Transcriptome Analysis Reveals Equine Endometrium’s Gene Expression Profile Around Embryo Fixation
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Mare Endometrial Tissue Samples
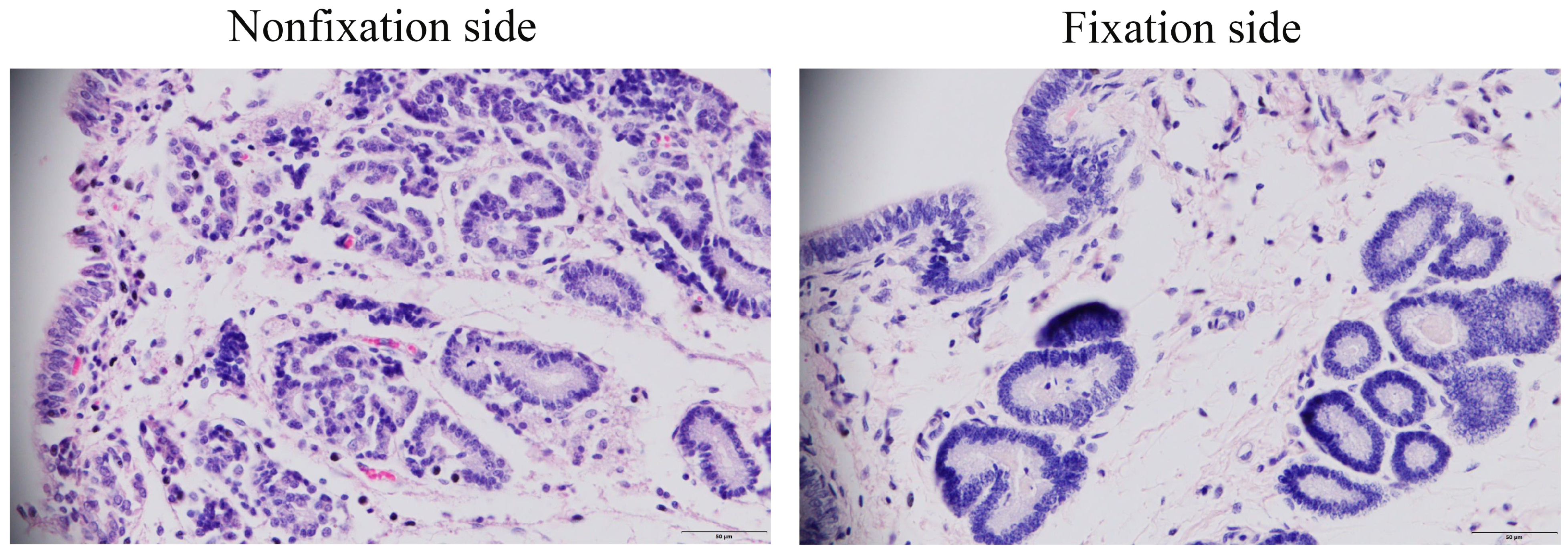
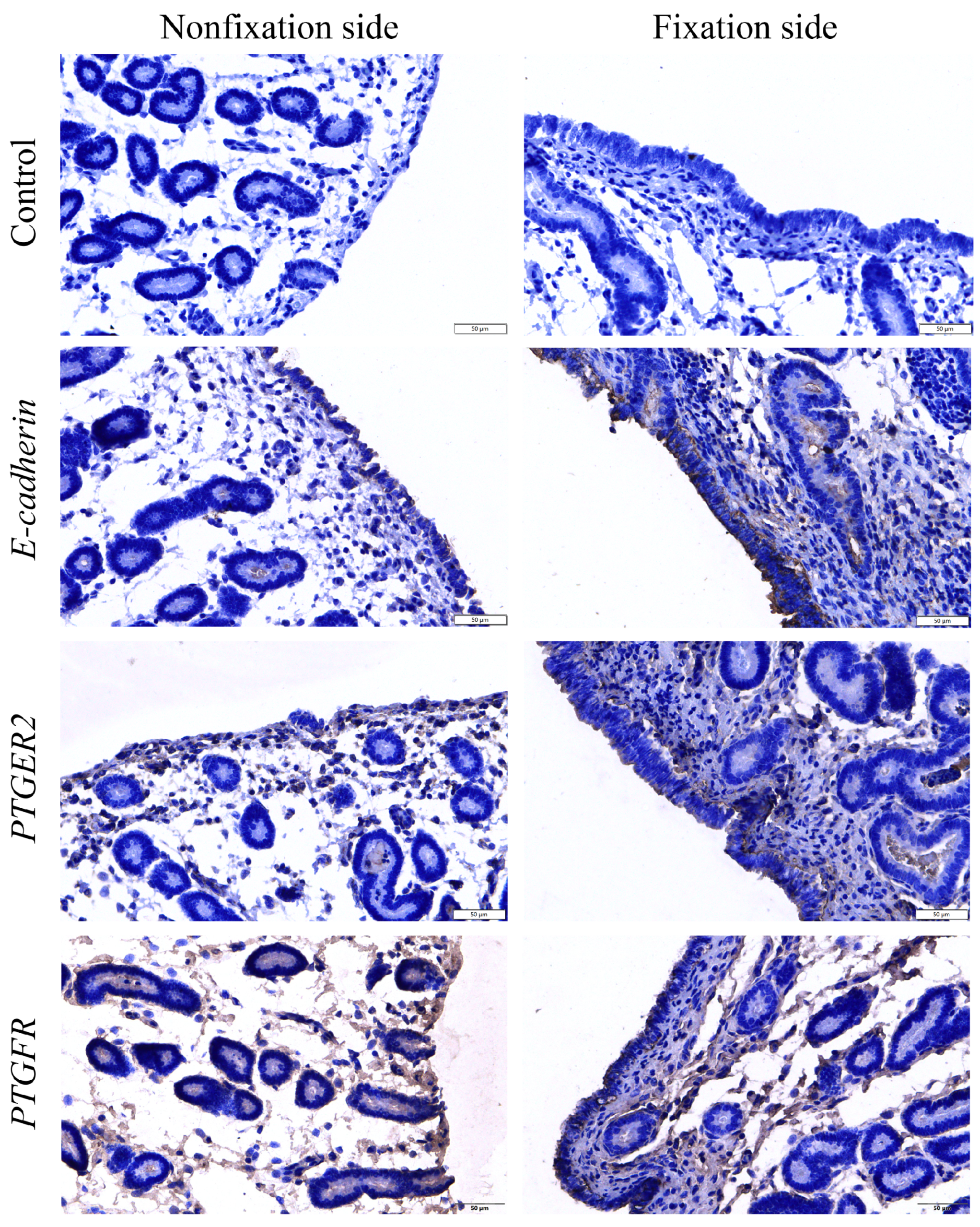
2.2. The Histomorphologic of Endometrial Tissue in Mares
2.3. RNA Sequencing
2.4. Quality Control and Alignment
2.5. Screening of DEGs
2.6. Enrichment Analysis of GO and KEGG Pathways
2.7. Quantitative Real-Time PCR (qRT-PCR) Assay
2.8. Immunohistochemistry
2.9. Statistical Analysis
3. Results
3.1. Comparison of Endometrial Histology
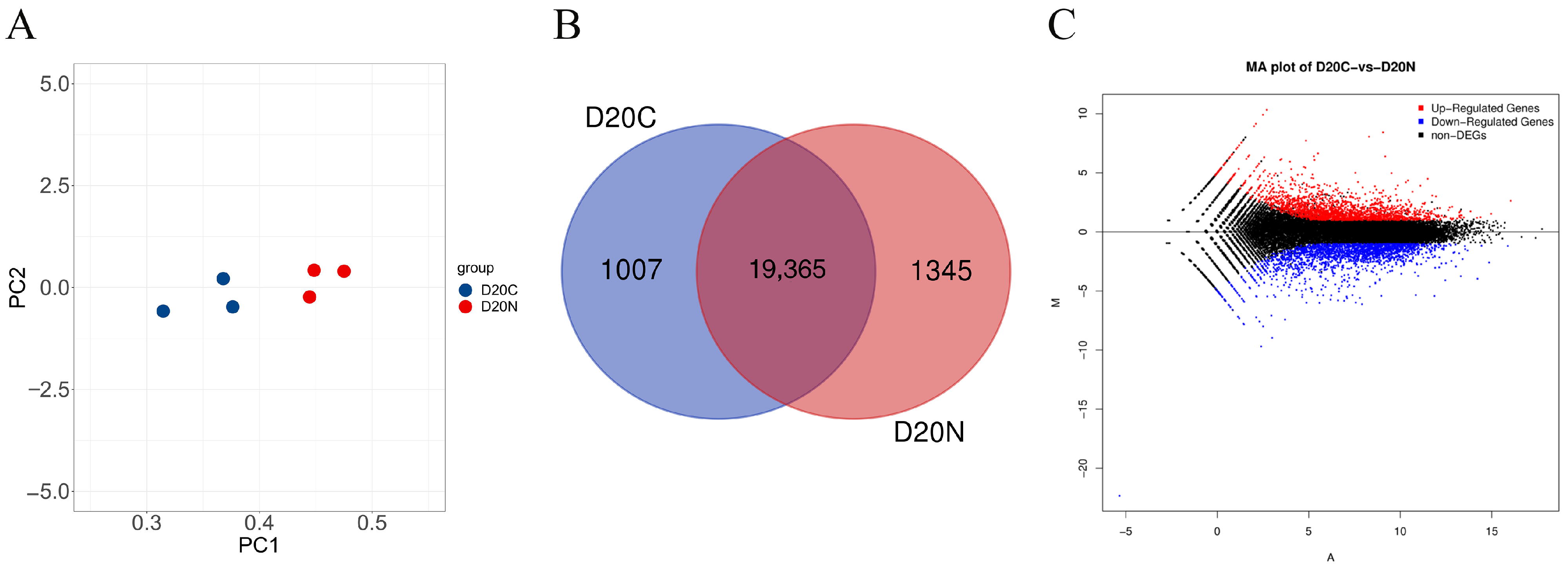
3.2. Basic Statistical Analysis of the RNA Sequencing Data
3.3. Differential Expression Analysis
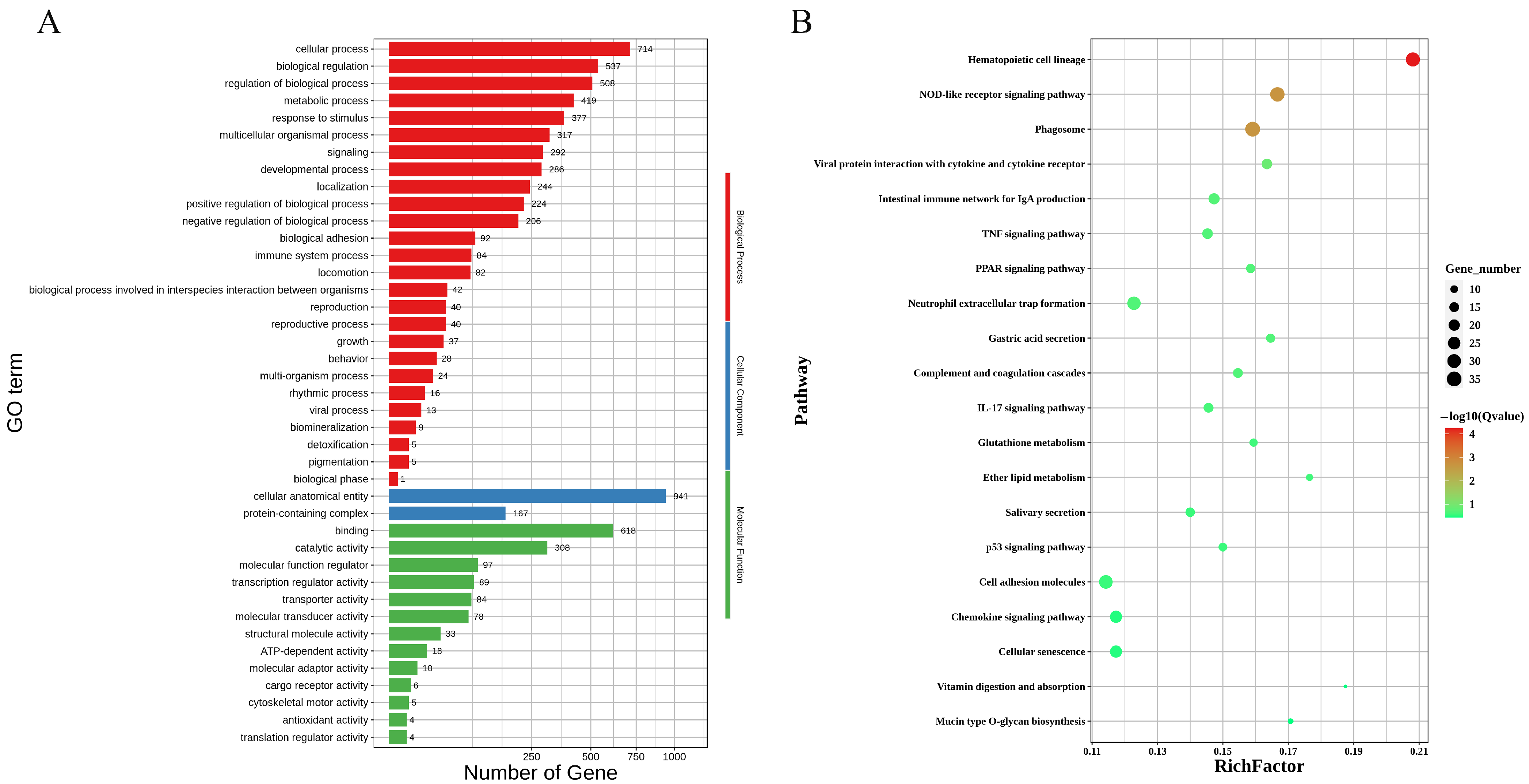
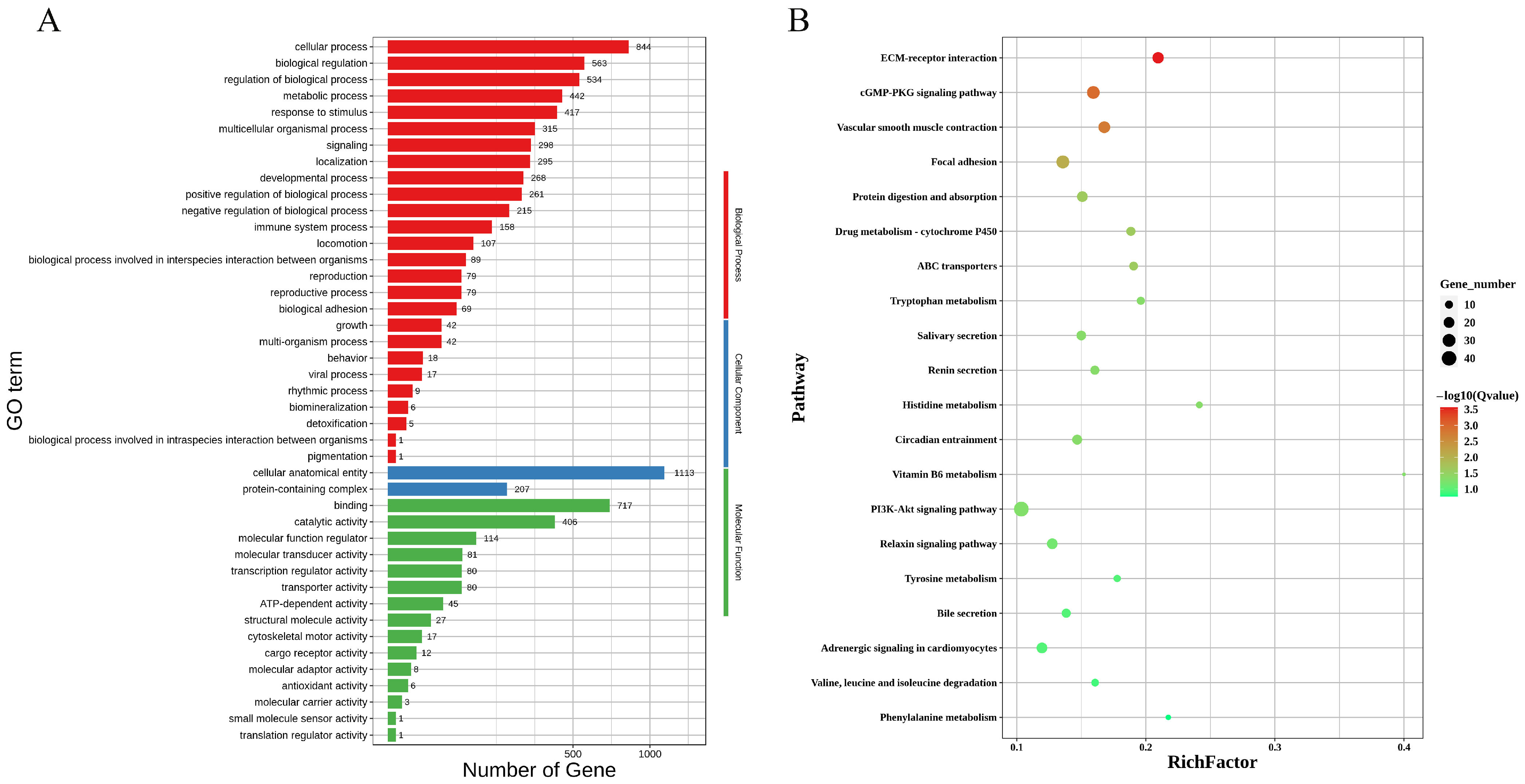
3.4. Gene Function Enrichment Analysis
3.5. Endometrium Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Navarro, S.; Giraudo, P.; Karseladze, A.I.; Smirnov, A.; Petrovichev, N.; Savelov, N.; Alvarado-Cabrero, I.; Llombart-Bosch, A. Immunophenotypic profile of biomarkers related to anti-apoptotic and neural development pathways in the Ewing’s family of tumors (EFT) and their therapeutic implications. Anticancer. Res. 2007, 27, 2457–2463. [Google Scholar] [PubMed]
- Allen, W.R.; Wilsher, S. A review of implantation and early placentation in the mare. Placenta 2009, 30, 1005–1015. [Google Scholar] [CrossRef]
- Gastal, M.O.; Gastal, E.L.; Kot, K.; Ginther, O.J. Factors related to the time of fixation of the conceptus in mares. Theriogenology 1996, 46, 1171–1180. [Google Scholar] [CrossRef]
- Cross, D.T.; Ginther, O.J. Uterine contractions in nonpregnant and early pregnant mares and jennies as determined by ultrasonography. J. Anim. Sci. 1988, 66, 250–254. [Google Scholar] [CrossRef]
- Carnevale, E.M.; Ginther, O.J. Relationships of age to uterine function and reproductive efficiency in mares. Theriogenology 1992, 37, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.G.; Ginther, O.J. Uterine contractile activity in mares during the estrous cycle and early pregnancy. Theriogenology 1990, 34, 47–56. [Google Scholar] [CrossRef]
- Dangudubiyyam, S.V.; Ginther, O.J. Relationship between more follicles in right than left ovary in recently born calves and right ovary propensity for ovulation in cattle. Reprod. Biol. 2019, 19, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Vergaro, P.; Tiscornia, G.; Rodriguez, A.; Santalo, J.; Vassena, R. Transcriptomic analysis of the interaction of choriocarcinoma spheroids with receptive vs. non-receptive endometrial epithelium cell lines: An in vitro model for human implantation. J. Assist. Reprod. Genet. 2019, 36, 857–873. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, A.M.; Macklon, N.S. The science of implantation emerges blinking into the light. Reprod. Biomed. Online 2013, 27, 453–460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hannan, N.J.; Jones, R.L.; White, C.A.; Salamonsen, L.A. The chemokines, CX3CL1, CCL14, and CCL4, promote human trophoblast migration at the feto-maternal interface. Biol. Reprod. 2006, 74, 896–904. [Google Scholar] [CrossRef]
- Hannan, N.J.; Paiva, P.; Meehan, K.L.; Rombauts, L.J.; Gardner, D.K.; Salamonsen, L.A. Analysis of fertility-related soluble mediators in human uterine fluid identifies VEGF as a key regulator of embryo implantation. Endocrinology 2011, 152, 4948–4956. [Google Scholar] [CrossRef]
- Greening, D.W.; Nguyen, H.P.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.; Saravelos, S.H.; Liu, Y.; Huang, J.; Zhang, J.; Li, T.C. HOXA-10 and E-cadherin expression in the endometrium of women with recurrent implantation failure and recurrent miscarriage. Fertil. Steril. 2017, 107, 136–143.e2. [Google Scholar] [CrossRef] [PubMed]
- Floridon, C.; Nielsen, O.; Holund, B.; Sunde, L.; Westergaard, J.G.; Thomsen, S.G.; Teisner, B. Localization of E-cadherin in villous, extravillous and vascular trophoblasts during intrauterine, ectopic and molar pregnancy. Mol. Hum. Reprod. 2000, 6, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Blechschmidt, K.; Mylonas, I.; Mayr, D.; Schiessl, B.; Schulze, S.; Becker, K.F.; Jeschke, U. Expression of E-cadherin and its repressor snail in placental tissue of normal, preeclamptic and HELLP pregnancies. Virchows Arch. 2007, 450, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Nollet, F.; Kools, P.; van Roy, F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 2000, 299, 551–572. [Google Scholar] [CrossRef] [PubMed]
- Paria, B.C.; Zhao, X.; Das, S.K.; Dey, S.K.; Yoshinaga, K. Zonula occludens-1 and E-cadherin are coordinately expressed in the mouse uterus with the initiation of implantation and decidualization. Dev. Biol. 1999, 208, 488–501. [Google Scholar] [CrossRef] [PubMed]
- Jha, R.K.; Titus, S.; Saxena, D.; Kumar, P.G.; Laloraya, M. Profiling of E-cadherin, β-catenin and Ca(2+) in embryo–uterine interactions at implantation. FEBS Lett. 2006, 580, 5653–5660. [Google Scholar] [CrossRef]
- Riethmacher, D.; Brinkmann, V.; Birchmeier, C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. USA 1995, 92, 855–859. [Google Scholar] [CrossRef]
- Coutifaris, C.; Kao, L.C.; Sehdev, H.M.; Chin, U.; Babalola, G.O.; Blaschuk, O.W.; Strauss, J.F. E-cadherin expression during the differentiation of human trophoblasts. Development 1991, 113, 767–777. [Google Scholar] [CrossRef] [PubMed]
- van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.G.; Mansel, R.E. E-cadherin complex and its abnormalities in human breast cancer. Surg. Oncol. 2000, 9, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Shirane, A.; Wada-Hiraike, O.; Tanikawa, M.; Seiki, T.; Hiraike, H.; Miyamoto, Y.; Sone, K.; Hirano, M.; Oishi, H.; Oda, K.; et al. Regulation of SIRT1 determines initial step of endometrial receptivity by controlling E-cadherin expression. Biochem. Biophys. Res. Commun. 2012, 424, 604–610. [Google Scholar] [CrossRef]
- Ginther, O.J. Equine embryo mobility. A game changer. Theriogenology 2021, 174, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Boerboom, D.; Brown, K.A.; Vaillancourt, D.; Poitras, P.; Goff, A.K.; Watanabe, K.; Doré, M.; Sirois, J. Expression of key prostaglandin synthases in equine endometrium during late diestrus and early pregnancy. Biol. Reprod. 2004, 70, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Hyland, J.H.; Manns, J.G.; Humphrey, W.D. Prostaglandin production by ovine embryos and endometrium in vitro. J. Reprod. Fertil. 1982, 65, 299–304. [Google Scholar] [CrossRef]
- Hwang, D.H.; Pool, S.H.; Rorie, R.W.; Boudreau, M.; Godke, R.A. Transitional changes in arachidonic acid metabolism by bovine embryos at different developmental stages. Prostaglandins 1988, 35, 387–402. [Google Scholar] [CrossRef]
- Watson, E.D.; Sertich, P.L. Prostaglandin production by horse embryos and the effect of co-culture of embryos with endometrium from pregnant mares. J. Reprod. Fertil. 1989, 87, 331–336. [Google Scholar] [CrossRef]
- Maitre, J.L.; Heisenberg, C.P. Three functions of cadherins in cell adhesion. Curr. Biol. 2013, 23, R626–R633. [Google Scholar] [CrossRef] [PubMed]
- McCracken, J.A.; Custer, E.E.; Lamsa, J.C. Luteolysis: A neuroendocrine-mediated event. Physiol. Rev. 1999, 79, 263–323. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.S.; Rueda, B.R. The corpus luteum: An ovarian structure with maternal instincts and suicidal tendencies. Front. Biosci. 2002, 7, d1949–d1978. [Google Scholar] [CrossRef] [PubMed]
- Sirois, J.; Dore, M. The late induction of prostaglandin G/H synthase-2 in equine preovulatory follicles supports its role as a determinant of the ovulatory process. Endocrinology 1997, 138, 4427–4434. [Google Scholar] [CrossRef] [PubMed]
- Kargman, S.; Charleson, S.; Cartwright, M.; Frank, J.; Riendeau, D.; Mancini, J.; Evans, J.; O’Neill, G. Characterization of Prostaglandin G/H Synthase 1 and 2 in rat, dog, monkey, and human gastrointestinal tracts. Gastroenterology 1996, 111, 445–454. [Google Scholar] [CrossRef]
- Stout, T.A.; Allen, W.R. Prostaglandin E0 and F2α production by equine conceptuses and concentrations in conceptus fluids and uterine flushings recovered from early pregnant and dioestrous mares. Reproduction 2002, 123, 261–268. [Google Scholar] [CrossRef]
- Piotrowska-Tomala, K.K.; Jonczyk, A.W.; Skarzynski, D.J.; Szostek-Mioduchowska, A.Z. Luteinizing hormone and ovarian steroids affect in vitro prostaglandin production in the equine myometrium and endometrium. Theriogenology 2020, 153, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Budik, S.; Walter, I.; Leitner, M.C.; Ertl, R.; Aurich, C. Expression of Enzymes Associated with Prostaglandin Synthesis in Equine Conceptuses. Animals 2021, 11, 1180. [Google Scholar] [CrossRef]
- Weber, J.A.; Freeman, D.A.; Vanderwall, D.K.; Woods, G.L. Prostaglandin E2 secretion by oviductal transport-stage equine embryos. Biol. Reprod. 1991, 45, 540–543. [Google Scholar] [CrossRef]
- Weber, J.A.; Freeman, D.A.; Vanderwall, D.K.; Woods, G.L. Prostaglandin E2 hastens oviductal transport of equine embryos. Biol. Reprod. 1991, 45, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Stout, T.A. Embryo–maternal communication during the first 4 weeks of equine pregnancy. Theriogenology 2016, 86, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Klein, C. Early pregnancy in the mare: Old concepts revisited. Domest. Anim. Endocrinol. 2016, 56, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Allen, W.R. Fetomaternal interactions and influences during equine pregnancy. Reproduction 2001, 121, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Goff, A.K.; Pontbriand, D.; Sirois, J. Oxytocin stimulation of plasma 15-keto-13,14-dihydro prostaglandin F-2 alpha during the oestrous cycle and early pregnancy in the mare. J. Reprod. Fertil. Suppl. 1987, 35, 253–260. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulaangerel, T.; Mu, S.; Sodyelalt, J.; Yi, M.; Zhao, B.; Hao, A.; Wen, X.; Han, B.; Bou, G. Transcriptome Analysis Reveals Equine Endometrium’s Gene Expression Profile Around Embryo Fixation. Genes 2025, 16, 181. https://doi.org/10.3390/genes16020181
Ulaangerel T, Mu S, Sodyelalt J, Yi M, Zhao B, Hao A, Wen X, Han B, Bou G. Transcriptome Analysis Reveals Equine Endometrium’s Gene Expression Profile Around Embryo Fixation. Genes. 2025; 16(2):181. https://doi.org/10.3390/genes16020181
Chicago/Turabian StyleUlaangerel, Tseweendolmaa, Siqin Mu, Jolanqiqige Sodyelalt, Minna Yi, Bilig Zhao, Asiya Hao, Xin Wen, Baoxiang Han, and Gerelchimeg Bou. 2025. "Transcriptome Analysis Reveals Equine Endometrium’s Gene Expression Profile Around Embryo Fixation" Genes 16, no. 2: 181. https://doi.org/10.3390/genes16020181
APA StyleUlaangerel, T., Mu, S., Sodyelalt, J., Yi, M., Zhao, B., Hao, A., Wen, X., Han, B., & Bou, G. (2025). Transcriptome Analysis Reveals Equine Endometrium’s Gene Expression Profile Around Embryo Fixation. Genes, 16(2), 181. https://doi.org/10.3390/genes16020181






