Reliable Network on Breast Cancer Data
3.4. Planet Reveals Regulatory Mechanisms in Mouse Aging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GAT | Graph Attention Encoder |
| GRN | Gene Regulatory Network |
| scRNA-seq | single-cell RNA sequencing |
| TF | Transcription Factors |
| TriHAT | Triple Hybrid-Attention Transformer |
References
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Hedlund, E.; Deng, Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol. Asp. Med. 2018, 59, 36–46. [Google Scholar] [CrossRef]
- Pratapa, A.; Jalihal, A.P.; Law, J.N.; Bharadwaj, A.; Murali, T. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 2020, 17, 147–154. [Google Scholar] [CrossRef]
- Badia-i-Mompel, P.; Wessels, L.; Müller-Dott, S.; Trimbour, R.; Ramirez Flores, R.O.; Argelaguet, R.; Saez-Rodriguez, J. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 2023, 24, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Scharpf, R.B.; Bravo, H.C.; Simcha, D.; Langmead, B.; Johnson, W.E.; Geman, D.; Baggerly, K.; Irizarry, R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010, 11, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Marbach, D.; Costello, J.C.; Küffner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Kellis, M.; Collins, J.J. Wisdom of crowds for robust gene network inference. Nat. Methods 2012, 9, 796–804. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Matsumoto, H.; Kiryu, H.; Furusawa, C.; Ko, M.S.; Ko, S.B.; Gouda, N.; Hayashi, T.; Nikaido, I. SCODE: An efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics 2017, 33, 2314–2321. [Google Scholar] [CrossRef]
- Chen, G.; Liu, Z.-P. Graph attention network for link prediction of gene regulations from single-cell RNA-sequencing data. Bioinformatics 2022, 38, 4522–4529. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Bar-Joseph, Z. Deep learning for inferring gene relationships from single-cell expression data. Proc. Natl. Acad. Sci. USA 2019, 116, 27151–27158. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Zhou, J.; Lian, Q.; Li, H.; Zhao, D.; Zeng, J.; Ma, J. Modeling gene regulatory networks using neural network architectures. Nat. Comput. Sci. 2021, 1, 491–501. [Google Scholar] [CrossRef]
- Ma, A.; Wang, X.; Li, J.; Wang, C.; Xiao, T.; Liu, Y.; Cheng, H.; Wang, J.; Li, Y.; Chang, Y. Single-cell biological network inference using a heterogeneous graph transformer. Nat. Commun. 2023, 14, 964. [Google Scholar] [CrossRef]
- Kamimoto, K.; Stringa, B.; Hoffmann, C.M.; Jindal, K.; Solnica-Krezel, L.; Morris, S.A. Dissecting cell identity via network inference and in silico gene perturbation. Nature 2023, 614, 742–751. [Google Scholar] [CrossRef]
- Wang, L.; Trasanidis, N.; Wu, T.; Dong, G.; Hu, M.; Bauer, D.E.; Pinello, L. Dictys: Dynamic gene regulatory network dissects developmental continuum with single-cell multiomics. Nat. Methods 2023, 20, 1368–1378. [Google Scholar] [CrossRef]
- Yuan, Q.; Duren, Z. Inferring gene regulatory networks from single-cell multiome data using atlas-scale external data. Nat. Biotechnol. 2025, 43, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Guo, Z.; Liu, J.; Wang, Y.; Chen, M.; Wang, D.; Xu, D.; Cheng, J. Diffusion models in bioinformatics and computational biology. Nat. Rev. Bioeng. 2024, 2, 136–154. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Z.; Song, Y.; Hong, S.; Xu, R.; Zhao, Y.; Zhang, W.; Cui, B.; Yang, M.-H. Diffusion models: A comprehensive survey of methods and applications. ACM Comput. Surv. 2023, 56, 1–39. [Google Scholar] [CrossRef]
- Vignac, C.; Krawczuk, I.; Siraudin, A.; Wang, B.; Cevher, V.; Frossard, P. Digress: Discrete denoising diffusion for graph generation. arXiv 2022, arXiv:2209.14734. [Google Scholar]
- Wang, C.; Liu, Z.-P. Diffusion-based generation of gene regulatory networks from scRNA-seq data with DigNet. Genome Res. 2025, 35, 340–354. [Google Scholar] [CrossRef]
- Song, J.; Meng, C.; Ermon, S. Denoising diffusion implicit models. arXiv 2020, arXiv:2010.02502. [Google Scholar]
- Brody, S.; Alon, U.; Yahav, E. How attentive are graph attention networks? arXiv 2021, arXiv:2105.14491. [Google Scholar]
- Perez, E.; Strub, F.; De Vries, H.; Dumoulin, V.; Courville, A. Film: Visual reasoning with a general conditioning layer. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Dibaeinia, P.; Sinha, S. SERGIO: A single-cell expression simulator guided by gene regulatory networks. Cell Syst. 2020, 11, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef]
- Huang, M.; Wang, J.; Torre, E.; Dueck, H.; Shaffer, S.; Bonasio, R.; Murray, J.I.; Raj, A.; Li, M.; Zhang, N.R. SAVER: Gene expression recovery for single-cell RNA sequencing. Nat. Methods 2018, 15, 539–542. [Google Scholar] [CrossRef]
- Zhang, Z.; Schaefer, C.; Jiang, W.; Lu, Z.; Lee, J.; Sziraki, A.; Abdulraouf, A.; Wick, B.; Haeussler, M.; Li, Z. A panoramic view of cell population dynamics in mammalian aging. Science 2024, 387, eadn3949. [Google Scholar] [CrossRef]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Favera, R.D.; Califano, A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef]
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef]
- Aubin-Frankowski, P.-C.; Vert, J.-P. Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference. Bioinformatics 2020, 36, 4774–4780. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.E.; Stumpf, M.P.; Babtie, A.C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 2017, 5, 251–267.e253. [Google Scholar] [CrossRef] [PubMed]
- Papili Gao, N.; Ud-Dean, S.M.; Gandrillon, O.; Gunawan, R. SINCERITIES: Inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Bioinformatics 2018, 34, 258–266. [Google Scholar] [CrossRef]
- Haury, A.-C.; Mordelet, F.; Vera-Licona, P.; Vert, J.-P. TIGRESS: Trustful inference of gene regulation using stability selection. BMC Syst. Biol. 2012, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Mogilenko, D.A.; Shpynov, O.; Andhey, P.S.; Arthur, L.; Swain, A.; Esaulova, E.; Brioschi, S.; Shchukina, I.; Kerndl, M.; Bambouskova, M. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity 2021, 54, 99–115.e112. [Google Scholar] [CrossRef] [PubMed]
- Nah, J.; Seong, R.H. Krüppel-like factor 4 regulates the cytolytic effector function of exhausted CD8 T cells. Sci. Adv. 2022, 8, eadc9346. [Google Scholar] [CrossRef]
- Oellerich, M.F.; Potente, M. FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ. Res. 2012, 110, 1238–1251. [Google Scholar] [CrossRef]
- Lin, P.-H.; Lin, L.-T.; Li, C.-J.; Kao, P.-G.; Tsai, H.-W.; Chen, S.-N.; Wen, Z.-H.; Wang, P.-H.; Tsui, K.-H. Combining bioinformatics and experiments to identify CREB1 as a key regulator in senescent granulosa cells. Diagnostics 2020, 10, 295. [Google Scholar] [CrossRef]
- Goto, M.; Takahashi, H.; Yoshida, R.; Itamiya, T.; Nakano, M.; Nagafuchi, Y.; Harada, H.; Shimizu, T.; Maeda, M.; Kubota, A. Age-associated CD4+ T cells with B cell–promoting functions are regulated by ZEB2 in autoimmunity. Sci. Immunol. 2024, 9, eadk1643. [Google Scholar] [CrossRef]
- Martinez, G.J.; Pereira, R.M.; Äijö, T.; Kim, E.Y.; Marangoni, F.; Pipkin, M.E.; Togher, S.; Heissmeyer, V.; Zhang, Y.C.; Crotty, S. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 2015, 42, 265–278. [Google Scholar] [CrossRef]
- van der Kraan, P.M.; Blaney Davidson, E.N.; van den Berg, W.B. A role for age-related changes in TGFβ signaling in aberrant chondrocyte differentiation and osteoarthritis. Arthritis Res. Ther. 2010, 12, 201. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, C.; Wang, X.; Liao, H.; Liu, X.; Liu, H.; Zhang, M.; Zhang, L.; Wang, H. Knockdown of RUVBL2 improves hnRNPA2/B1-stress granules dynamics to inhibit perioperative neurocognitive disorders in aged mild cognitive impairment rats. Aging Cell 2025, 24, e14418. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016, 17, 250–258. [Google Scholar] [CrossRef]
- Lee, J.S.; Lee, W.-W.; Kim, S.H.; Kang, Y.; Lee, N.; Shin, M.S.; Kang, S.W.; Kang, I. Age-associated alteration in naive and memory Th17 cell response in humans. Clin. Immunol. 2011, 140, 84–91. [Google Scholar] [CrossRef]
- Ly, D.H.; Lockhart, D.J.; Lerner, R.A.; Schultz, P.G. Mitotic misregulation and human aging. Science 2000, 287, 2486–2492. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, L.; Zhong, Y.; Zhou, K.; Hou, Y.; Wang, Z.; Zhang, Z.; Xie, J.; Wang, C.; Chen, D. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021, 184, 404–421.e416. [Google Scholar] [CrossRef] [PubMed]

| Method | Modeling | Data Types | Time Series? |
|---|---|---|---|
| ARACNE | Correlation | Bulk RNA-seq | |
| CLR | Correlation | Bulk RNA-seq | |
| DeepSEM | Deep generative models | scRNA-seq | |
| DigNet | Deep generative models | scRNA-seq | |
| GENIE3 | Decision tree ensembles | Bulk RNA-seq | |
| GRISLI | Differential equations | scRNA-seq | Yes |
| PIDC | Information theory | scRNA-seq | |
| SCENIC | Decision tree ensembles | scRNA-seq | |
| SINCERITIES | Regression model | scRNA-seq | Yes |
| SCODE | Differential equations | scRNA-seq | Yes |
| Tigress | Regression model | Bulk RNA-seq | |
| MI | Information theory | Baseline | |
| PCC | Correlation | Baseline | |
| Base | w/o G | w/o C | w/o P | wo G/C | w/o G/P | w/o C/P | w/o GCP | |
|---|---|---|---|---|---|---|---|---|
| AUROC | 0.9136 | 0.9068 | 0.9050 | 0.9074 | 0.9071 | 0.8997 | 0.9084 | 0.9049 |
| AUPRC | 0.8063 | 0.7862 | 0.7847 | 0.7855 | 0.7955 | 0.7804 | 0.7889 | 0.7850 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Yu, N.; Zhang, D.; Wang, C. Attention-Guided Probabilistic Diffusion Model for Generating Cell-Type-Specific Gene Regulatory Networks from Gene Expression Profiles. Genes 2025, 16, 1255. https://doi.org/10.3390/genes16111255
Xu S, Yu N, Zhang D, Wang C. Attention-Guided Probabilistic Diffusion Model for Generating Cell-Type-Specific Gene Regulatory Networks from Gene Expression Profiles. Genes. 2025; 16(11):1255. https://doi.org/10.3390/genes16111255
Chicago/Turabian StyleXu, Shiyu, Na Yu, Daoliang Zhang, and Chuanyuan Wang. 2025. "Attention-Guided Probabilistic Diffusion Model for Generating Cell-Type-Specific Gene Regulatory Networks from Gene Expression Profiles" Genes 16, no. 11: 1255. https://doi.org/10.3390/genes16111255
APA StyleXu, S., Yu, N., Zhang, D., & Wang, C. (2025). Attention-Guided Probabilistic Diffusion Model for Generating Cell-Type-Specific Gene Regulatory Networks from Gene Expression Profiles. Genes, 16(11), 1255. https://doi.org/10.3390/genes16111255







