The Complete Mitochondrial Genome of Stromateus stellatus (Scombriformes: Stromateidae): Organization, Gene Arrangement, and Phylogenetic Position Within the Suborder Stromateoidei
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and mtDNA Extraction
2.2. Mitochondrial Genome Sequencing, Assembly, and Annotation
2.3. Phylogenetic Analysis
3. Results
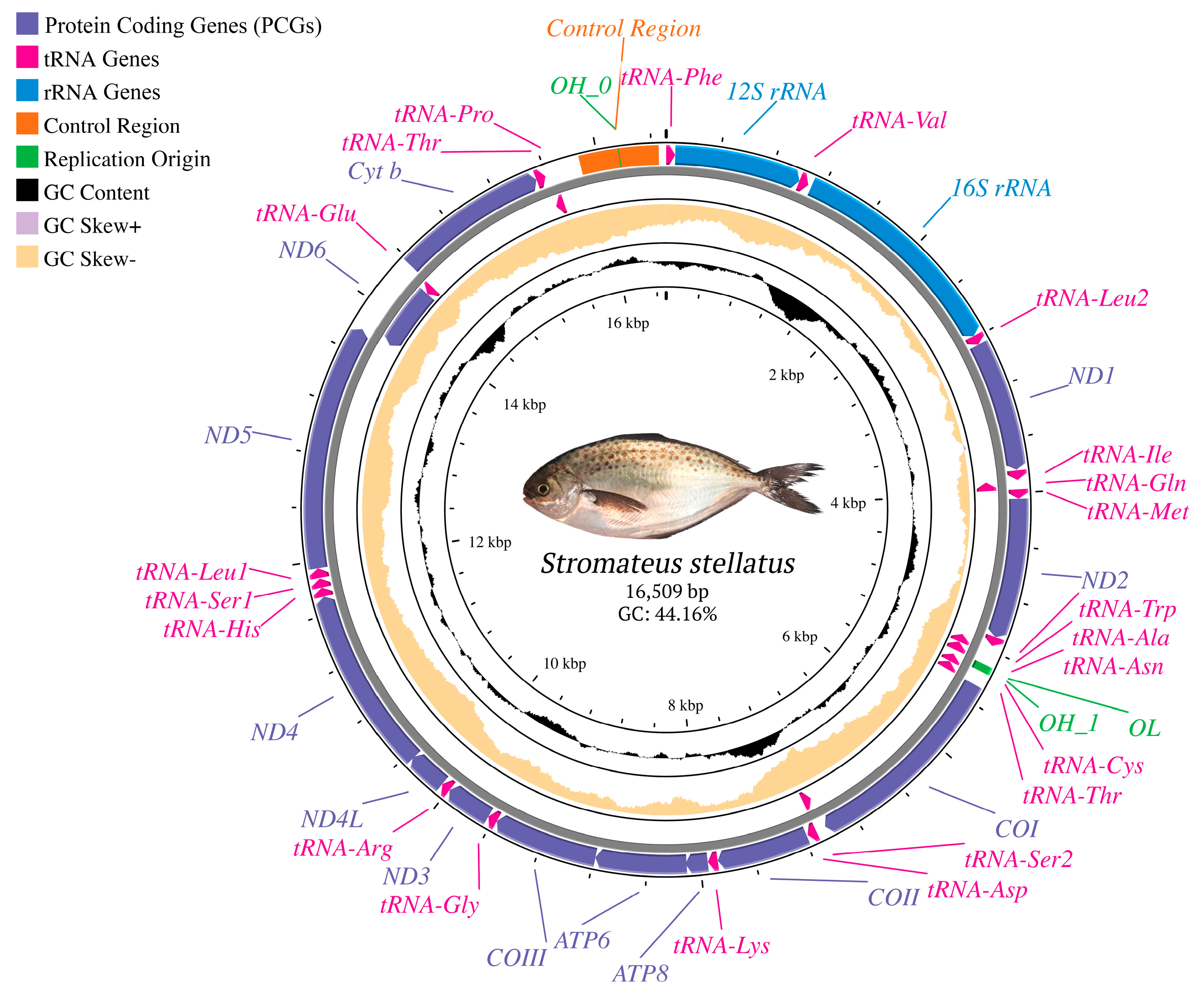
3.1. Features and Gene Content of the Mitochondrial Genome of S. stellatus
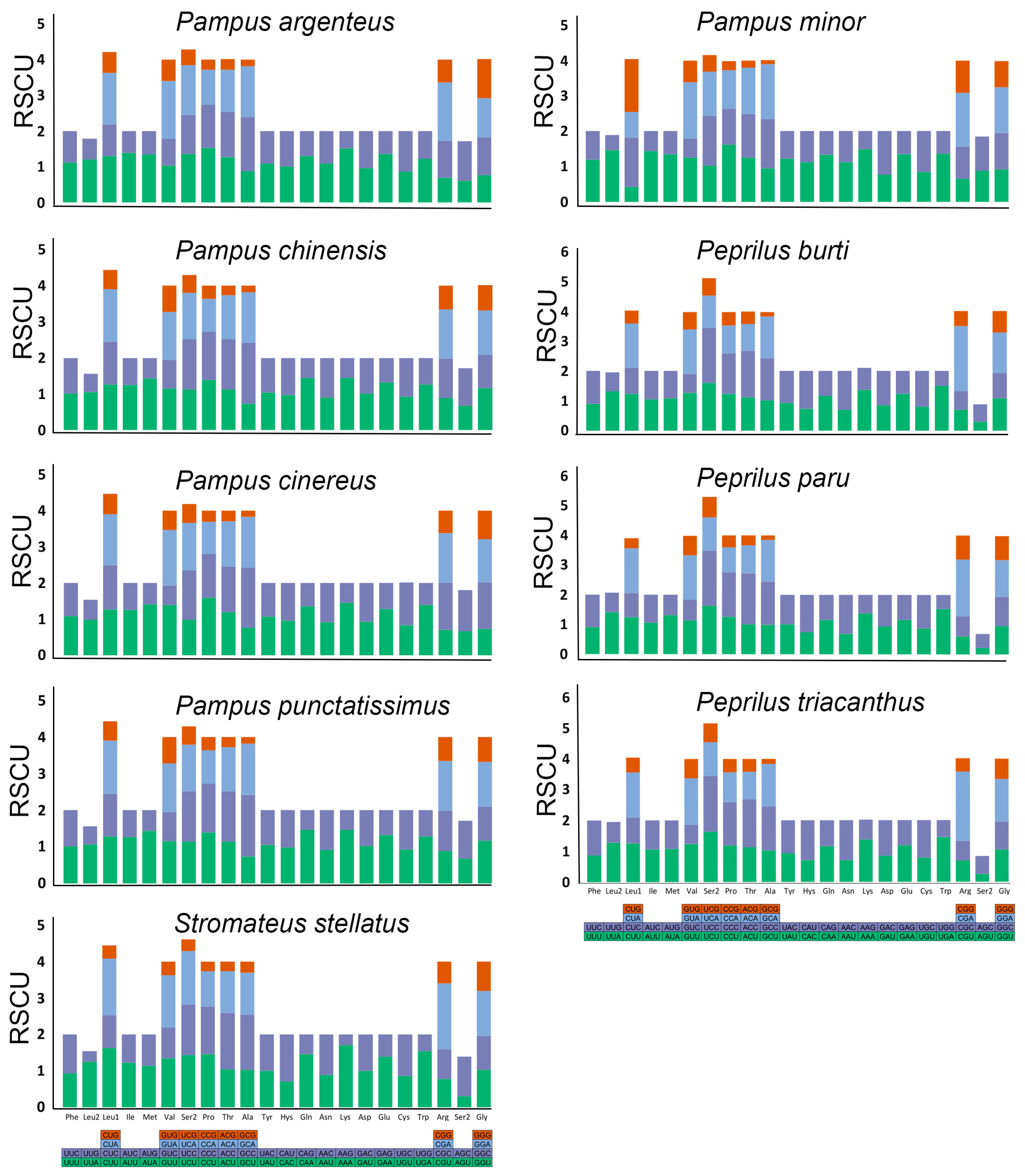
3.2. Codon Usage and Comparative RSCU Among Stromateid Species
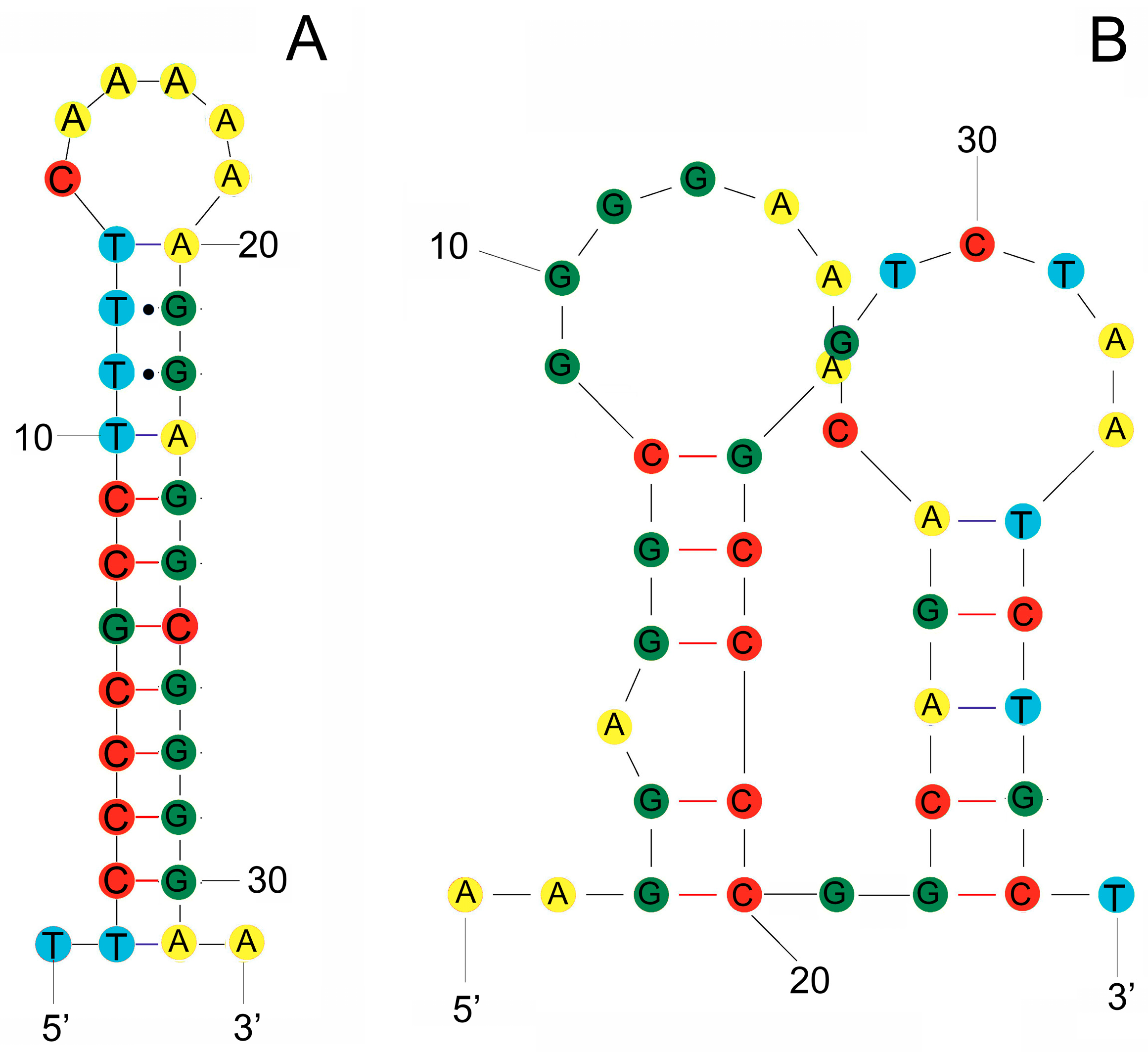
3.3. rRNA and tRNA Genes of S. stellatus
3.4. Non-Coding, Overlapping, and Intergenic Regions
3.5. Mitochondrial Gene Rearrangement in Stromateidae
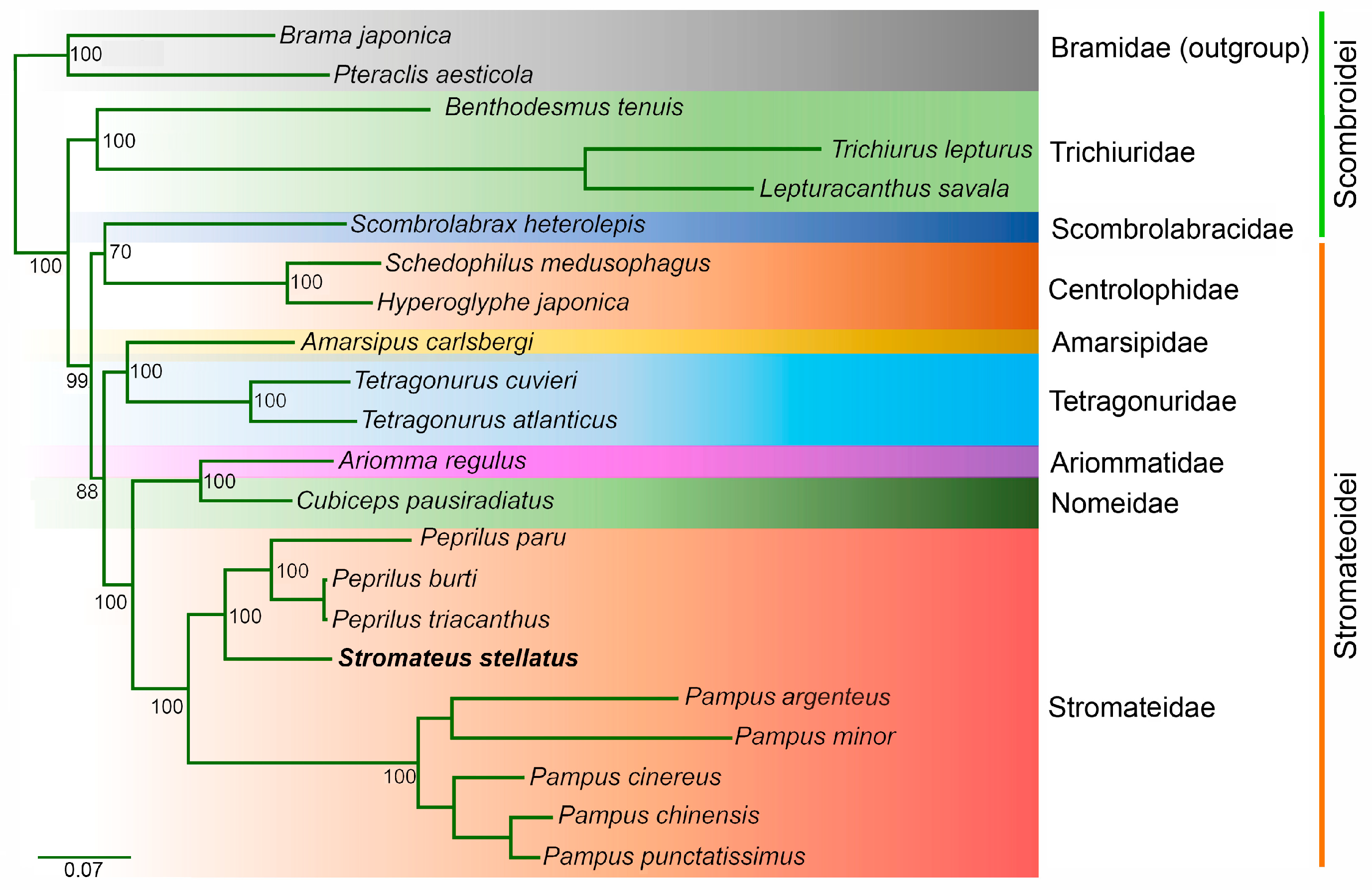
3.6. Phylogenetic Relationships
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Board, W.E. World Register of Marine Species. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=125567 (accessed on 30 August 2025).
- Pequeño, G. Lista sistemática revisada y comentada: Addendum. Rev. Biol. Mar. Oceanogr. 1997, 32, 77–94. Available online: https://rbmo.uv.cl/escaneados/322-77.pdf (accessed on 13 August 2025).
- Pastana, M.N.L.; Johnson, G.D.; Datovo, A. Comprehensive phenotypic phylogenetic analysis supports the monophyly of stromateiform fishes (Teleostei: Percomorphacea). Zool. J. Linn. Soc. 2021, 195, 841–963. [Google Scholar] [CrossRef]
- Arcila, D.; Hughes, L.C.; Meléndez-Vazquez, B.; Baldwin, C.C.; White, W.T.; Carpenter, K.E.; Williams, J.T.; Santos, M.D.; Pogonoski, J.J.; Miya, M. Testing the utility of alternative metrics of branch support to address the ancient evolutionary radiation of tunas, stromateoids, and allies (Teleostei: Pelagiaria). Syst. Biol. 2021, 70, 1123–1144. [Google Scholar] [CrossRef]
- Chirichigno, N.; Cornejo, R. Catálogo Comentado de Los Peces Marinos del Perú (Publicación Especial). Instituto del Mar del Perú: el Callao, Perú, 2001. [Google Scholar]
- Herrera, G.A.; Zavala-Muñoz, F.; Landaeta, M.F. Observations on the ontogeny of butterfish Stromateus stellatus larvae (Pisces: Stromateidae) off central Chile. Rev. Biol. Mar. y Oceanogr. 2018, 53 (Suppl. S1), 89–98. [Google Scholar] [CrossRef]
- Sato, T.; Seo HwanSook, S.H.; Endo, Y.; Fujimoto, K. Diacyl glyceryl ether as the major muscle lipid in Stromateus stellatus and its hydrolyzability by lipase and oral acute toxicity on mice. Nippon Suisan Gakkaishi 2002, 68, 569–575. [Google Scholar] [CrossRef]
- Ramilo-Fernández, G.; Sotelo, C.G. Characterization and potential strategies for the valorisation of the Southwest Atlantic butterfish (Stromateus brasiliensis). J. Food Sci. Technol. 2020, 57, 2994–3003. [Google Scholar] [CrossRef]
- Sun, P.; Yu, J.; Tang, B.; Liu, Z. Gene variation and population structure of Pampus chinensis in the China coast revealed by mitochondrial control region sequences. Mitochondrial DNA Part B 2021, 6, 2240–2245. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.; Huang, X.; Yuan, Z.; Zhang, S.; Xu, S.; Liu, J.; Wang, Y.; Wang, D.; Hu, J. Comparative Analysis of the Systematics and Evolution of the Pampus Genus of Fish (Perciformes: Stromateidae) Based on Osteology, Population Genetics and Complete Mitogenomes. Animals 2024, 14, 814. [Google Scholar] [CrossRef]
- Basurto, X.; Gutierrez, N.L.; Franz, N.; Mancha-Cisneros, M.d.M.; Gorelli, G.; Aguión, A.; Funge-Smith, S.; Harper, S.; Mills, D.J.; Nico, G.; et al. Illuminating the multidimensional contributions of small-scale fisheries. Nature 2025, 637, 875–884. [Google Scholar] [CrossRef]
- Desalle, R.; Schierwater, B.; Hadrys, H. MtDNA: The small workhorse of evolutionary studies. Front. Biosci. (Landmark Ed.) 2017, 22, 873–887. [Google Scholar] [CrossRef]
- Fourdrilis, S.; de Frias Martins, A.M.; Backeljau, T. Relation between mitochondrial DNA hyperdiversity, mutation rate and mitochondrial genome evolution in Melarhaphe neritoides (Gastropoda: Littorinidae) and other Caenogastropoda. Sci. Rep. 2018, 8, 17964. [Google Scholar] [CrossRef]
- Shtolz, N.; Mishmar, D. The metazoan landscape of mitochondrial DNA gene order and content is shaped by selection and affects mitochondrial transcription. Commun. Biol. 2023, 6, 93. [Google Scholar] [CrossRef]
- Ferreira, T.; Rodriguez, S. Mitochondrial DNA: Inherent Complexities Relevant to Genetic Analyses. Genes 2024, 15, 617. [Google Scholar] [CrossRef]
- Formenti, G.; Rhie, A.; Balacco, J.; Haase, B.; Mountcastle, J.; Fedrigo, O.; Brown, S.; Capodiferro, M.R.; Al-Ajli, F.O.; Ambrosini, R.; et al. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 2021, 22, 120. [Google Scholar] [CrossRef]
- Bar-Yaacov, D.; Hadjivasiliou, Z.; Levin, L.; Barshad, G.; Zarivach, R.; Bouskila, A.; Mishmar, D. Mitochondrial Involvement in Vertebrate Speciation? The Case of Mito-nuclear Genetic Divergence in Chameleons. Genome Biol. Evol. 2015, 7, 3322–3336. [Google Scholar] [CrossRef]
- Mthethwa, S.; Bester-van der Merwe, A.E.; Roodt-Wilding, R. Addressing the complex phylogenetic relationship of the Gempylidae fishes using mitogenome data. Ecol. Evol. 2023, 13, e10217. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, Y.; Mao, S.; Guo, J.; Bao, S.; Xu, X.; Li, J.; Shen, Y. The complete mitogenome reveals genetic diversity and differentiation among wild and farmed black carp (Mylopharyngodon piceus) populations. Aquac. Fish. 2025, 10, 953–960. [Google Scholar] [CrossRef]
- Vieira, A.R.; de Sousa, F.; Bilro, J.; Viegas, M.B.; Svanbäck, R.; Gordo, L.S.; Paulo, O.S. Mitochondrial genomes of the European sardine (Sardina pilchardus) reveal Pliocene diversification, extensive gene flow and pervasive purifying selection. Sci. Rep. 2024, 14, 30977. [Google Scholar] [CrossRef]
- Roy, S.; Parida, P.K.; Ramya, V.L.; Kumar, V.; Bhakta, D.; Behera, B.K.; Das, B.K. Whole mitochondrial genome sequencing and phylogenetic analysis of Gangetic mystus (Mystus cavasius). Mitochondrial DNA Part B 2024, 9, 1539–1543. [Google Scholar] [CrossRef]
- Clayton, D.A.; Shadel, G.S. Isolation of Mitochondria from Animal Tissue. Cold Spring Harb. Protoc. 2014, 2014, pdb.prot080010. [Google Scholar] [CrossRef]
- Sambrook, J.F.R.; David, W. Molecular Cloning: A Laboratory Manual (3-Volume Set); Cold Springs Harbour Press: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Gould, M.P.; Bosworth, C.M.; McMahon, S.; Grandhi, S.; Grimerg, B.T.; LaFramboise, T. PCR-Free Enrichment of Mitochondrial DNA from Human Blood and Cell Lines for High Quality Next-Generation DNA Sequencing. PLoS ONE 2015, 10, e0139253. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control [version 2; peer review: 4 approved]. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Near, T.J.; Thacker, C.E. Phylogenetic Classification of Living and Fossil Ray-Finned Fishes (Actinopterygii). Bull. Peabody Mus. Nat. Hist. 2024, 65, 3–302. [Google Scholar] [CrossRef]
- Campbell, M.A.; Sado, T.; Shinzato, C.; Koyanagi, R.; Okamoto, M.; Miya, M. Multilocus phylogenetic analysis of the first molecular data from the rare and monotypic Amarsipidae places the family within the Pelagia and highlights limitations of existing data sets in resolving pelagian interrelationships. Mol. Phylogenetics Evol. 2018, 124, 172–180. [Google Scholar] [CrossRef]
- Miya, M.; Friedman, M.; Satoh, T.P.; Takeshima, H.; Sado, T.; Iwasaki, W.; Yamanoue, Y.; Nakatani, M.; Mabuchi, K.; Inoue, J.G.; et al. Evolutionary Origin of the Scombridae (Tunas and Mackerels): Members of a Paleogene Adaptive Radiation with 14 Other Pelagic Fish Families. PLoS ONE 2013, 8, e73535. [Google Scholar] [CrossRef]
- Yagishita, N.; Miya, M.; Yamanoue, Y.; Shirai, S.M.; Nakayama, K.; Suzuki, N.; Satoh, T.P.; Mabuchi, K.; Nishida, M.; Nakabo, T. Mitogenomic evaluation of the unique facial nerve pattern as a phylogenetic marker within the percifom fishes (Teleostei: Percomorpha). Mol. Phylogenetics Evol. 2009, 53, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, N.; Lin, L.; Gao, T. The complete mitochondrial genome of Pampus echinogaster (Perciformes: Stromateidae). Mitochondrial DNA Part A 2016, 27, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, L.; Li, Y.; Zhang, Z.; Wang, Z.; Gao, T. The complete mitochondrial genome of Pampus minor (Liu & Li, 1998) (Perciformes: Stromateidae). Mitochondrial DNA Part B 2019, 4, 205–206. [Google Scholar] [CrossRef]
- Sun, D.; Cheng, Q.; Qiao, H.; Zhang, H.; Chen, Y. The complete mitochondrial genome sequence of Pampus chinensis (Perciformes: Stromateidae). Mitochondrial DNA Part A 2016, 27, 510–511. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, L.; Li, Y.; Zhang, Z.; Wang, Z.; Gao, T. Complete mitochondrial DNA genome of grey pomfret Pampus cinereus (Bloch, 1795) (Perciformes: Stromateidae). Mitochondrial DNA Part B 2018, 3, 683–684. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Wei, D.-D.; Shao, R.; Dou, W.; Wang, J.-J. The Complete Mitochondrial Genome of the Booklouse, Liposcelis decolor: Insights into Gene Arrangement and Genome Organization within the Genus Liposcelis. PLoS ONE 2014, 9, e91902. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Wang, M.; Li, D.; Tang, S.; Chen, X. Mitochondrial Genome of Eleutheronema rhadinum with an Additional Non-coding Region and Novel Insights Into the Phylogenetics. Front. Mar. Sci. 2021, 8, 746598. [Google Scholar] [CrossRef]
- Yang, J.; Li, A.; Liu, S. Structural Characteristics of Mitochondrial Genomes of Two Species of Mackerel and Phylogenetic Analysis of Scombridae Family. Biomolecules 2025, 15, 555. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.L. Mitochondrial genome organization and vertebrate phylogenetics. Genet. Mol. Biol. 2000, 23, 745–752. [Google Scholar] [CrossRef]
- Qian, L.; Wang, H.; Yan, J.; Pan, T.; Jiang, S.; Rao, D.; Zhang, B. Multiple independent structural dynamic events in the evolution of snake mitochondrial genomes. BMC Genom. 2018, 19, 354. [Google Scholar] [CrossRef]
- Broughton, R.E.; Milam, J.E.; Roe, B.A. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001, 11, 1958–1967. [Google Scholar] [CrossRef]
- Siddika, M.A.; Ahmed, K.A.; Alam, M.S.; Bushra, J.; Begum, R.A. Complete mitogenome and intra-family comparative mitogenomics showed distinct position of Pama Croaker Otolithoides pama. Sci. Rep. 2024, 14, 13820. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-S.; Kang, D.-W.; Kim, K.-Y.; Heo, J.S.; Song, H.-Y.; Yoon, J.-D. Characterization of the complete mitogenome of the endangered freshwater fish Gobiobotia naktongensis from the Geum River in South Korea: Evidence of stream connection with the Paleo-Huanghe. Genes Genom. 2022, 44, 945–956. [Google Scholar] [CrossRef]
- Jia, W.; Higgs, P.G. Codon Usage in Mitochondrial Genomes: Distinguishing Context-Dependent Mutation from Translational Selection. Mol. Biol. Evol. 2008, 25, 339–351. [Google Scholar] [CrossRef]
- Zhang, R.-Q.; Bai, Y.-W.; Hu, J.-T.; Wang, P.-Y.; Qi, Y.; Zhang, T.-X.; Jiao, H.-Y.; Lin, X.-L.; Yan, Z.-G. Complete mitochondrial genomes and phylogenetic analysis of native and non-native fishes in a national key wetland of China. Front. Environ. Sci. 2024, 12, 1415150. [Google Scholar] [CrossRef]
- DeVore, M.L.; Bazzini, A.A. Codon optimality influences homeostatic gene expression in zebrafish. G3 Genes Genomes Genet. 2024, 14, jkae247. [Google Scholar] [CrossRef]
- Doiuchi, R.; Nakabo, T. Molecular phylogeny of the stromateoid fishes (Teleostei: Perciformes) inferred from mitochondrial DNA sequences and compared with morphology-based hypotheses. Mol. Phylogenetics Evol. 2006, 39, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, M.; D’Elia, A.K.P.; Rocha, G.; Arantes, C.A.; Henning, F.; de Vasconcelos, A.T.R.; Solé-Cava, A.M. Mitochondrial genome structure and composition in 70 fishes: A key resource for fisheries management in the South Atlantic. BMC Genom. 2024, 25, 215. [Google Scholar] [CrossRef] [PubMed]

| Family | Species Name | GenBank Accession Number | References |
|---|---|---|---|
| Amarsipidae | Amarsipus carlsbergi | NC_037474 | [36] |
| Ariommatidae | Ariomma regulus | PV357913 | Unpublished |
| Bramidae | Brama japonica | KT908039 | Unpublished |
| Pteraclis aesticola | AP012499 | [37] | |
| Centrolophidae | Schedophilus medusophagus | MT410878 | Unpublished |
| Hyperoglyphe japonica | AP006037 | [38] | |
| Nomeidae | Cubiceps pausiradiatus | AP006038 | [38] |
| Tetragonuridae | Tetragonurus cuvieri | AP012514 | [37] |
| Tetragonurus atlanticus | AP012515 | [37] | |
| Trichiuridae | Benthodesmus tenuis | AP012522 | [37] |
| Lepturacanthus savala | OP724236 | Unpublished | |
| Trichiurus lepturus | NC_018791 | Unpublished | |
| Scombrolabracidae | Scombrolabrax heterolepis | OP035059 | Unpublished |
| Stromateidae | Pampus argenteus | KF373560 | [39] |
| Pampus minor | MH037007 | [40] | |
| Pampus cinereus | OR538383 | [10] | |
| Pampus chinensis | KJ418377 | [41] | |
| Pampus punctatissimus | OR538387 | [10] | |
| Peprilus paru | OP056882 | Unpublished | |
| Peprilus burti | AP012947 | [37] | |
| Peprilus triacanthus | AP012518 | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angulo, F.E.; Pedrero-Pacheco, R.; Nuñez, J.J. The Complete Mitochondrial Genome of Stromateus stellatus (Scombriformes: Stromateidae): Organization, Gene Arrangement, and Phylogenetic Position Within the Suborder Stromateoidei. Genes 2025, 16, 1256. https://doi.org/10.3390/genes16111256
Angulo FE, Pedrero-Pacheco R, Nuñez JJ. The Complete Mitochondrial Genome of Stromateus stellatus (Scombriformes: Stromateidae): Organization, Gene Arrangement, and Phylogenetic Position Within the Suborder Stromateoidei. Genes. 2025; 16(11):1256. https://doi.org/10.3390/genes16111256
Chicago/Turabian StyleAngulo, Fernanda E., Rodrigo Pedrero-Pacheco, and José J. Nuñez. 2025. "The Complete Mitochondrial Genome of Stromateus stellatus (Scombriformes: Stromateidae): Organization, Gene Arrangement, and Phylogenetic Position Within the Suborder Stromateoidei" Genes 16, no. 11: 1256. https://doi.org/10.3390/genes16111256
APA StyleAngulo, F. E., Pedrero-Pacheco, R., & Nuñez, J. J. (2025). The Complete Mitochondrial Genome of Stromateus stellatus (Scombriformes: Stromateidae): Organization, Gene Arrangement, and Phylogenetic Position Within the Suborder Stromateoidei. Genes, 16(11), 1256. https://doi.org/10.3390/genes16111256






