Genome-Wide Identification and Tissue-Specific Expression Profiling of Goji CER Gene Family
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. The Cuticular Wax Crystallization Patterns
2.3. The Cuticular Wax Load Analysis
2.4. Identification and Physicochemical Characterization of CER Gene Family Members
2.5. Chromosome Localization and Synteny Relationship Analysis
2.6. Conserved Domain and Motif Analysis
2.7. Cis-Acting Element Analysis
2.8. Phylogenetic Analysis
2.9. qRT–PCR Assay
2.10. Data Processing
3. Results
3.1. Differences in Wax Ultrastructure and Load Across Organs of Different Goji Berry Cultivars
3.2. Identification and Physicochemical Properties of the Goji CER Gene Family Members
3.3. Chromosomal Localization and Intra-Genomic Synteny Analysis of the Goji CER Gene Family Members
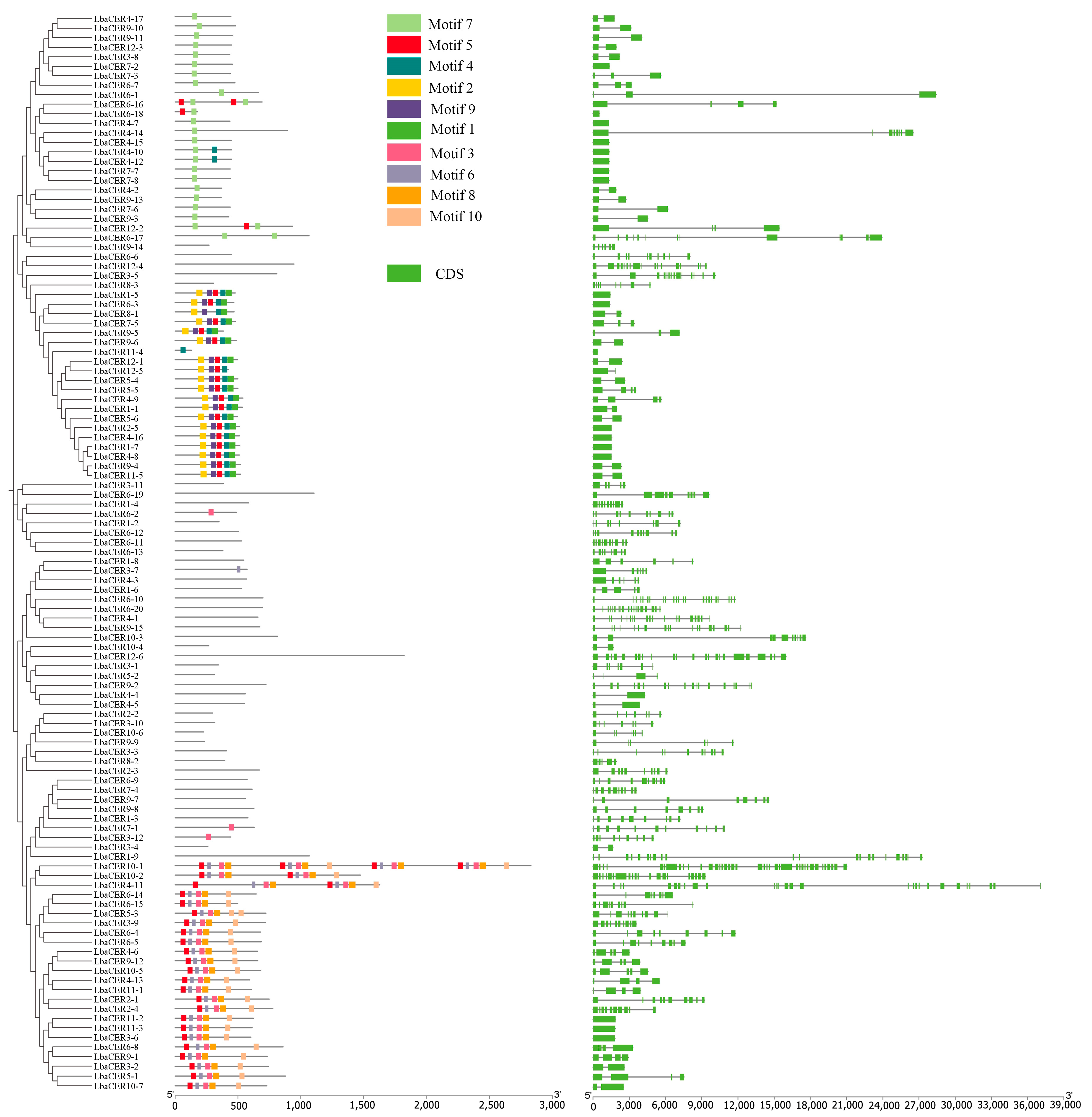
3.4. Protein Conserved Motifs and Domain Analysis of Goji CER Gene Family Members
3.5. Cis-Acting Elements Analysis of Goji CER Gene Family Members
3.6. Synteny Analysis of Goji CER Gene Family Members
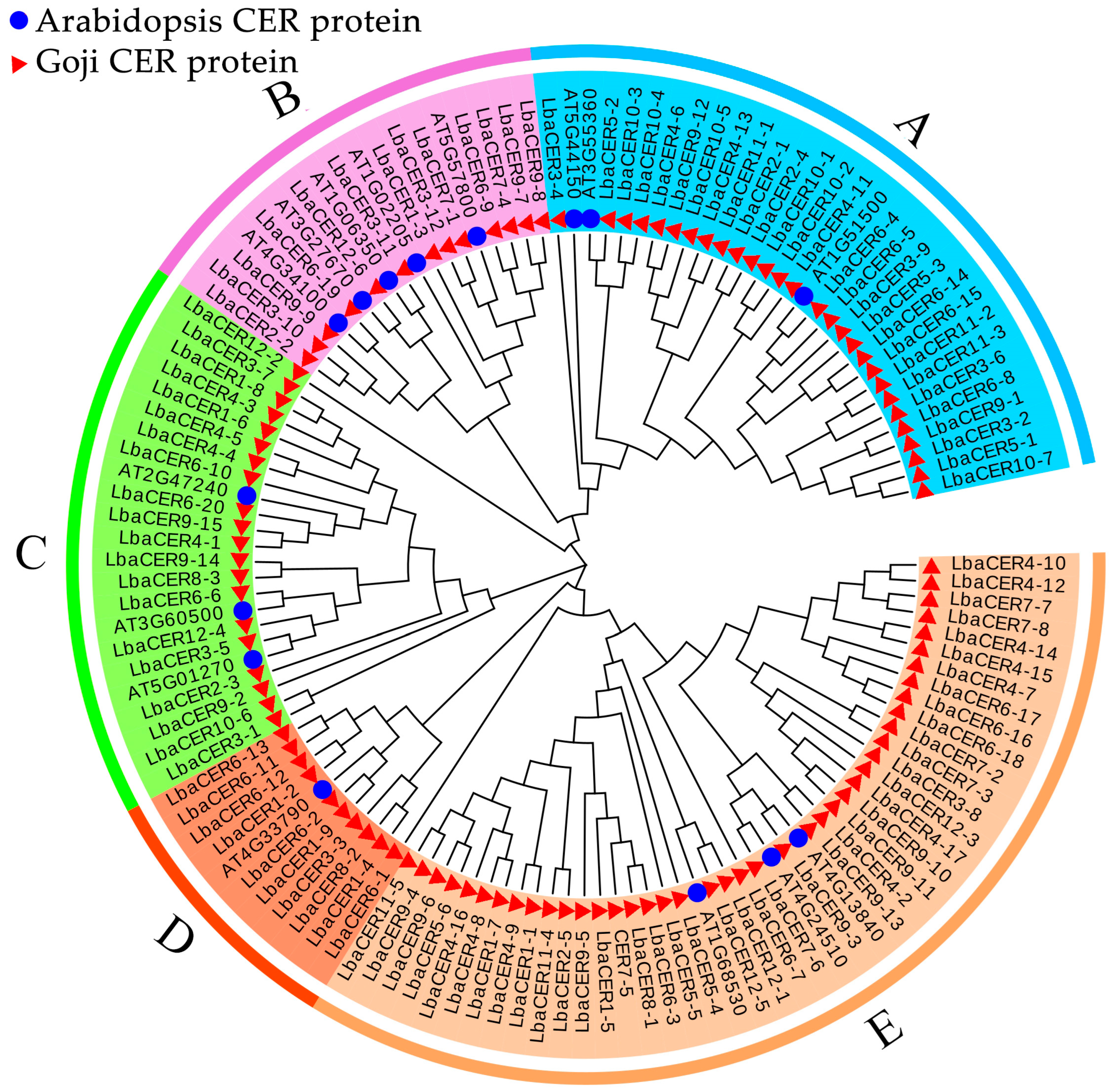
3.7. Phylogenetic Analysis of Goji CER Proteins with Arabidopsis
3.8. Expression Analysis of Five CER Genes in Different Goji Cultivars and Organs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CER | Eceriferum |
| UV | Ultraviolet Radiation |
| VLCFAs | Very-long-chain fatty acids |
| FAR | Fatty acyl reductase |
| MAH | Medium chain alkane hydroxylase |
| SEM | Scanning electron microscopy |
| qRT–PCR | Quantitative real-time polymerase chain reaction |
| MeJA | Methyl jasmonate |
| ABA | Abscisic acid |
| PEG | Polyethylene glycol |
| VLC | Very long chain |
References
- Yang, Z.; Dai, G.; Qin, K.; Wu, J.; Wang, Z.; Wang, C. Comprehensive Evaluation of Germplasm Resources in Various Goji Cultivars Based on Leaf Anatomical Traits. Forests 2025, 16, 187. [Google Scholar] [CrossRef]
- Hõrak, H. Learning from the Experts: Drought Resistance in Desert Plants. New Phytol. 2017, 216, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Rao, S.; Du, C.; Liu, L.; Dai, G.; Chen, J. Strategies Used by Two Goji Species, Lycium ruthenicum and Lycium barbarum, to Defend against Salt Stress. Sci. Hortic. 2022, 306, 111430. [Google Scholar] [CrossRef]
- Zhang, Z.; He, K.; Zhang, T.; Tang, D.; Li, R.; Jia, S. Physiological Responses of Goji Berry (Lycium barbarum L.) to Saline-Alkaline Soil from Qinghai Region, China. Sci. Rep. 2019, 9, 12057. [Google Scholar] [CrossRef]
- Teixeira, S.; Luís, I.; Oliveira, M.; Abreu, I.; Batista, R. Goji Berries Superfood—Contributions for the Characterisation of Proteome and Ige-Binding Proteins. Food Agric. Immunol. 2019, 30, 262–280. [Google Scholar] [CrossRef]
- Amagase, H.; Farnsworth, N.R. A Review of Botanical Characteristics, Phytochemistry, Clinical Relevance in Efficacy and Safety of Lycium barbarum Fruit (Goji). Food Res. Int. 2011, 44, 1702–1717. [Google Scholar] [CrossRef]
- Jeffree, C.E. The Fine Structure of the Plant Cuticle. In Annual Plant Reviews Volume 23: Biology of the Plant Cuticle; Wiley-Blackwell: Hoboken, NJ, USA, 2006; pp. 11–125. [Google Scholar]
- Steinmüller, D.; Tevini, M. Action of Ultraviolet Radiation (UV-B) Upon Cuticular Waxes in Some Crop Plants. Planta 1985, 164, 557–564. [Google Scholar] [CrossRef]
- Moreno, A.G.; Cózar, A.; Prieto, P.; Domínguez, E.; Heredia, A. Radiationless Mechanism of Uv Deactivation by Cuticle Phenolics in Plants. Nat. Commun. 2022, 13, 1786. [Google Scholar] [CrossRef]
- Barthlott, W.; Neinhuis, C. Purity of the Sacred Lotus, or Escape from Contamination in Biological Surfaces. Planta 1997, 202, 1–8. [Google Scholar] [CrossRef]
- Khanal, B.P.; Grimm, E.; Finger, S.; Blume, A.; Knoche, M. Intracuticular Wax Fixes and Restricts Strain in Leaf and Fruit Cuticles. N. Phytol. 2013, 200, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Peng, Y.; Hou, G.; Yang, M.; He, C.; She, M.; Li, X.; Li, M.; Chen, Q.; Lin, Y. A High Epicuticular Wax Strawberry Mutant Reveals Enhanced Resistance to Tetranychus urticae Koch and Botrytis cinerea. Sci. Hortic. 2024, 324, 112636. [Google Scholar] [CrossRef]
- Buschhaus, C.; Jetter, R. Composition and Physiological Function of the Wax Layers Coating Arabidopsis Leaves: Β-amyrin Negatively Affects the Intracuticular Water Barrier. Plant Physiol. 2012, 160, 1120–1129. [Google Scholar] [CrossRef]
- Jiang, B.; Liu, R.; Fang, X.; Tong, C.; Gao, H. Effects of Salicylic Acid Treatment on Fruit Quality and Wax Composition of Blueberry (Vaccinium virgatum Ait). Food Chem. 2021, 5, 130757. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Advances in the Understanding of Cuticular Waxes in Arabidopsis thaliana and Crop Species. Plant Cell Rep. 2015, 34, 4. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The Formation and Function of Plant Cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Aarts, M.G.; Keijzer, C.J.; Stiekema, W.J.; Pereira, A. Molecular Characterization of the Cer1 Gene of Arabidopsis Involved in Epicuticular Wax Biosynthesis and Pollen Fertility. Plant Cell 1995, 7, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Sturaro, M.; Hartings, H.; Schmelzer, E.; Velasco, R.; Salamini, F.; Motto, M. Cloning and Characterization of Glossy1, a Maize Gene Involved in Cuticle Membrane and Wax Production. Plant Physiol. 2005, 138, 478–489. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber Eceriferum1 (Cscer1), Which Influences the Cuticle Properties and Drought Tolerance of Cucumber, Plays a Key Role in Vlc Alkanes Biosynthesis. Plant Mol. Biol. 2015, 87, 219–233. [Google Scholar] [CrossRef]
- Li, T.; Sun, Y.; Liu, T.; Wu, H.; An, P.; Shui, Z.; Wang, J.; Zhu, Y.; Li, C.; Wang, Y.; et al. Tacer1-1a Is Involved in Cuticular Wax Alkane Biosynthesis in Hexaploid Wheat and Responds to Plant Abiotic Stresses. Plant Cell Environ. 2019, 42, 3077–3091. [Google Scholar] [CrossRef]
- Xiong, C.; Xie, Q.; Yang, Q.; Sun, P.; Gao, S.; Li, H.; Zhang, J.; Wang, T.; Ye, Z.; Yang, C. Woolly, Interacting with MYB Transcription Factor MYB31, Regulates Cuticular Wax Biosynthesis by Modulating Cer6 Expression in Tomato. Plant J. 2020, 103, 323–337. [Google Scholar] [CrossRef]
- Muhammad Ahmad, H.; Wang, X.; Fiaz, S.; Mahmood Ur, R.; Azhar Nadeem, M.; Aslam Khan, S.; Ahmar, S.; Azeem, F.; Shaheen, T.; Mora-Poblete, F. Comprehensive Genomics and Expression Analysis of Eceriferum (Cer) Genes in Sunflower (Helianthus annuus). Saudi J. Biol. Sci. 2021, 28, 6884–6896. [Google Scholar] [CrossRef]
- Li, N.; Li, X.Z.; Song, Y.Q.; Yang, S.T.; Li, L.L. Genome-Wide Identification, Characterization, and Expression Profiling of the Eceriferum (Cer) Gene Family in Ziziphus jujube. Russ. J. Plant Physiol. 2021, 68, 828–837. [Google Scholar] [CrossRef]
- Zhao, S.; Nie, X.; Liu, X.; Wang, B.; Liu, S.; Qin, L.; Xing, Y. Genome-Wide Identification of the Cer Gene Family and Significant Features in Climate Adaptation of Castanea mollissima. Int. J. Mol. Sci. 2022, 23, 16202. [Google Scholar] [CrossRef] [PubMed]
- Panahi, B.; Hamid, R.; Jacob, F. Genome-Wide Characterization of the Eceriferum (Cer) Gene Family in Barley (Hordeum vulgare L.). Sci. Rep. 2025, 15, 20674. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wei, K.; Zhang, Y.; Chang, X.; Yang, W.; Yao, Q.; Xiao, H. Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns under Different Treatments in Pepper (Capsicum annuum L.). Horticulturae 2025, 11, 571. [Google Scholar] [CrossRef]
- Bernard, A.; Joubès, J. Arabidopsis Cuticular Waxes: Advances in Synthesis, Export and Regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef]
- Zhong, M.-S.; Jiang, H.; Cao, Y.; Wang, Y.-X.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. Mdcer2 Conferred to Wax Accumulation and Increased Drought Tolerance in Plants. Plant Physi Biochem. 2020, 149, 277–285. [Google Scholar] [CrossRef]
- Haslam, T.M.; Haslam, R.; Thoraval, D.; Pascal, S.; Delude, C.; Domergue, F.; Fernández, A.M.; Beaudoin, F.; Napier, J.A.; Kunst, L.; et al. Eceriferum2-Like Proteins Have Unique Biochemical and Physiological Functions in Very-Long-Chain Fatty Acid Elongation. Plant Physiol. 2015, 167, 682–692. [Google Scholar] [CrossRef]
- Yang, X.; Feng, T.; Li, S.; Zhao, H.; Zhao, S.; Ma, C.; Jenks, M.A.; Lü, S. Cer16 Inhibits Post-Transcriptional Gene Silencing of Cer3 to Regulate Alkane Biosynthesis. Plant Physiol. 2020, 182, 1211–1221. [Google Scholar] [CrossRef]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCg11/WBC11, an Atp Binding Cassette (ABC) Transporter That Is Required for Cuticular Lipid Secretion. Plant J. 2007, 52, 485–498. [Google Scholar] [CrossRef]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in Cer6, a Gene Identical to Cut1, Differentially Affect Long-Chain Lipid Content on the Surface of Pollen and Stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis Eceriferum9 Involvement in Cuticle Formation and Maintenance of Plant Water Status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kunst, L. Superkiller Complex Components Are Required for the Rna Exosome-Mediated Control of Cuticular Wax Biosynthesis in Arabidopsis Inflorescence Stems. Plant Physiol. 2016, 171, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis Cer8 Encodes Long-Chain Acyl-Coa Synthetase 1 (Lacs1) That Has Overlapping Functions with Lacs2 in Plant Wax and Cutin Synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Oshima, Y.; Ariga, H.; Kajino, T.; Koyama, T.; Yaguchi, Y.; Tanaka, K.; Yotsui, I.; Sakata, Y.; Taji, T. Eceriferum 10 Encoding an Enoyl-Coa Reductase Plays a Crucial Role in Osmotolerance and Cuticular Wax Loading in Arabidopsis. Front. Plant Sci. 2022, 13, 898317. [Google Scholar] [CrossRef]
- Shi, L.; Dean, G.H.; Zheng, H.; Meents, M.J.; Haslam, T.M.; Haughn, G.W.; Kunst, L. Eceriferum11/C-Terminal Domain Phosphatase-Like2 Affects Secretory Trafficking. Plant Physiol. 2019, 181, 901–915. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, H.; Kosma, D.K.; Tomasi, P.; Dyer, J.M.; Li, R.; Liu, X.; Wang, Z.; Parsons, E.P.; Jenks, M.A.; et al. The Acyl Desaturase Cer17 Is Involved in Producing Wax Unsaturated Primary Alcohols and Cutin Monomers. Plant Physiol. 2017, 173, 1109–1124. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, L.; Jia, J.; Song, L.; Wang, J. Management of Drought Risk under Global Warming. Theor. Appl. Climatol. 2016, 125, 187–196. [Google Scholar] [CrossRef]
- Qi, C.; Jiang, H.; Zhao, X.; Mao, K.; Liu, H.; Li, Y.; Hao, Y. The Characterization, Authentication, and Gene Expression Pattern of the Mdcer Family in Malus domestica. Hortic. Plant J. 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Wu, H.; Liu, L.; Chen, Y.; Liu, T.; Jiang, Q.; Wei, Z.; Li, C.; Wang, Z. Tomato Slcer1–1 Catalyzes the Synthesis of Wax Alkanes, Increasing Drought Tolerance and Fruit Storability. Hortic. Res. 2022, 9, uhac004. [Google Scholar] [CrossRef]
- Grünhofer, P.; Herzig, L.; Zhang, Q.; Vitt, S.; Stöcker, T.; Malkowsky, Y.; Brügmann, T.; Fladung, M.; Schreiber, L. Changes in Wax Composition but Not Amount Enhance Cuticular Transpiration. Plant Cell Environ. 2024, 47, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.; Zhang, X.; Wang, J.; Hou, T.; He, J.; Li, J. Integration of Transcriptome and Metabolome Reveals Wax Serves a Key Role in Preventing Leaf Water Loss in Goji (Lycium barbarum). Int. J. Mol. Sci. 2024, 25, 10939. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, X.; Zhang, S.-D.; Zhang, X.; Feng, L.-D.; He, J. An Increased Wax Load on the Leaves of Goji Plants (Lycium barbarum) Results in Increased Resistance to Powdery Mildew. Chem. Biol. Technol. Agric. 2024, 11, 62. [Google Scholar] [CrossRef]
- Wang, P.; Wang, J.; Zhang, H.; Wang, C.; Zhao, L.; Huang, T.; Qing, K. Chemical Composition, Crystal Morphology, and Key Gene Expression of the Cuticular Waxes of Goji (Lycium barbarum L.) Berries. J. Agri. Food Chem. 2021, 69, 7874–7883. [Google Scholar] [CrossRef]
- Li, N.; Song, Y.; Li, J.; Hao, R.; Feng, X.; Li, L. Transcriptome and Genome Re-Sequencing Analysis Reveals Differential Expression Patterns and Sequence Variation in Pericarp Wax Metabolism-Related Genes in Ziziphus jujuba (Chinese Jujube). Sci. Hortic. 2021, 288, 110415. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, J.; Xia, X.; Xu, Y.; Chen, S. Comparative Analysis of Leaf Trichomes, Epidermal Wax and Defense Enzymes Activities in Response to Puccinia horiana in Chrysanthemum and Ajania Species. Hortic. Plant J. 2020, 6, 8. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. Mg2c: A User-Friendly Online Tool for Drawing Genetic Maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Tao, Y.X.; Peer, A.F.V.; Huang, Q.H.; Shao, Y.P.; Xie, B.G. Identification of Novel and Robust Internal Control Genes from Volvariella volvacea That Are Suitable for RT-qPCR in Filamentous Fungi. Sci. Rep. 2016, 6, 29236. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.; Wang, M.; Bi, Q.; Cui, Y.; Wang, L. Transcriptome and Physiological Analyses Provide Insights into the Leaf Epicuticular Wax Accumulation Mechanism in Yellowhorn. Hortic. Res. 2021, 8, 134. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Regulatory Mechanisms Underlying Cuticular Wax Biosynthesis. J. Exp. Bot. 2022, 73, 2799–2816. [Google Scholar] [CrossRef]
- Yang, H.; Mei, W.; Wan, H.; Xu, R.; Cheng, Y. Comprehensive Analysis of Kcs Gene Family in Citrinae Reveals the Involvement of Cskcs2 and Cskcs11 in Fruit Cuticular Wax Synthesis at Ripening. Plant Sci. 2021, 310, 110972. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Sun, Y.; Wang, Y.; Li, T.; Chai, G.; Jiang, W.; Shan, L.; Li, C.; Xiao, E.; et al. Far5, a Fatty Acyl-Coenzyme a Reductase, Is Involved in Primary Alcohol Biosynthesis of the Leaf Blade Cuticular Wax in Wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.; Ghorbanzadeh, Z.; Jacob, F.; Nekouei, M.K.; Zeinalabedini, M.; Mardi, M.; Sadeghi, A.; Ghaffari, M.R. Decoding Drought Resilience: A Comprehensive Exploration of the Cotton Eceriferum (Cer) Gene Family and Its Role in Stress Adaptation. BMC Plant Biol. 2024, 24, 468. [Google Scholar] [CrossRef]
- Finocchiaro, F.; Terzi, V.; Delbono, S. Barley: From Molecular Basis of Quality to Advanced Genomics-Based Breeding. In Compendium of Crop Genome Designing for Nutraceuticals; Springer Nature: Singapore, 2023; pp. 1–38. [Google Scholar]
- Rizwan, H.M.; Waheed, A.; Ma, S.; Li, J.; Arshad, M.B.; Irshad, M.; Li, B.; Yang, X.; Ali, A.; Ahmed Ma, A.; et al. Comprehensive Genome-Wide Identification and Expression Profiling of Eceriferum (Cer) Gene Family in Passion Fruit (Passiflora edulis) under Fusarium kyushuense and Drought Stress Conditions. Front. Plant Sci. 2022, 13, 898307. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Azhar, M.T.; Rana, I.A.; Azeem, F.; Ali, M.A.; Nawaz, M.A.; Chung, G.; Atif, R.M. Genome-Wide Identification and Expression Analyses of Wrky Transcription Factor Family Members from Chickpea (Cicer arietinum L.) Reveal Their Role in Abiotic Stress-Responses. Genes Genom. 2019, 41, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Sun, H.; Wang, C.; Yang, L.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y.; Liu, F.; Ji, J. Genome-Wide Identification of the Eceriferum (Cer) Gene Family in Cabbage and Critical Role of Bocer4.1 in Wax Biosynthesis. Plant Physiol Biochem. 2025, 222, 109718. [Google Scholar] [CrossRef]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The Ap2/Erf Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes 2020, 11, 1464. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Xu, B.; Shi, Y.; Wu, Y.; Meng, Y.; Jin, Y. Role of Rna Secondary Structures in Regulating Dscam Alternative Splicing. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194381. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-Regulatory Elements: Molecular Mechanisms and Evolutionary Processes Underlying Divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bai, C.; Luo, N.; Jiang, Y.; Wang, Y.; Liu, Y.; Chen, C.; Wang, Y.; Gan, Q.; Jin, S.; et al. Brassica Napus Bnac9.Dewax1 Negatively Regulates Wax Biosynthesis Via Transcriptional Suppression of Bncer1-2. Int. J. Mol. Sci. 2023, 24, 4287. [Google Scholar] [CrossRef]
- Xu, L.; Hao, J.; Lv, M.; Liu, P.; Ge, Q.; Zhang, S.; Yang, J.; Niu, H.; Wang, Y.; Xue, Y.; et al. A Genome-Wide Association Study Identifies Genes Associated with Cuticular Wax Metabolism in Maize. Plant Physiol. 2024, 194, 2616–2630. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Areb1, Areb2, and Abf3 Are Master Transcription Factors That Cooperatively Regulate Abre-Dependent Aba Signaling Involved in Drought Stress Tolerance and Require Aba for Full Activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- De Silva, W.S.I.; Perera, M.M.N.; Perera, K.L.N.S.; Wickramasuriya, A.M.; Jayasekera, G.A.U. In Silico Analysis of Osr40c1 Promoter Sequence Isolated from Indica Variety Pokkali. Rice Sci. 2017, 24, 228–234. [Google Scholar] [CrossRef]






| Gene ID | Gene Name | Primer Type | Primer Sequences (5′ to 3′) |
|---|---|---|---|
| Lba01g00543 | LbaCER1–1 | F | CATCACCACGCTAGGACCATTG |
| R | TTTCACCTTGGCTTTCAACACTTTC | ||
| Lba01g01071 | LbaCER1–5 | F | CCTAGCCCTTCTCTTTCTTCCATTATC |
| R | ACAACCCATTCCACTTATCGTATAGC | ||
| Lba02g02625 | LbaCER2–5 | F | CTGGTGTGATAGCGGTTGATCTTG |
| R | TCTCCGTACTAACAACAACAGCATAC | ||
| Lba03g02510 | LbaCER3–11 | F | CAAGTTGAATTGTTATGGACTGGAAGG |
| R | GCATTCACAGTTGGTATTGGCATTAC | ||
| Lba03g02612 | LbaCER3–12 | F | TCGTGCTTATTATGTGGCTGAAGTC |
| R | GTTTGGTTGAGTTTCCCTCTTATGTTG | ||
| Loc132636280 | RH37 | F R | GCAGGCAAGTCAGGATTAGCA CGCATAACGAGTCAACCATTCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Q.; Li, J.; Jing, L.; Zhang, F.; Liu, B.; Guo, L. Genome-Wide Identification and Tissue-Specific Expression Profiling of Goji CER Gene Family. Genes 2025, 16, 1257. https://doi.org/10.3390/genes16111257
Yu Q, Li J, Jing L, Zhang F, Liu B, Guo L. Genome-Wide Identification and Tissue-Specific Expression Profiling of Goji CER Gene Family. Genes. 2025; 16(11):1257. https://doi.org/10.3390/genes16111257
Chicago/Turabian StyleYu, Qian, Jie Li, Lijuan Jing, Feng Zhang, Bohua Liu, and Liuwei Guo. 2025. "Genome-Wide Identification and Tissue-Specific Expression Profiling of Goji CER Gene Family" Genes 16, no. 11: 1257. https://doi.org/10.3390/genes16111257
APA StyleYu, Q., Li, J., Jing, L., Zhang, F., Liu, B., & Guo, L. (2025). Genome-Wide Identification and Tissue-Specific Expression Profiling of Goji CER Gene Family. Genes, 16(11), 1257. https://doi.org/10.3390/genes16111257






