Broad Target Metabolomics Revealed the Key Regulatory Mechanisms of the Effects of Trace Element Water-Soluble Fertilizer on the Growth of Corylus heterophylla × Corylus avellana Seedlings
Abstract
1. Introduction
2. Plant Materials and Methods
3. Experimental Methods
3.1. Measurement of Photosynthetic Characteristics
3.2. Broad Target Metabolomics
3.3. Determination of Flavonoid Content
3.4. Quantitative Real-Time PCR
3.5. Data Preprocessing and Statistical Analysis
4. Results
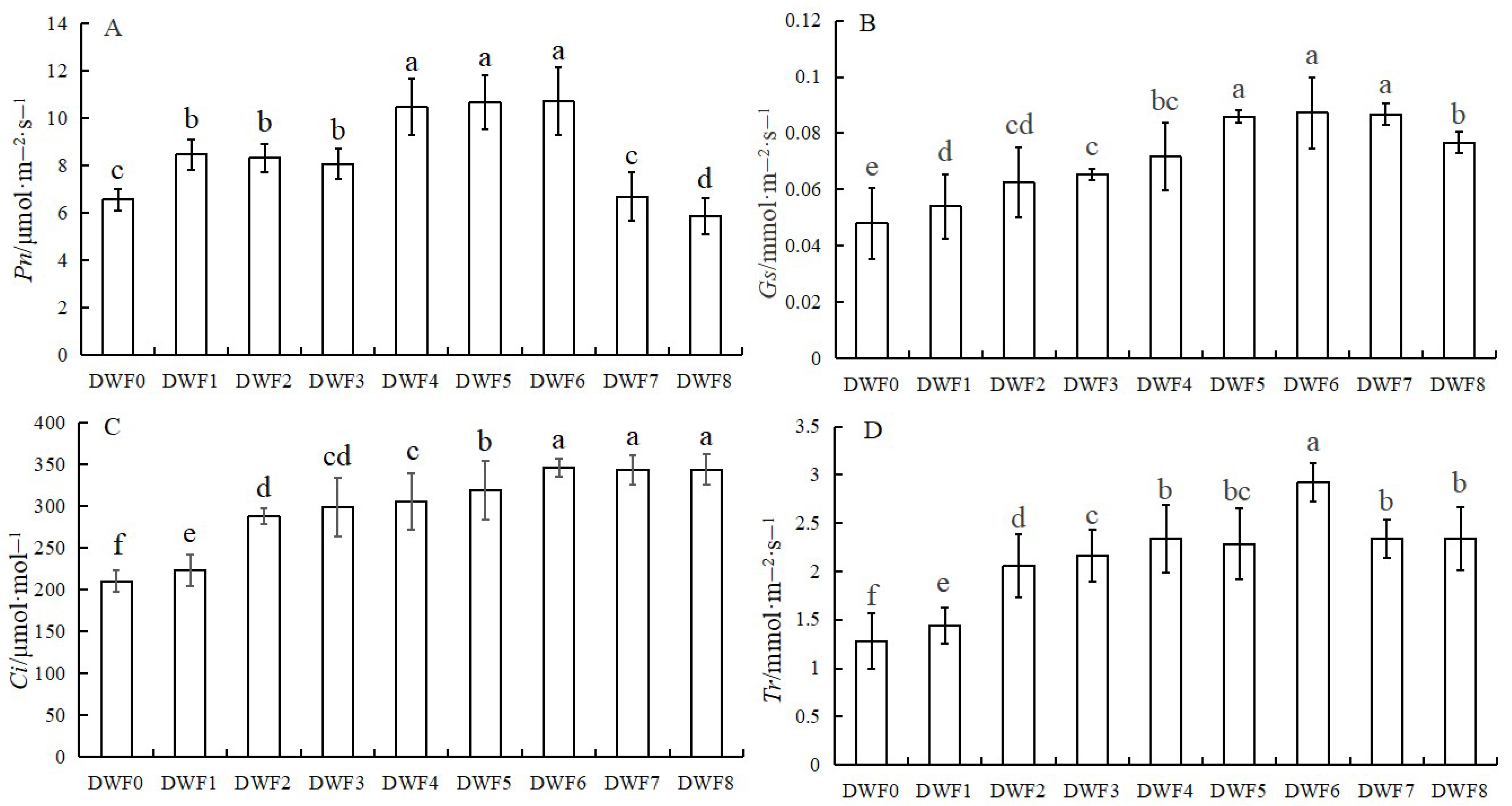
4.1. Effects of Different Doses of Trace Element Water-Soluble Fertilizers on Photosynthetic Parameters of Hazelnut Leaves
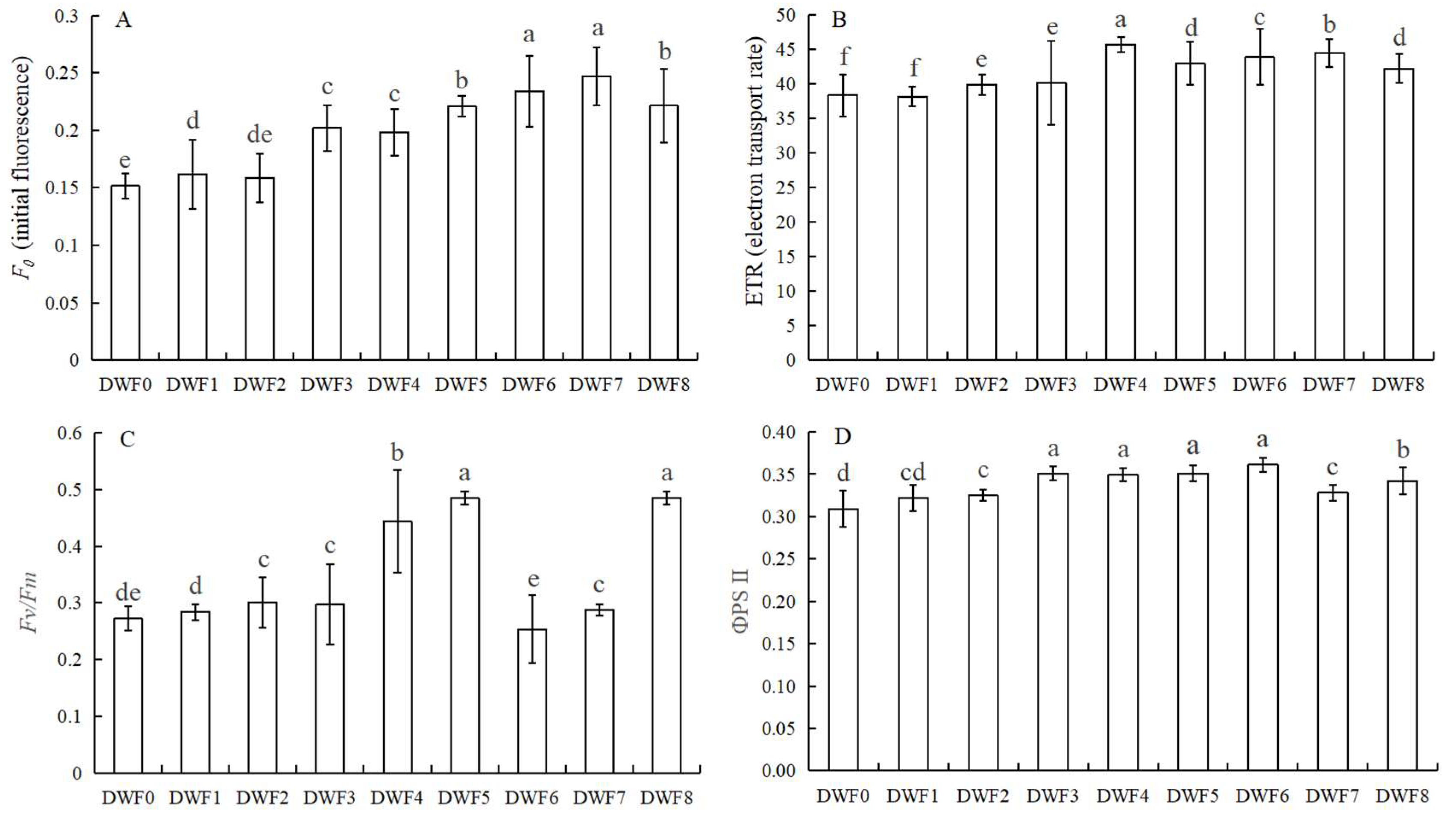
4.2. Effects of Different Doses of Trace Element Water-Soluble Fertilizers on the Fluorescence Parameters of Hazelnut Leaves
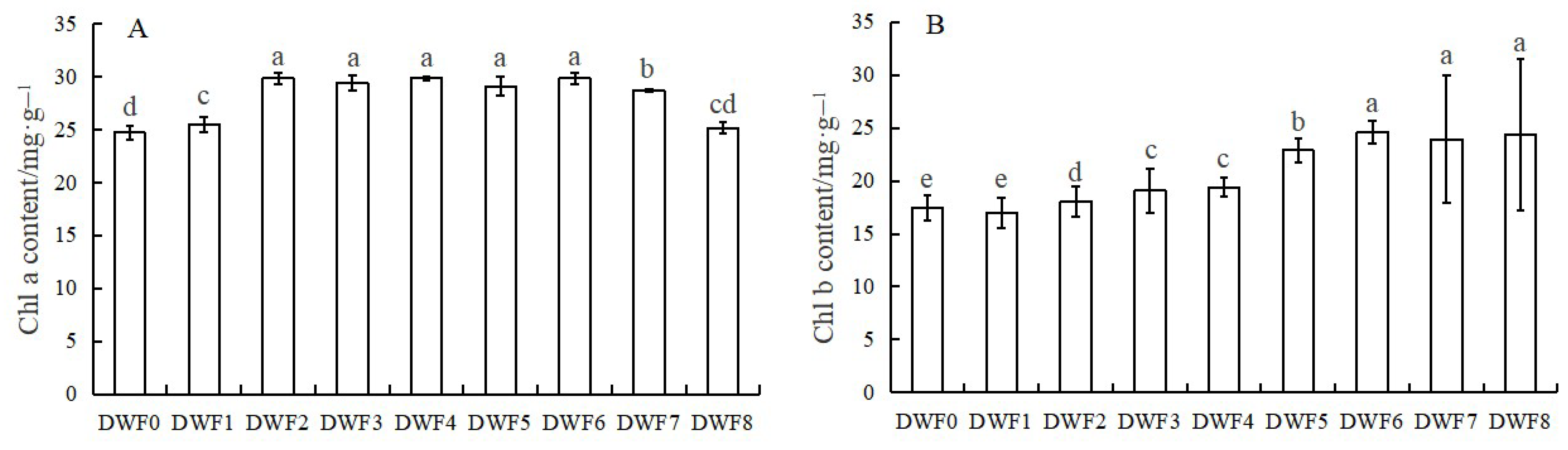
4.3. Effects of Different Doses of Trace Element Water-Soluble Fertilizers on Photosynthetic Pigments in Hazelnut Leaves
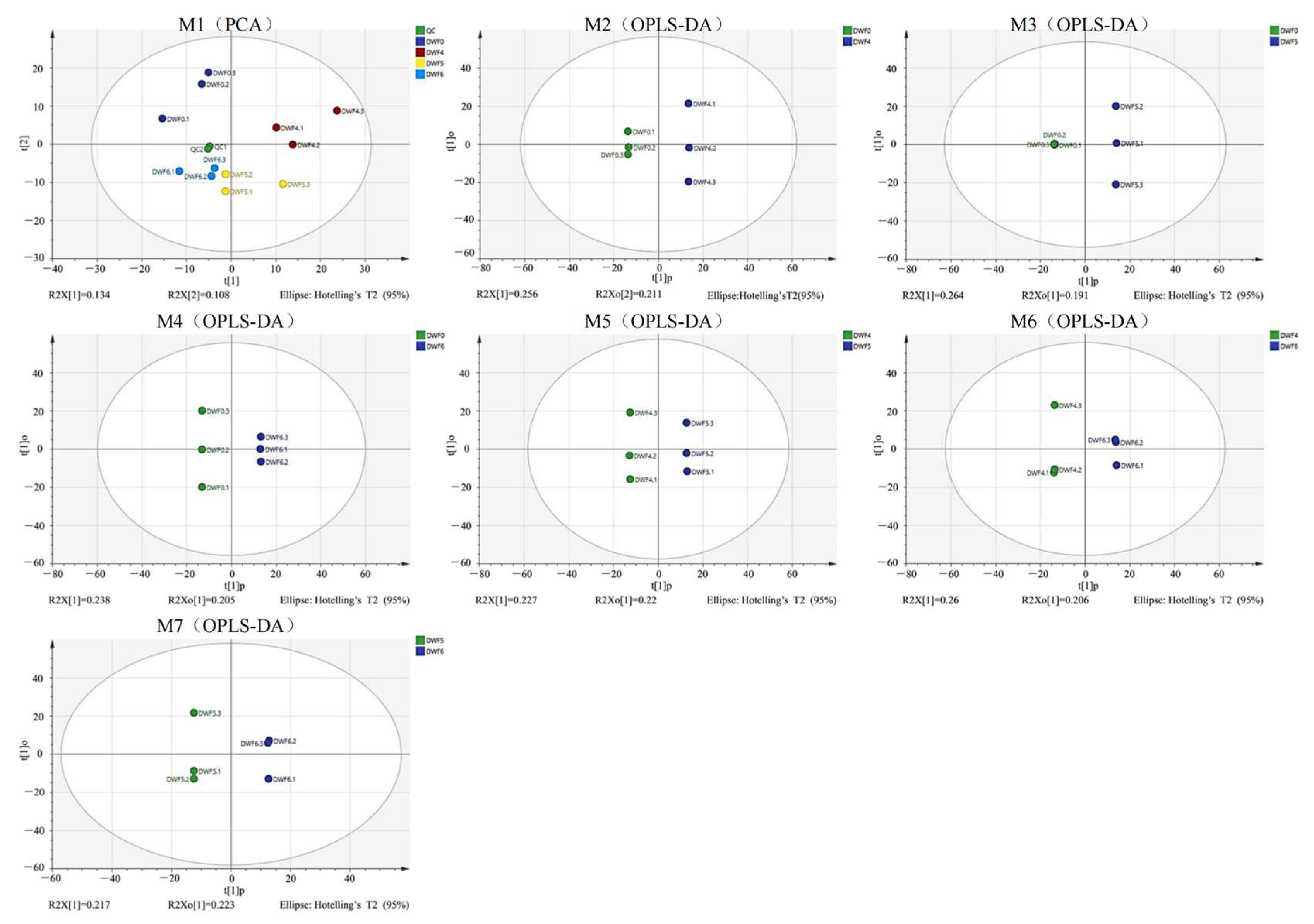
4.4. Metabolic Trajectory of Hazelnut Leaves Treated with Different Doses of Trace Element Water-Soluble Fertilizers
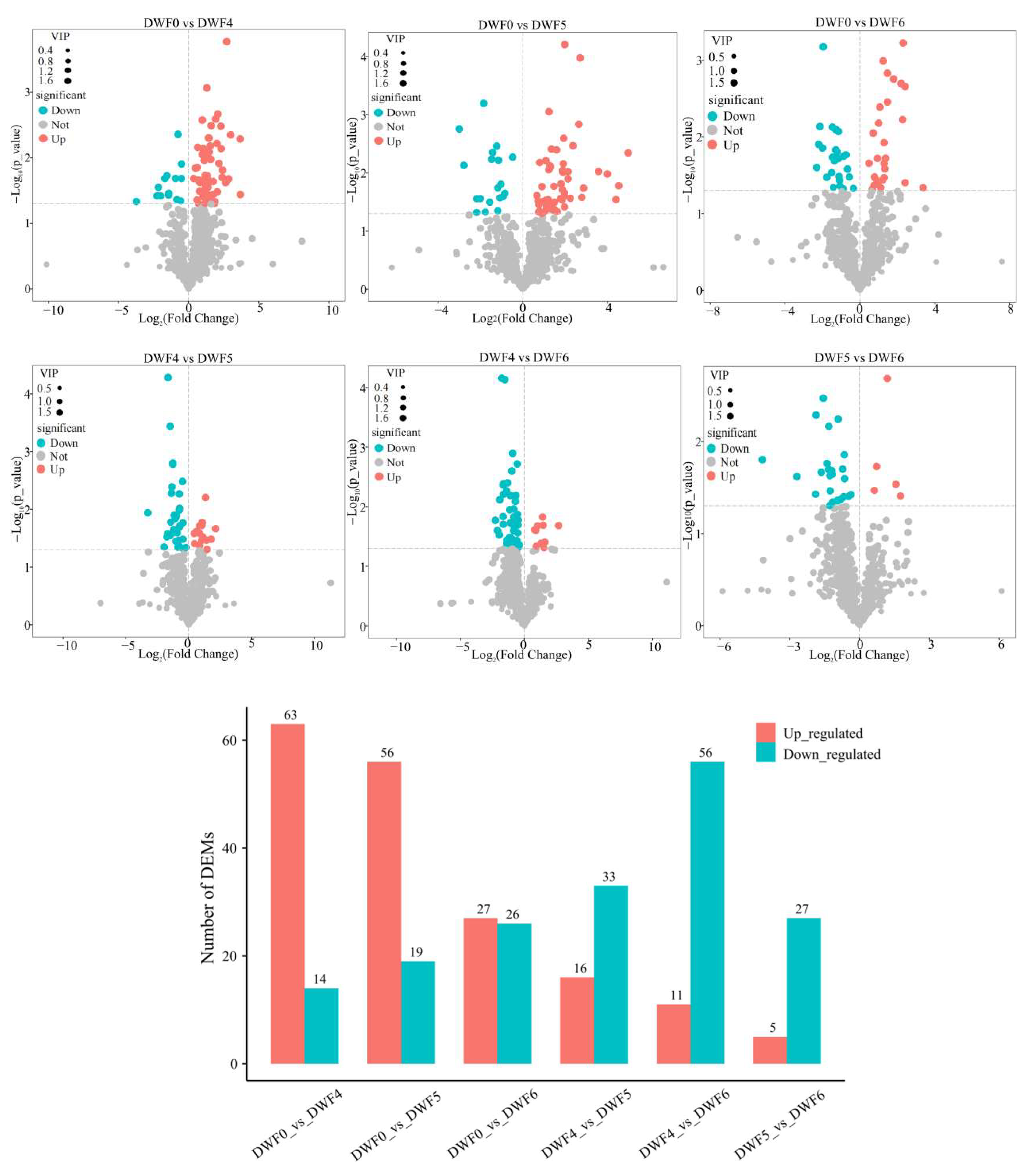
4.5. Metabolite Analysis of Hazelnut Leaves
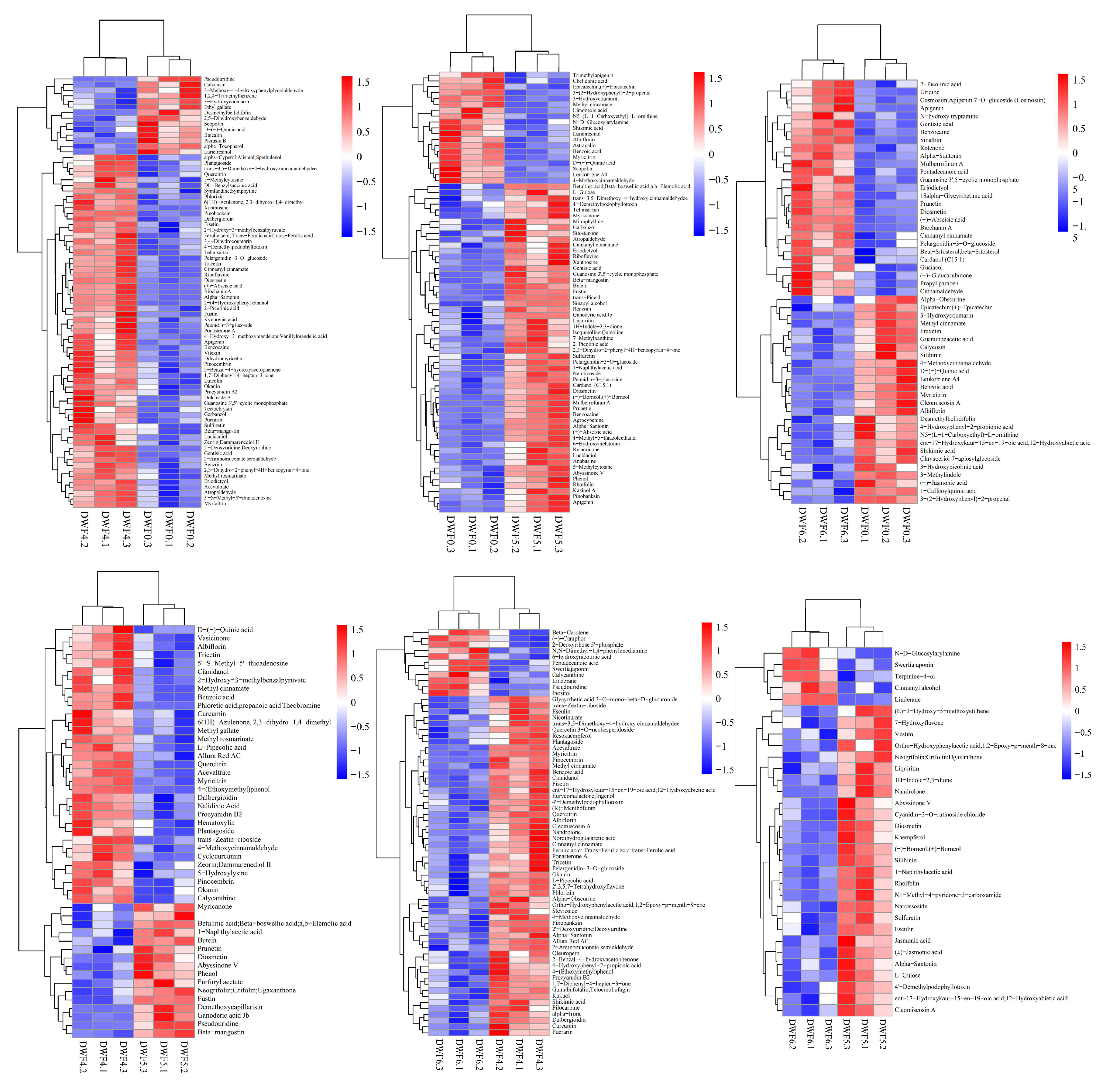
4.6. Hierarchical Clustering Analysis of Metabolic Components in Leaves of Hazelnut at Different Doses of Trace Element Water-Soluble Fertilizers
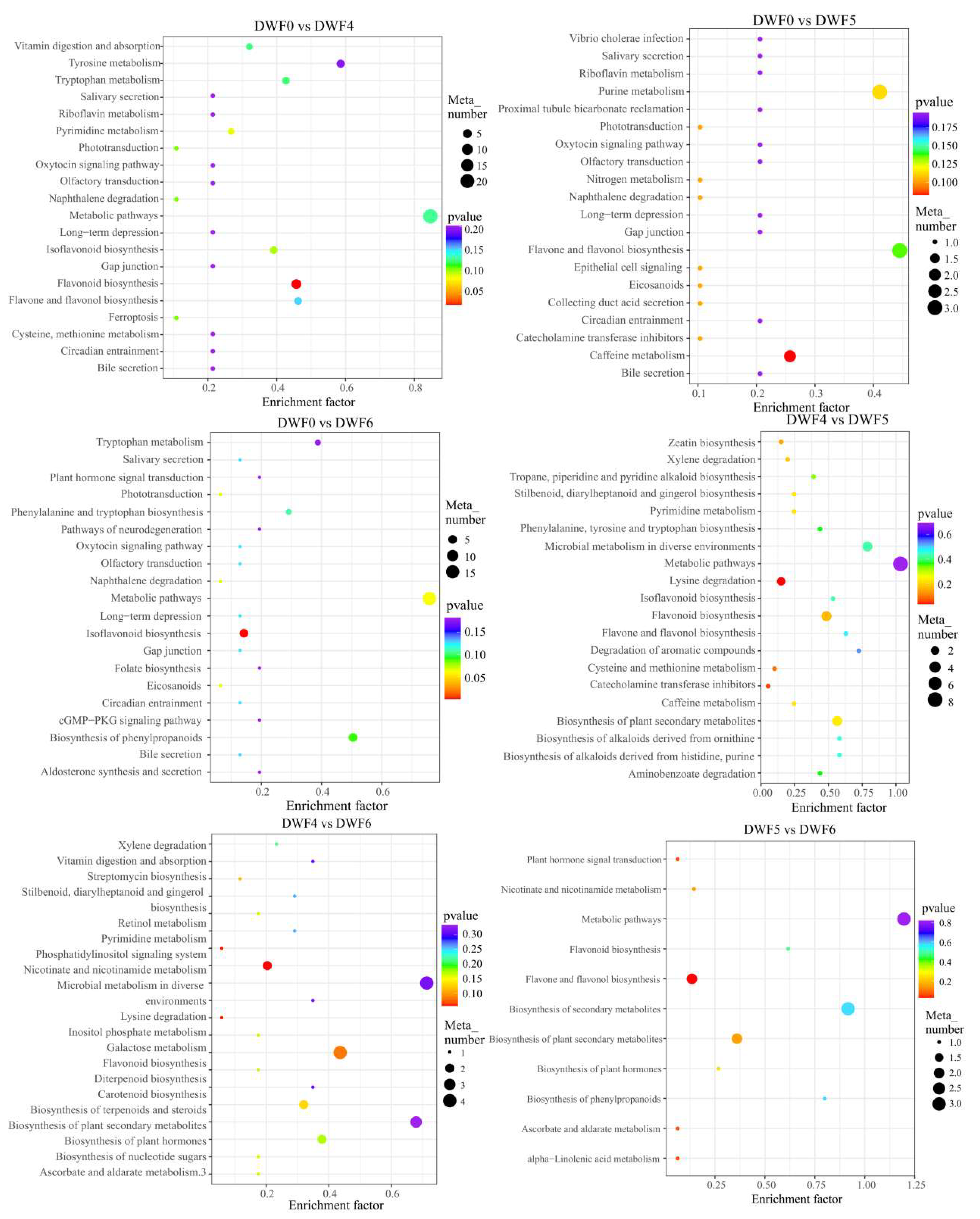
4.7. Pathway Mapping and Metabolite-to-Metabolite Network Visualization
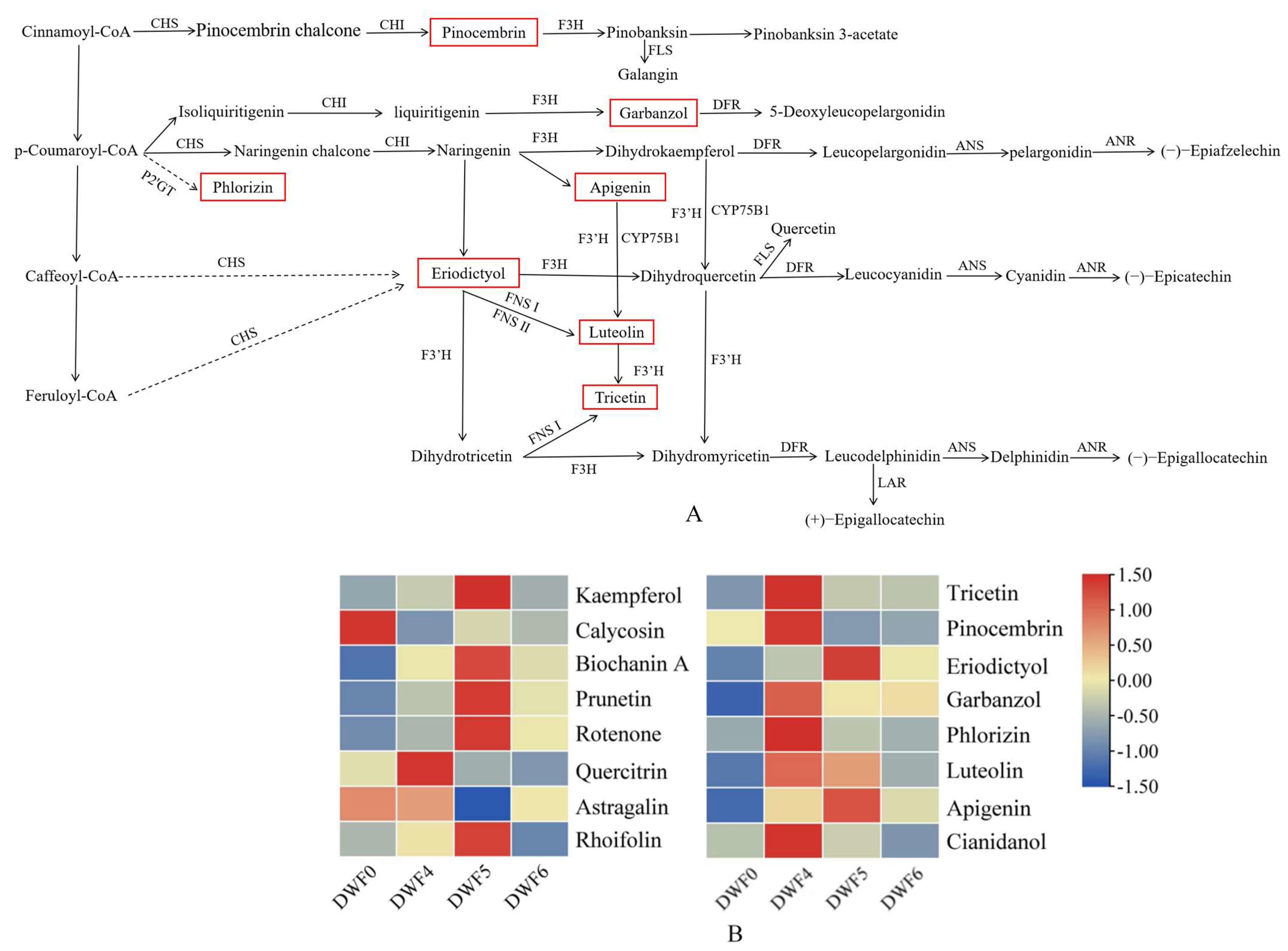
4.8. Cluster Analysis of Differential Flavonoid Metabolites
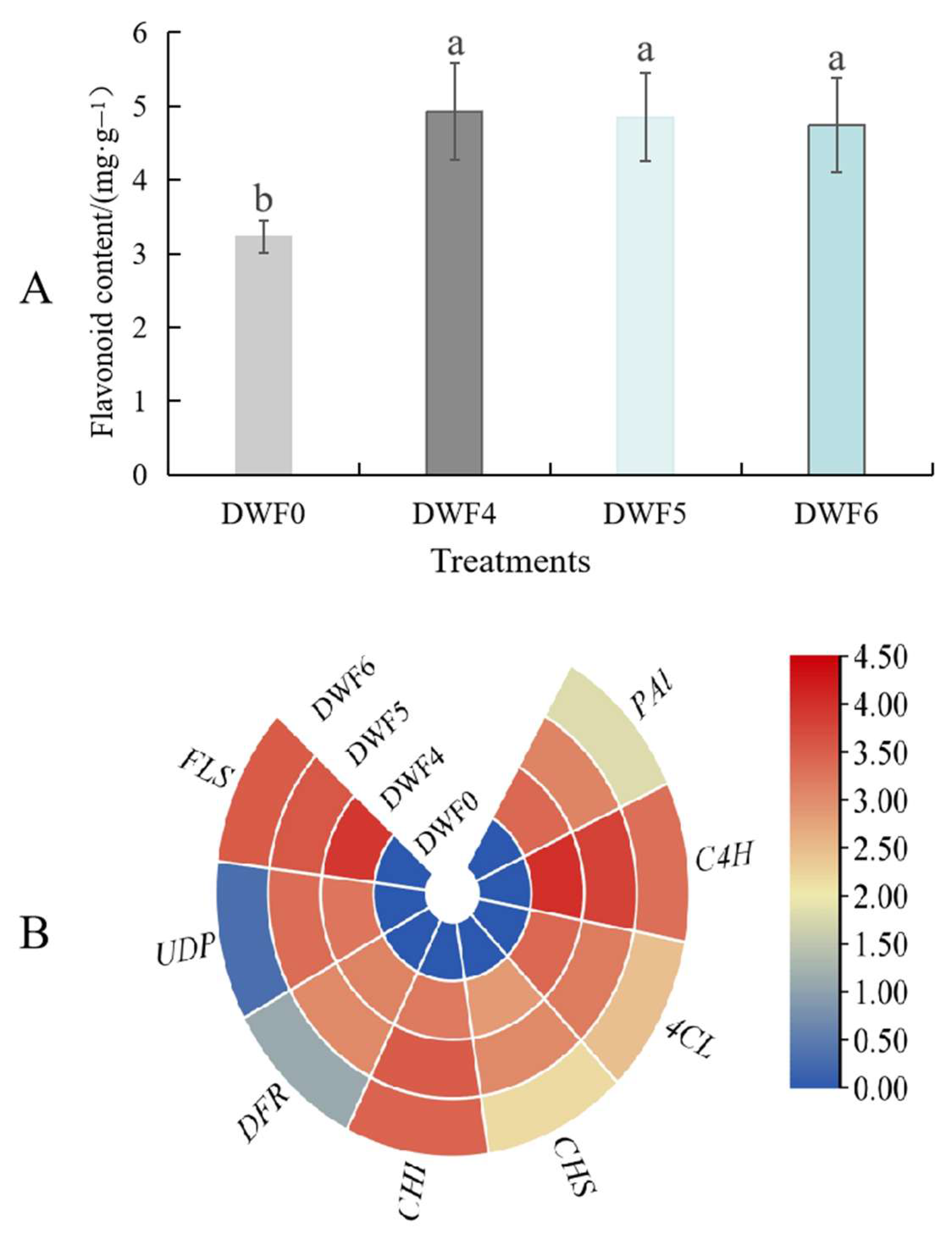
4.9. Effects of Different Fertilization Treatments on Flavonoid Content and Expression Levels of Key Genes in Hazelnut Leaves
5. Discussions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, J.; Wang, X.; Liu, H.; Lin, Z. Hazelnut and its by-products: A comprehensive review of nutrition, phytochemical profile, extraction, bioactivities and applications. Food Chem. 2023, 413, 135576. [Google Scholar] [PubMed]
- Chen, X.; Zhang, J.; Liu, Q.; Guo, W.; Zhao, T.; Ma, Q.; Wang, G. Transcriptome sequencing and identification of cold tolerance genes in hardy Corylus species (C. heterophylla Fisch) floral buds. PLoS ONE 2014, 9, e108604. [Google Scholar]
- Jakopic, J.; Petkovsek, M.M.; Likozar, A.; Solar, A.; Stampar, F.; Veberic, R. HPLC-MS identification of phenols in hazenut (Corylus avellana L.) kernels. Food Chem. 2011, 124, 1100–1106. [Google Scholar]
- Shaaban, M.M. Injection fertilization: A full nutritional technique for fruit trees saves 90–95% of fertilizers and maintains a clean environment. Fruit Veg. Cereal Sci. Biotechnol. 2009, 3, 22–27. [Google Scholar]
- Oztolan-Erol, N.; Helmstetter, A.J.; İnan, A.; Buggs, R.J.; Lucas, S.J. Unraveling genetic diversity amongst European hazelnut (Corylus avellana L.) varieties in Turkey. Front. Plant Sci. 2021, 12, 661274. [Google Scholar]
- Zinnanti, C.; Schimmenti, E.; Borsellino, V.; Paolini, G.; Severini, S. Economic performance and risk of farming systems specialized in perennial crops: An analysis of Italian hazelnut production. Agric. Syst. 2019, 176, 102645. [Google Scholar]
- Orlandi, F.; Ranfa, A.; Ruga, L.; Fornaciari, M. Hazelnut phenological phases and environmental effects in two central Italy areas. J. Agric. Meteorol. 2019, 75, 137–143. [Google Scholar]
- Silvestri, C.; Bacchetta, L.; Bellincontro, A.; Cristofori, V. Advances in cultivar choice, hazelnut orchard management, and nut storage to enhance product quality and safety: An overview. J. Sci. Food Agric. 2021, 101, 27–43. [Google Scholar]
- Thompson, K.R.; Chambers, D.H.; Chambers, E., IV. Sensory characteristics of ice cream produced in the USA and Italy. J. Sens. Stud. 2009, 24, 396–414. [Google Scholar]
- Sahu, K.C. The rise of Ferrero in India: The sweet truth. Int. J. Res. Financ. Mark. 2016, 6, 33–38. [Google Scholar]
- Fideghelli, C.; De Salvador, F.R. World hazelnut situation and perspectives. Vii Int. Congr. Hazelnut 2008, 845, 39–52. [Google Scholar]
- Özenç, N.; Bender Özenç, D.; Duyar, Ö. Nutritional composition of hazelnut (Corylus avellana L.) as influenced by basic fertilization. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2014, 64, 710–721. [Google Scholar]
- Özdemir, M.; Açkurt, F.; Kaplan, M.; Yıldız, M.; Löker, M.; Gürcan, T.; Biringen, G.; Okay, A.; Seyhan, F.G. Evaluation of new Turkish hybrid hazelnut (Corylus avellana L.) varieties: Fatty acid composition, a-tocopherol content, mineral composition and stability. Food Chem. 2001, 73, 411–415. [Google Scholar]
- Dündar, M.S.; Bahçıvan, E.; Muslu, C. Influence of variety and geographical region on mineral contents of hazelnut (Corylus avellana L.) varieties. Acta Chim. Slov. 2002, 49, 537–544. [Google Scholar]
- Amaral, J.S.; Casal, S.; Alves, M.R.; Seabra, R.M.; Oliveira, B.B.P. Tocopherol and tocotrienol content of hazelnut cultivars grown in Portugal. J. Agric. Food Chem. 2006, 54, 1329–1336. [Google Scholar] [PubMed]
- Seyhan, F.; Özay, G.; Saklar, S.; Ertaş, E.; Satır, G.; Alaşalvar, C. Chemical changes of three native Turkish hazelnut varieties (Corylus avellana L.) during fruit development. Food Chem. 2007, 105, 590–596. [Google Scholar]
- Köksal, A.İ.; Artık, N.; Şimşek, A.; Güneş, N. Nutrient composition of hazelnut (Corylus avellana L.) varieties cultivated in Turkey. Food Chem. 2006, 99, 509–515. [Google Scholar]
- Cosmulescu, S.; Botu, M.; Trandafır, I. The mineral source for human nutrition of nuts in different hazelnut (Corylus avellana L.) cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 250–254. [Google Scholar]
- Özkutlu, F.; Turan, M.; Kebapci, T.; Ete Aydemir, Ö.; Kocaman, A. Optimization of urea fertilization with various doses and applications on the yield and quality of the hazelnut. Appl. Fruit Sci. 2024, 26, 963–971. [Google Scholar]
- Özenç, N.; Bender Özenç, D. Effect of iron fertilization on nut traits and nutrient composition of ‘Tombul’ hazelnut (Corylus avellana L.) and its potential value for human nutrition. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2014, 64, 633–643. [Google Scholar]
- Lin, L.; Niu, Z.; Jiang, C.; Yu, L.; Wang, H.; Qiao, M. Infuences of open-central canopy on photosynthetic parameters and fruit quality of apples (Malus domestica) in the Loess Plateau of China. Hortic. Plant J. 2022, 8, 133–142. [Google Scholar]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar]
- Robotti, E.; Marengo, E. Chemometric Multivariate Tools for Candidate Biomarker Identification: LDA, PLS-DA, SIMCA, Ranking-PCA. Methods Protoc. 2016, 384, 237–267. [Google Scholar]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures(OPLS). J. Chemom. A J. Chemom. Soc. 2002, 16, 119–128. [Google Scholar]
- Sun, J.; Huang, D.; Zhang, Y.; Tao, J. Research progress of woody oil crops in China: A review. Seed Biol. 2024, 3, e008. [Google Scholar]
- Yang, J.Y.; Zheng, W.; Tian, Y.; Wu, Y.; Zhou, D.W. Effects of various mixed salt-alkaline stresses on growth, photosynthesis, and photosynthetic pigment concentrations of Medicago ruthenica seedlings. Photosynthetica 2011, 49, 275–284. [Google Scholar]
- Mahesh, R.; Raja, N.A. Effect of Water Soluble Fertilizers and Normal Fertilizers at Different Levels on Chlorophyll Content, Leaf Area Index and Cane Yield of Sugarcane Under Subsurface Drip Fertigation. Trends Biosci. 2015, 8, 1091–1094. [Google Scholar]
- Jia, X.M.; Wang, H.; Svetla, S.; Zhu, Y.F.; Hu, Y.; Cheng, L.; Wang, Y.X. Comparative physiological responses and adaptive strategies of apple Malus halliana to salt, alkali and saline-alkali stress. Sci. Hortic. 2019, 245, 154–162. [Google Scholar]
- Guo, X.; Li, S.; Wang, D.; Huang, Z.; Sarwar, N.; Mubeen, K.; Hussain, M. Effects of water and fertilizer coupling on the physiological characteristics and growth of rabbiteye blueberry. PLoS ONE 2021, 16, e0254013. [Google Scholar]
- Yang, Z.; Tian, J.; Feng, K.; Gong, X.; Liu, J. Application of a hyperspectral imaging system to quantify leaf-scale chlorophyll, nitrogen and chlorophyll fluorescence parameters in grapevine. Plant Physiol. Biochem. 2021, 166, 723–737. [Google Scholar]
- Muguruma, Y.; Tsutsui, H.; Noda, T.; Akatsu, H.; Inoue, K. Widely targeted metabolomics of Alzheimer’s disease postmortem cerebrospinal fluid based on 9-fluorenylmethyl chloroformate derivatized ultra-high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. B 2018, 1091, 53–66. [Google Scholar]
- Nisar, M.F.; Wan, C.; Manzoor, Z.; Waqas, Y.; Niaz, K.; Ayaz, M. Glycosidic derivatives of flavonoids. Phytochem. Classif. Glycosidic Deriv. Flavonoids 2020, 3, 57–84. [Google Scholar]
- Zhuang, W.B.; Li, Y.H.; Shu, X.C.; Pu, Y.T.; Wang, X.J.; Wang, T.; Wang, Z. The classification, molecular structure and biological biosynthesis of flavonoids, and their roles in biotic and abiotic stresses. Molecules 2023, 28, 3599. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Jian, H.; Xue, L.; Wang, Y.; Cao, Y.; Wan, M.; Lv, D.; Li, J.; Lu, K.; Xu, X.; Liu, L. Characterization of cold stress responses in different rape seed ecotypes based on metabolomics and transcriptomics analyses. PeerJ 2020, 8, e8704. [Google Scholar]
- Zhang, H.; Tao, H.; Yang, H.; Zhang, L.; Feng, G.; An, Y.; Wang, L. MdSCL8 as a negative regulator participates in ALA-I nduced FLS1 to promote flavonol accumulation in apples. Int. J. Mol. Sci. 2022, 23, 2033. [Google Scholar] [PubMed]
- Watkins, J.M.; Chapman, J.M.; Muday, G.K. Abscisic acid-induced reactive oxygen specie s are modulated by flavonols to control stomata aperture. Plant Physiol. 2017, 175, 1807–1825. [Google Scholar]
- Lelli, V.; Molinari, R.; Merendino, N.; Timperio, A.M. Detection and comparison of bioactive compounds in different extracts of two hazelnut skin varieties, tonda gentile romana and tonda di giffoni, using a metabolomics approach. Metabolites 2021, 11, 296. [Google Scholar] [CrossRef]
- Chang, P.Y.; Hsieh, M.J.; Hsieh, Y.S.; Chen, P.N.; Yang, J.S.; Lo, F.C.; Yang, S.F.; Lu, K.H. Tricetin inhibits human osteosarcoma cells metastasis by transcriptionally repressing MMP-9 via p38 and Akt pathways. Environ. Toxicol. 2017, 32, 2032–2040. [Google Scholar] [CrossRef] [PubMed]
- Chao, R.; Chow, J.M.; Hsieh, Y.H.; Chen, C.K.; Lee, W.J.; Hsieh, F.K.; Yu, N.Y.; Chou, M.C.; Cheng, C.W.; Yang, S.F.; et al. Tricetin suppresses the migration/invasion of human glioblastoma multiforme cells by inhibiting matrix metalloproteinase-2 through modulation of the expression and transcriptional activity of specificity protein 1. Expert Opin. Ther. Targets 2015, 19, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Uen, Y.H.; Chen, Y.; Liang, H.L.; Kuo, P.L. Tricetin, a dietary flavonoid, inhibits proliferation of human breast adenocarcinoma mcf-7 cells by blocking cell cycle progression and inducing apoptosis. J. Agric. Food Chem. 2009, 57, 8688–8695. [Google Scholar] [CrossRef]
- Grzelak-Błaszczyk, K.; Milala, J.; Kosmala, M.; Kołodziejczyk, K.; Sójka, M.; Czarnecki, A.; Klewicki, R.; Juśkiewicz, J.; Fotschki, B.; Jurgoński, A. Onion quercetin monoglycosides alter microbial activity and increase antioxidant capacity. J. Nutr. Biochem. 2018, 56, 81–88. [Google Scholar] [CrossRef]
- Kumar, R.; Vijayalakshmi, S.; Nadanasabapathi, S. Health benefits of quercetin. Def. Life Sci. J. 2017, 2, 142–151. [Google Scholar] [CrossRef]
- Takahashi, S.; Muguruma, H.; Osakabe, N.; Inoue, H.; Ohsawa, T. Electrochemical determination with a long-length carbon nanotube electrode of quercetin glucosides in onion, apple peel, and tartary buckwheat. Food Chem. 2019, 300, 125189. [Google Scholar] [CrossRef]
- Falla, N.M.; Demasi, S.; Caser, M.; Scariot, V. Phytochemical profile and antioxidant properties of italian green tea, a new high quality niche product. Horticulturae 2021, 7, 91. [Google Scholar] [CrossRef]
- Luciano, A.; Picariello, L.; Forino, M.; Moio, L.; Gambuti, A. Anthocyanins and nucleation seeds are key factors affecting quercetin precipitation in red wines. J. Sci. Food Agric. 2024, 104, 5163–5175. [Google Scholar] [CrossRef]
- Chen, Q.; Gu, Y.; Tan, C.; Sundararajan, B.; Li, Z.; Wang, D.; Zhou, Z. Comparative effects of five polymethoxyflavones purified from Citrus tangerina on inflammation and cancer. Front. Nutr. 2022, 9, 963662. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Q.; Wang, T.; Liu, J.; Chen, D. Comparison of the antioxidant effects of quercitrin and isoquercitrin: Understanding the role of the 6″-OH group. Molecules 2016, 21, 1246. [Google Scholar] [CrossRef]
| Group | Fertilization Treatment (g/L): A + B + C | Group | Fertilization Treatment (g/L): A + B + C |
|---|---|---|---|
| DWF0 | 0 +0 + 0 | DWF5 | 1 + 1 + 0.5 |
| DWF1 | 1 + 0.1 + 0.5 | DWF6 | 1 + 1.5 + 0.5 |
| DWF2 | 1 + 0.2 + 0.5 | DWF7 | 1 + 2 + 0.5 |
| DWF3 | 1 + 0.4 + 0.5 | DWF8 | 1 + 2.5 + 0.5 |
| DWF4 | 1 + 0.5 + 0.5 |
| Gene Name | Primer Sequence (5′-3′) | |
|---|---|---|
| Forward Primer | Reverse Primer | |
| P-coumaroyl coenzyme A ligase (4CL) | TTAGCAAGCTCGGAATTCGGAAGG | CTCGGCGGCGGTGAAGAAAG |
| Flavonol synthase (FLS) | GCGTGAGGTGGCGGATAAACTG | GCGTTAGATCAGGCTGAGGACATG |
| Uridine diphosphate (UDP) | TGTTTCCGAATCTCCTGCCCATTG | ACCGAGCGAGTTGCTTGTTGATC |
| Dihydroflavonol 4-reductase (DFR) | GCTCTGCGTGCGATGCTACC | GCCTCCAAGTTCTCGTCCACATC |
| Chalcone isomerase (CHI) | GGCTCCACCAATACCTTGTTCCTC | ACGGCGGTATCCTCCAAGTACAC |
| Chalcone synthase (CHS) | AGACATAGTGGTGGCGGAGGTG | CGAGGTGGGTGATCTTGGATTTGG |
| Cinnamate 4-hydroxylase (C4H) | GTGTACGGTGAGCATTGGAGGAAG | CGCCTCTGGGTTCTTCTTCACATC |
| Phenylalanine ammonia-lyase (PAL) | CATATCCCAGGTGGCAGCGATTG | CGCCCTGCTTGGTGCTCTTG |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | TGAGGGCAAGGTGAAGGGTATCTT | TCAAGTCAACCACACGCGTACTGT |
| Treatments | Metabolites | VIP | p-Value | Fold Change |
|---|---|---|---|---|
| DWF0 vs. DWF4 | Diosmetin | 1.72376 | 1.69 × 10−4 | 6.56 × 100 |
| Pinobanksin | 1.72496 | 8.58 × 10−4 | 2.46 × 100 | |
| Telmisartan | 1.68743 | 2.15 × 10−3 | 4.21 × 100 | |
| Dalbergioidin | 1.69662 | 2.54 × 10−3 | 3.82 × 100 | |
| 2-Aminomuconate semialdehyde | 1.68287 | 2.65 × 10−3 | 1.96 × 100 | |
| Cinnamyl cinnamate | 1.67804 | 3.22 × 10−3 | 3.04 × 100 | |
| Alpha-Santonin | 1.7137 | 3.29 × 10−3 | 4.91 × 100 | |
| alpha-Tocopherol | 1.67998 | 4.38 × 10−3 | 5.92 × 10−1 | |
| 2-(4-Hydroxyphenyl)ethanol | 1.71552 | 4.46 × 10−3 | 8.00 × 100 | |
| Riboflavin | 1.6886 | 4.98 × 10−3 | 2.66 × 100 | |
| Phlorizin | 1.66599 | 4.98 × 10−3 | 2.74 × 100 | |
| Xanthosine | 1.70745 | 5.14 × 10−3 | 1.27 × 101 | |
| Pelargonidin-3-O-glucoside | 1.64755 | 6.03 × 10−3 | 4.03 × 100 | |
| trans-3,5-Dimethoxy-4-hydroxy cinnamaldehyde | 1.62291 | 6.59 × 10−3 | 2.89 × 100 | |
| 3,4-Dihydrocoumarin | 1.64793 | 6.91 × 10−3 | 1.57 × 100 | |
| DWF0 vs. DWF5 | Butein | 1.71868 | 6.14 × 10−5 | 3.96 × 100 |
| Fustin | 1.72626 | 1.04 × 10−4 | 6.55 × 100 | |
| 3-(2-Hydroxyphenyl)-2-propenal | 1.70069 | 6.32 × 10−4 | 2.77 × 10−1 | |
| Benzocaine | 1.69823 | 8.81 × 10−4 | 2.36 × 100 | |
| Beta-mangostin | 1.70101 | 1.45 × 10−3 | 6.29 × 100 | |
| 3-Hydroxycoumarin | 1.70265 | 1.74 × 10−3 | 1.25 × 10−1 | |
| Alpha-Santonin | 1.70075 | 2.53 × 10−3 | 3.83 × 100 | |
| Guanosine 3′,5′-cyclic monophosphate | 1.67239 | 3.40 × 10−3 | 5.20 × 100 | |
| Astragalin | 1.66927 | 3.45 × 10−3 | 4.27 × 10−1 | |
| Anabasine | 1.65347 | 3.88 × 10−3 | 2.56 × 100 | |
| Peonidin-3-glucoside | 1.65909 | 4.01 × 10−3 | 3.03 × 100 | |
| Methyl cinnamate | 1.66713 | 4.44 × 10−3 | 3.73 × 10−1 | |
| Agrocybenine | 1.71853 | 4.51 × 10−3 | 3.18 × 101 | |
| Epicatechin; (+)-Epicatechin | 1.64442 | 5.34 × 10−3 | 7.18 × 10−1 | |
| Benzoic acid | 1.65635 | 5.79 × 10−3 | 3.60 × 10−1 | |
| DWF0 vs. DWF6 | Biochanin A | 1.78505 | 5.99 × 10−4 | 4.95 × 100 |
| 3-(2-Hydroxyphenyl)-2-propenal | 1.76743 | 6.69 × 10−4 | 2.56 × 10−1 | |
| Benzocaine | 1.75314 | 1.02 × 10−3 | 2.36 × 100 | |
| (+)-Abscisic acid | 1.7912 | 1.48 × 10−3 | 2.76 × 100 | |
| Prunetin | 1.74471 | 1.76 × 10−3 | 3.48 × 100 | |
| Sinalbin | 1.77845 | 2.02 × 10−3 | 4.61 × 100 | |
| Diosmetin | 1.7488 | 2.2 × 10−3 | 5.32 × 100 | |
| Alpha-Santonin | 1.73205 | 3.51 × 10−3 | 2.76 × 100 | |
| 18alpha-Glycyrrhetinic acid | 1.72805 | 4.10 × 10−3 | 2.09 × 100 | |
| Guanosine 3′,5′-cyclic monophosphate | 1.72264 | 5.96 × 10−3 | 4.86 × 100 | |
| Rotenone | 1.71587 | 6.63 × 10−3 | 2.00 × 100 | |
| 3-Hydroxycoumarin | 1.77257 | 7.33 × 10−3 | 2.27 × 10−1 | |
| Albiflorin | 1.70167 | 7.47 × 10−3 | 3.61 × 10−1 | |
| Myricitrin | 1.75625 | 8.05 × 10−3 | 4.11 × 10−1 | |
| Methyl cinnamate | 1.72535 | 8.50 × 10−3 | 4.50 × 10−1 | |
| DWF4 vs. DWF5 | Myricitrin | 1.81102 | 5.22 × 10−5 | 3.22 × 10−1 |
| Acevaltrate | 1.79949 | 3.63 × 10−4 | 3.62 × 10−1 | |
| Methyl cinnamate | 1.77422 | 1.58 × 10−3 | 4.20 × 10−1 | |
| Allura Red AC | 1.76653 | 1.65 × 10−3 | 4.20 × 10−1 | |
| Quercitrin | 1.75775 | 3.28 × 10−3 | 7.12 × 10−1 | |
| Benzoic acid | 1.76006 | 4.08 × 10−3 | 3.98 × 10−1 | |
| Nalidixic Acid | 1.75741 | 5.26 × 10−3 | 3.87 × 10−1 | |
| Methyl rosmarinate | 1.71246 | 5.41 × 10−3 | 6.00 × 10−1 | |
| Beta-mangostin | 1.76631 | 6.23 × 10−3 | 2.54 × 100 | |
| trans-Zeatin-riboside | 1.67633 | 9.62 × 10−3 | 6.11 × 10−1 | |
| Okanin | 1.72165 | 1.03 × 10−2 | 5.96 × 10−1 | |
| Phloretic acid; Ethylparaben; 3-(2-Hydroxyphenyl)propanoic acid | 1.78558 | 1.15 × 10−2 | 1.05 × 10−1 | |
| Dalbergioidin | 1.71805 | 1.26 × 10−2 | 4.51 × 10−1 | |
| Albiflorin | 1.68098 | 1.29 × 10−2 | 5.10 × 10−1 | |
| 6(1H)-Azulenone, 2,3-dihydro-1,4-dimethyl | 1.65178 | 1.45 × 10−2 | 4.74 × 10−1 | |
| DWF4 vs. DWF6 | Myricitrin | 1.72507 | 6.98 × 10−5 | 2.93 × 10−1 |
| Acevaltrate | 1.71887 | 7.38 × 10−5 | 3.46 × 10−1 | |
| Fisetin | 1.70209 | 1.27 × 10−3 | 5.30 × 10−1 | |
| Quercitrin | 1.68405 | 1.92 × 10−3 | 6.79 × 10−1 | |
| Methyl cinnamate | 1.66177 | 2.52 × 10−3 | 5.06 × 10−1 | |
| Gamabufotalin; Telocinobufagin | 1.66111 | 3.99 × 10−3 | 4.03 × 10−1 | |
| 2′-Deoxyuridine; Deoxyuridine | 1.64943 | 4.14 × 10−3 | 3.23 × 10−1 | |
| Glycyrrhetic acid 3-O-mono-beta-D-glucuronide | 1.64974 | 4.29 × 10−3 | 3.16 × 10−1 | |
| Albiflorin | 1.65853 | 5.41 × 10−3 | 3.76 × 10−1 | |
| Procyanidin B2 | 1.6236 | 6.20 × 10−3 | 3.24 × 10−1 | |
| (R)-Menthofuran | 1.63394 | 6.48 × 10−3 | 6.28 × 10−1 | |
| Cianidanol | 1.65171 | 6.50 × 10−3 | 4.68 × 10−1 | |
| Eurycomalactone; Ingenol | 1.64399 | 6.73 × 10−3 | 6.13 × 10−1 | |
| Allura Red AC | 1.64917 | 7.66 × 10−3 | 2.64 × 10−1 | |
| trans-Zeatin-riboside | 1.61102 | 8.19 × 10−3 | 6.18 × 10−1 | |
| DWF5 vs. DWF6 | Linderane | 1.82323 | 2.06 × 10−3 | 2.28 × 100 |
| 1-Naphthylacetic acid | 1.78242 | 3.38 × 10−3 | 3.39 × 10−1 | |
| Silibinin | 1.79779 | 5.15 × 10−3 | 2.74 × 10−1 | |
| 1H-Indole-2,3-dione | 1.77139 | 5.71 × 10−3 | 5.28 × 10−1 | |
| (±)-Jasmonic acid | 1.73667 | 6.82 × 10−3 | 4.00 × 10−1 | |
| Cleomiscosin A | 1.71992 | 1.39 × 10−2 | 6.35 × 10−1 | |
| (−)-Borneol; (+)-Borneol | 1.80678 | 1.57 × 10−2 | 5.51× 10−2 | |
| Narcissoside | 1.69806 | 1.73 × 10−2 | 3.79 × 10−1 | |
| Terpinine-4-ol | 1.73342 | 1.87 × 10−2 | 1.66 × 100 | |
| Neogrifolin; Grifolin; Ugaxanthone | 1.66903 | 1.99 × 10−2 | 6.04 × 10−1 | |
| Cyanidin-3-O-rutinoside chloride | 1.68873 | 2.01 × 10−2 | 3.97 × 10−1 | |
| L-Gulose | 1.70231 | 2.08 × 10−2 | 4.39 × 10−1 | |
| Rhoifolin | 1.73982 | 2.16 × 10−2 | 3.20 × 10−1 | |
| Diosmetin | 1.74586 | 2.27 × 10−2 | 4.44 × 10−1 | |
| Liquiritin | 1.65423 | 2.35 × 10−2 | 4.12 × 10−1 |
| Treatments | ID Annotation | Annotation | p-Value | −log(p-Value) |
|---|---|---|---|---|
| DWF0 vs. DWF4 | map00941 | Flavonoid biosynthesis | 2.67 × 10−2 | 1.57 × 100 |
| map00240 | Pyrimidine metabolism | 8.91 × 10−2 | 1.05 × 100 | |
| map00943 | Isoflavonoid biosynthesis | 9.94 × 10−2 | 1.00 × 100 | |
| map00626 | Naphthalene degradation | 1.06 × 10−1 | 9.74 × 10−1 | |
| map04744 | Phototransduction | 1.06 × 10−1 | 9.74 × 10−1 | |
| map04216 | Ferroptosis | 1.06 × 10−1 | 9.74 × 10−1 | |
| map00380 | Tryptophan metabolism | 1.23 × 10−1 | 9.10 × 10−1 | |
| map04977 | Vitamin digestion and absorption | 1.25 × 10−1 | 9.03 × 10−1 | |
| map01100 | Metabolic pathways | 1.28 × 10−1 | 8.93 × 10−1 | |
| map00944 | Flavone and flavonol biosynthesis | 1.49 × 10−1 | 8.28 × 10−1 | |
| DWF0 vs. DWF5 | map00232 | Caffeine metabolism | 8.35 × 10−2 | 1.08 × 100 |
| map07216 | Catecholamine transferase inhibitors | 1.02 × 10−1 | 9.90 × 10−1 | |
| map04966 | Collecting duct acid secretion | 1.02 × 10−1 | 9.90 × 10−1 | |
| map05120 | Epithelial cell signaling in Helicobacter pylori infection | 1.02 × 10−1 | 9.90 × 10−1 | |
| map07034 | Eicosanoids | 1.02 × 10−1 | 9.90 × 10−1 | |
| map00626 | Naphthalene degradation | 1.02 × 10−1 | 9.90 × 10−1 | |
| map00910 | Nitrogen metabolism | 1.02 × 10−1 | 9.90 × 10−1 | |
| map04744 | Phototransduction | 1.02 × 10−1 | 9.90 × 10−1 | |
| map00230 | Purine metabolism | 1.13 × 10−1 | 9.47 × 10−1 | |
| map00944 | Flavone and flavonol biosynthesis | 1.37 × 10−1 | 8.64 × 10−1 | |
| DWF0 vs. DWF6 | map00943 | Isoflavonoid biosynthesis | 2.37 × 10−4 | 3.63 × 100 |
| map01100 | Metabolic pathways | 6.17 × 10−2 | 1.21 × 100 | |
| map07034 | Eicosanoids | 6.46 × 10−2 | 1.19 × 100 | |
| map00626 | Naphthalene degradation | 6.46 × 10−2 | 1.19 × 100 | |
| map04744 | Phototransduction | 6.46 × 10−2 | 1.19 × 100 | |
| map01061 | Biosynthesis of phenylpropanoids | 8.98 × 10−2 | 1.05 × 100 | |
| map00400 | Phenylalanine, tyrosine, and tryptophan biosynthesis | 1.09 × 10−1 | 9.64 × 10−1 | |
| map04713 | Circadian entrainment | 1.25 × 10−1 | 9.02 × 10−1 | |
| map04976 | Bile secretion | 1.25 × 10−1 | 9.02 × 10−1 | |
| map04921 | Oxytocin signaling pathway | 1.25 × 10−1 | 9.02 × 10−1 | |
| DWF4 vs. DWF5 | map00310 | Lysine degradation | 2.87 × 10−2 | 1.54 × 100 |
| map07216 | Catecholamine transferase inhibitors | 4.79 × 10−2 | 1.32 × 100 | |
| map00270 | Cysteine and methionine metabolism | 9.37 × 10−2 | 1.03 × 100 | |
| map00908 | Zeatin biosynthesis | 1.37 × 10−1 | 8.62 × 10−1 | |
| map00941 | Flavonoid biosynthesis | 1.64 × 10−1 | 7.86 × 10−1 | |
| map00622 | Xylene degradation | 1.79 × 10−1 | 7.46 × 10−1 | |
| map00240 | Pyrimidine metabolism | 2.19 × 10−1 | 6.59 × 10−1 | |
| map00945 | Stilbenoid, diarylheptanoid, and gingerol biosynthesis | 2.19 × 10−1 | 6.59 × 10−1 | |
| map00232 | Caffeine metabolism | 2.19 × 10−1 | 6.59 × 10−1 | |
| map01060 | Biosynthesis of plant secondary metabolites | 2.28 × 10−1 | 6.42 × 10−1 | |
| DWF4 vs. DWF6 | map00760 | Nicotinate and nicotinamide metabolism | 5.64 × 10−2 | 1.25 × 100 |
| map00562 | Inositol phosphate metabolism | 5.82 × 10−2 | 1.23 × 100 | |
| map04070 | Phosphatidylinositol signaling system | 5.82 × 10−2 | 1.23 × 100 | |
| map00941 | Flavonoid biosynthesis | 8.33 × 10−2 | 1.08 × 100 | |
| map00521 | Streptomycin biosynthesis | 1.13 × 10−1 | 9.46 × 10−1 | |
| map01062 | Biosynthesis of terpenoids and steroids | 1.29 × 10−1 | 8.90 × 10−1 | |
| map00052 | Galactose metabolism | 1.65 × 10−1 | 7.82 × 10−1 | |
| map01250 | Biosynthesis of nucleotide sugars | 1.65 × 10−1 | 7.82 × 10−1 | |
| map00053 | Ascorbate and aldarate metabolism | 1.65 × 10−1 | 7.82 × 10−1 | |
| map00830 | Retinol metabolism | 1.65 × 10−1 | 7.82 × 10−1 | |
| DWF5 vs. DWF6 | map00944 | Flavone and flavonol biosynthesis | 2.47 × 10−2 | 1.61 × 100 |
| map00592 | alpha-Linolenic acid metabolism | 6.04 × 10−2 | 1.22 × 100 | |
| map04075 | Plant hormone signal transduction | 6.04 × 10−2 | 1.22 × 100 | |
| map00053 | Ascorbate and aldarate metabolism | 6.04 × 10−2 | 1.22 × 100 | |
| map00760 | Nicotinate and nicotinamide metabolism | 1.36 × 10−1 | 8.66 × 10−1 | |
| map01060 | Biosynthesis of plant secondary metabolites | 1.53 × 10−1 | 8.15 × 10−1 | |
| map01070 | Biosynthesis of plant hormones | 2.40 × 10−1 | 6.19 × 10−1 | |
| map00941 | Flavonoid biosynthesis | 4.80 × 10−1 | 3.19 × 10−1 | |
| map01110 | Biosynthesis of secondary metabolites | 5.74 × 10−1 | 2.41 × 10−1 | |
| map01061 | Biosynthesis of phenylpropanoids | 5.79 × 10−1 | 2.37 × 10−1 |
| Treatments | Metabolic Pathway | ||
|---|---|---|---|
| Flavonoid Biosynthesis | Isoflavonoid Biosynthesis | Flavone and Flavonol Biosynthesis | |
| DWF0 vs. DWF4 | Tricetin Pinocembrin Eriodictyol Garbanzol Phlorizin Luteolin Apigenin | Calycosin Biochanin A Apigenin | Quercitrin Luteolin Apigenin |
| DWF0 vs. DWF5 | Eriodictyol Garbanzol Apigenin | Prunetin Apigenin | Astragalin Rhoifolin Apigenin |
| DWF0 vs. DWF6 | Eriodictyol Apigenin | Calycosin Rotenone Prunetin Biochanin A Apigenin | Apigenin |
| DWF4 vs. DWF5 | Tricetin Pinocembrin Cianidanol | Prunetin | Quercitrin |
| DWF4 vs. DWF6 | Tricetin Pinocembrin Cianidanol Phlorizin | Quercitrin | |
| DWF5 vs. DWF6 | Kaempferol | Rhoifolin Kaempferol | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Ma, C.; Zhao, M.; Liu, Z.; Zhang, D.; Lu, J.; Hao, J.; Wu, L. Broad Target Metabolomics Revealed the Key Regulatory Mechanisms of the Effects of Trace Element Water-Soluble Fertilizer on the Growth of Corylus heterophylla × Corylus avellana Seedlings. Genes 2025, 16, 373. https://doi.org/10.3390/genes16040373
Chen W, Ma C, Zhao M, Liu Z, Zhang D, Lu J, Hao J, Wu L. Broad Target Metabolomics Revealed the Key Regulatory Mechanisms of the Effects of Trace Element Water-Soluble Fertilizer on the Growth of Corylus heterophylla × Corylus avellana Seedlings. Genes. 2025; 16(4):373. https://doi.org/10.3390/genes16040373
Chicago/Turabian StyleChen, Weiqing, Chao Ma, Mengjiong Zhao, Zaiguo Liu, De Zhang, Juan Lu, Jing Hao, and Lei Wu. 2025. "Broad Target Metabolomics Revealed the Key Regulatory Mechanisms of the Effects of Trace Element Water-Soluble Fertilizer on the Growth of Corylus heterophylla × Corylus avellana Seedlings" Genes 16, no. 4: 373. https://doi.org/10.3390/genes16040373
APA StyleChen, W., Ma, C., Zhao, M., Liu, Z., Zhang, D., Lu, J., Hao, J., & Wu, L. (2025). Broad Target Metabolomics Revealed the Key Regulatory Mechanisms of the Effects of Trace Element Water-Soluble Fertilizer on the Growth of Corylus heterophylla × Corylus avellana Seedlings. Genes, 16(4), 373. https://doi.org/10.3390/genes16040373





