Genome-Wide Identification of the BREVIS RADIX Gene Family in Foxtail Millet: Function, Evolution, and Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Analysis of BRX Family Genes in Millet
2.2. Analysis of Phylogeny and Chromosomal Positioning of BRX Family Genes
2.3. Analysis of Conserved Domains and Motifs
2.4. Cis-Acting Regulatory Elements and miRNA Prediction
2.5. Synteny Analysis and Calculation of Nonsynonymous to Synonymous Substitution Ratios
2.6. Forecasting the Three-Dimensional Structure of the SiBRX Protein
2.7. Expression Profiling of SiBRX Genes
2.8. Plant Resources and Treatment Approaches
2.9. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
2.10. Statistical Analyses
3. Results
3.1. Identification of the BRX Gene Family in Foxtail Millet and Green Foxtail
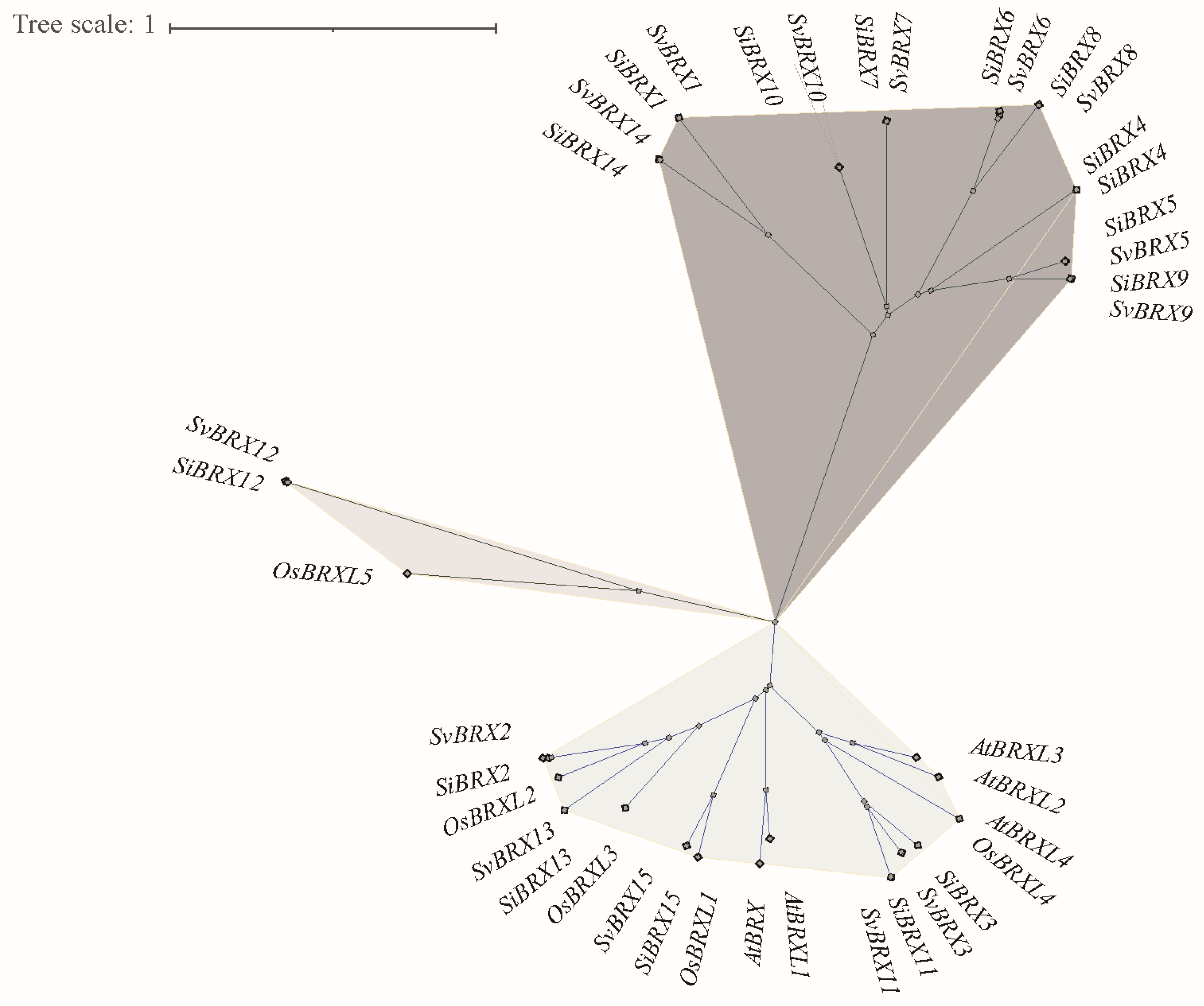
3.2. Phylogenetic Analysis of the BRX Gene Family
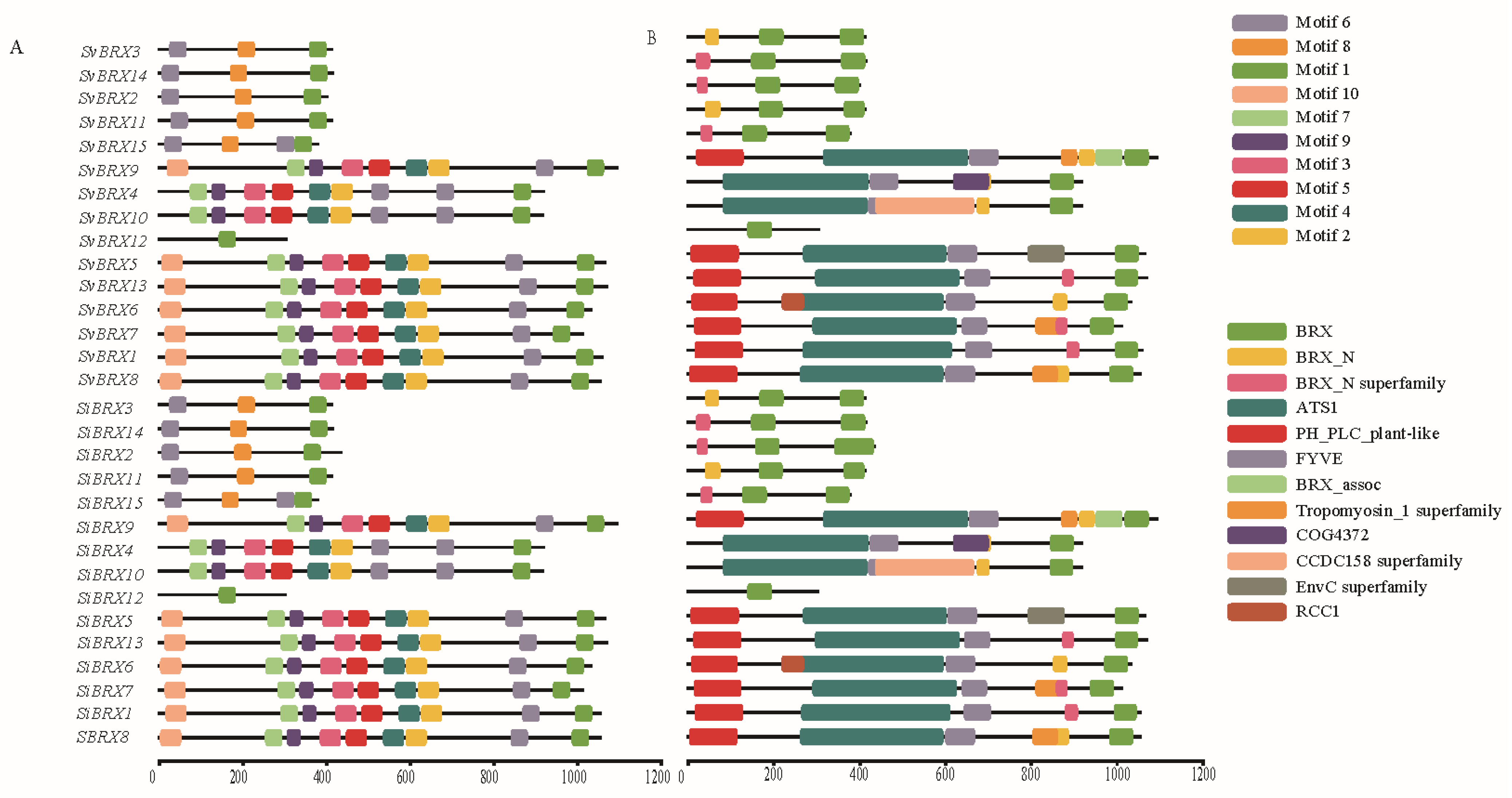
3.3. Conserved Motif and Domain Analyses of SiBRX Genes
3.4. Estimating the 3D Structures of SiBRX Proteins
3.5. miRNA Prediction
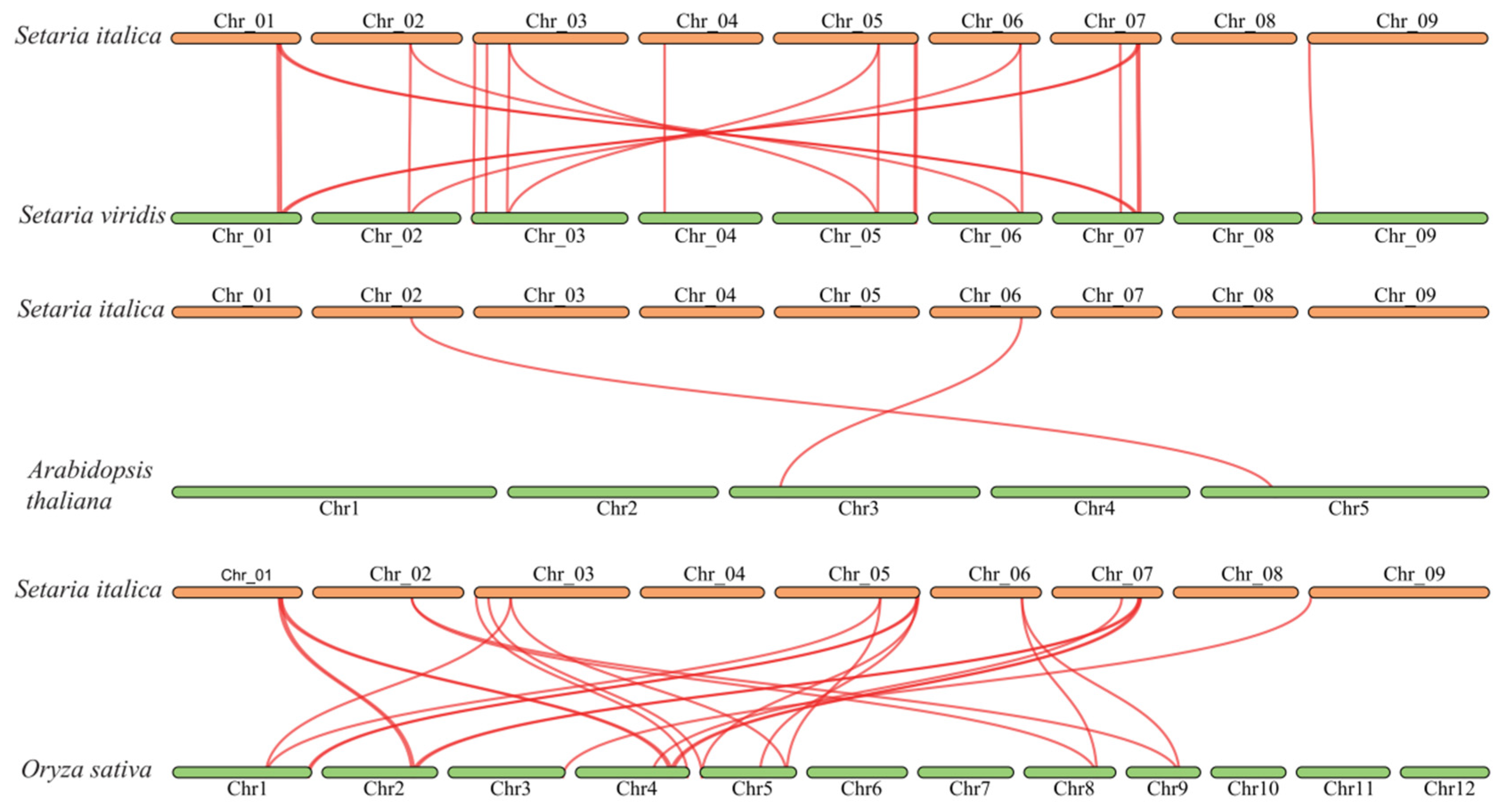
3.6. Gene Duplication and Collinearity Analysis of SiBRX Genes
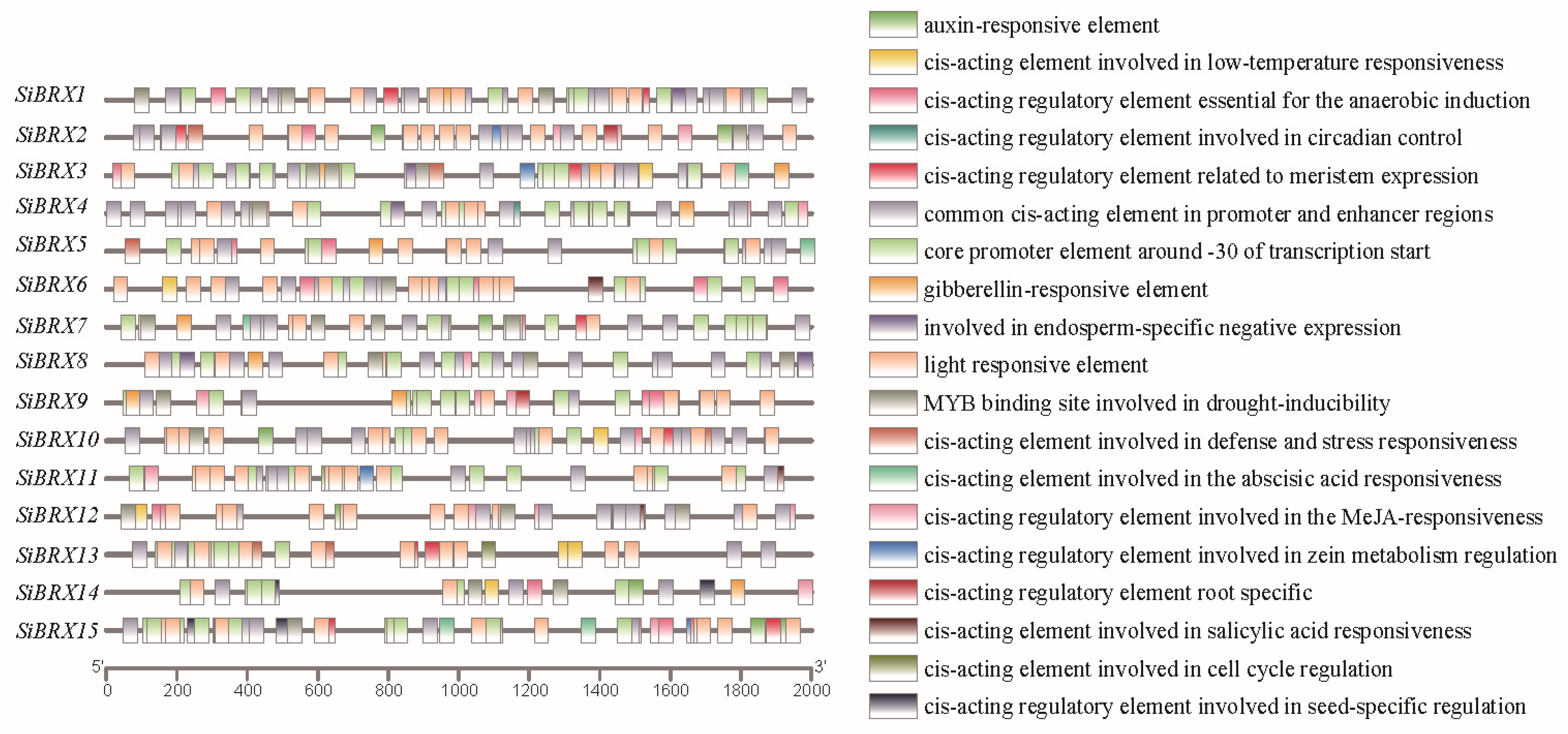
3.7. Analysis of Cis-Regulatory Elements in SiBRX Genes
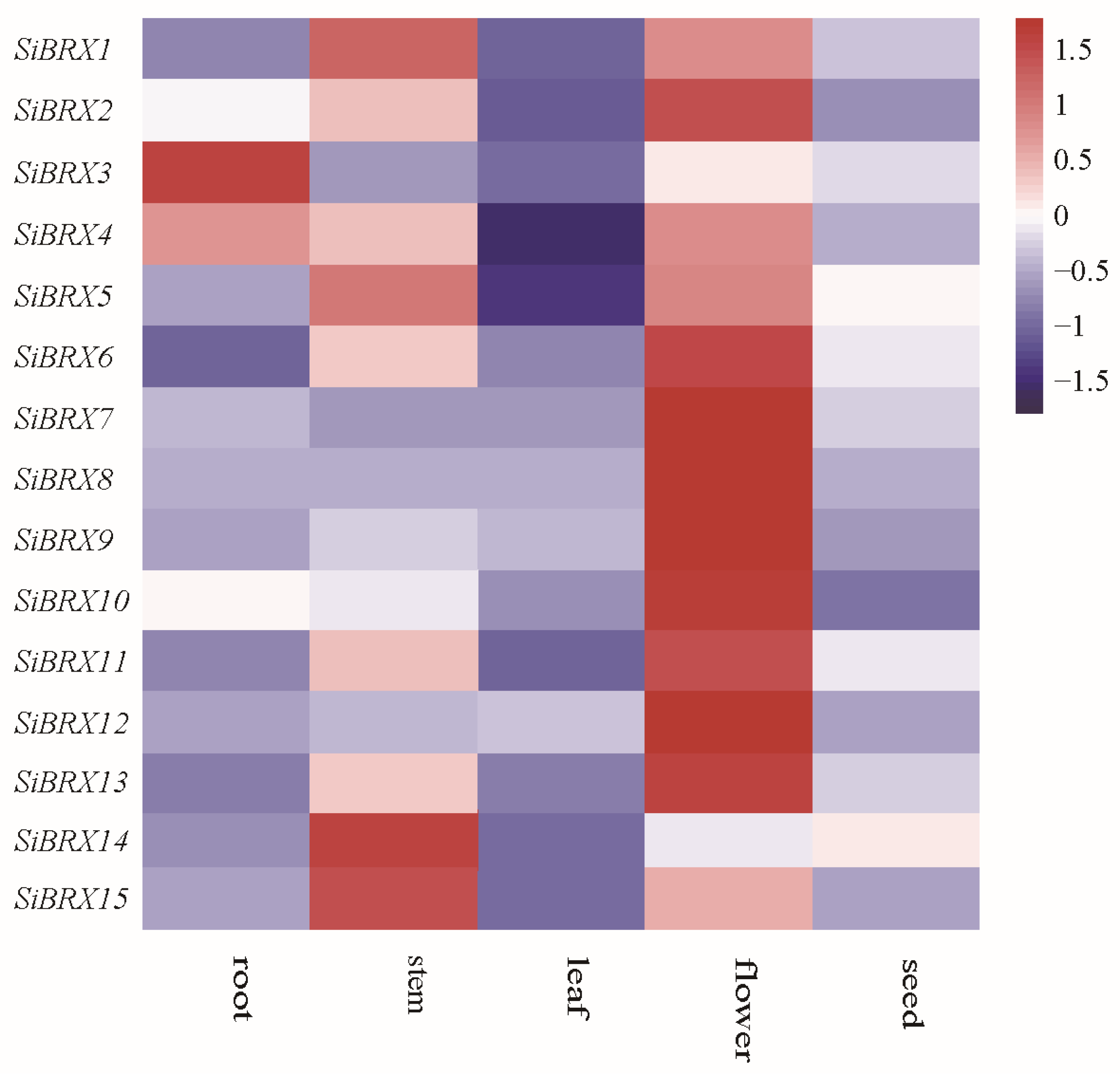
3.8. Expression Patterns of SiBRX Genes
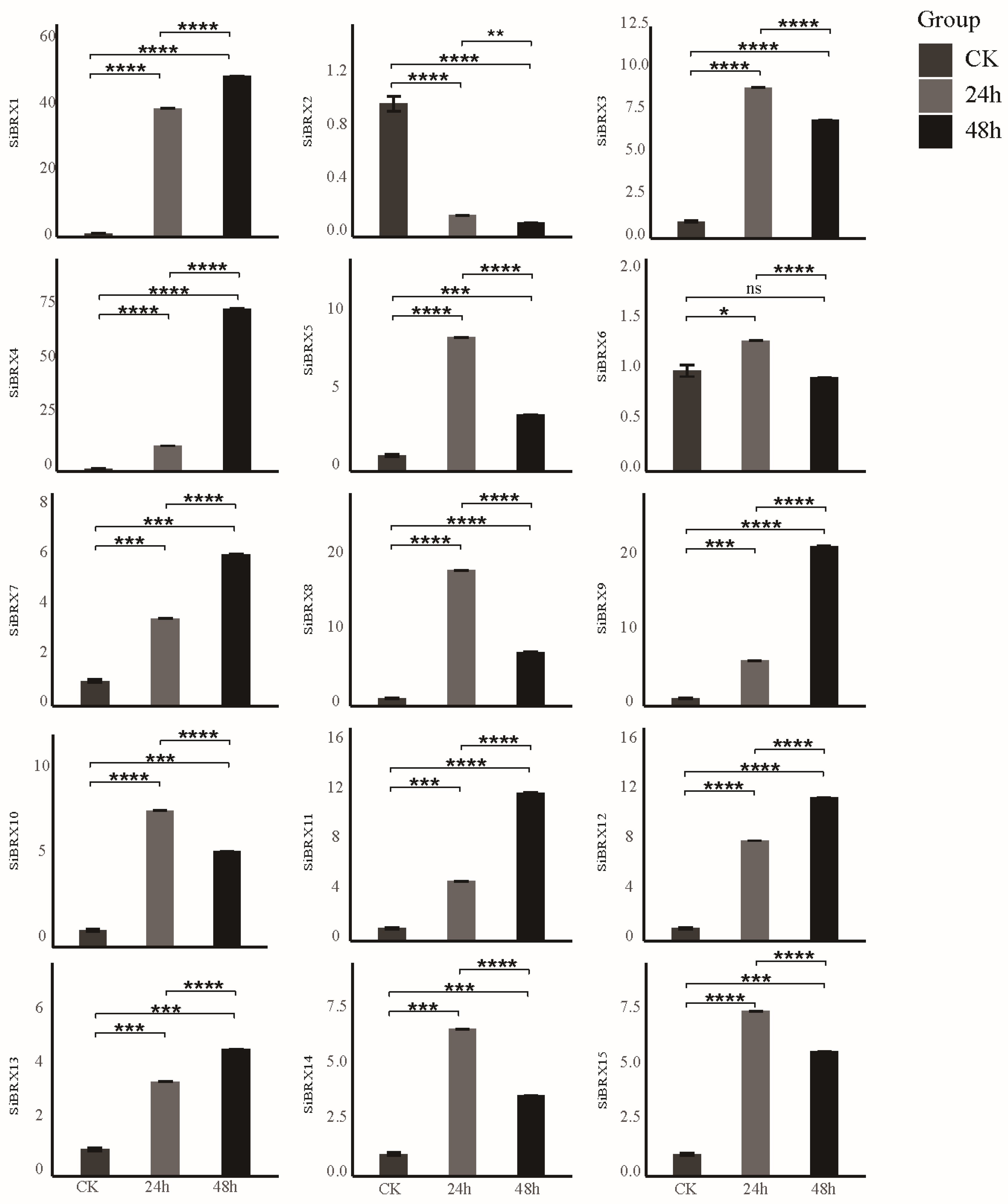
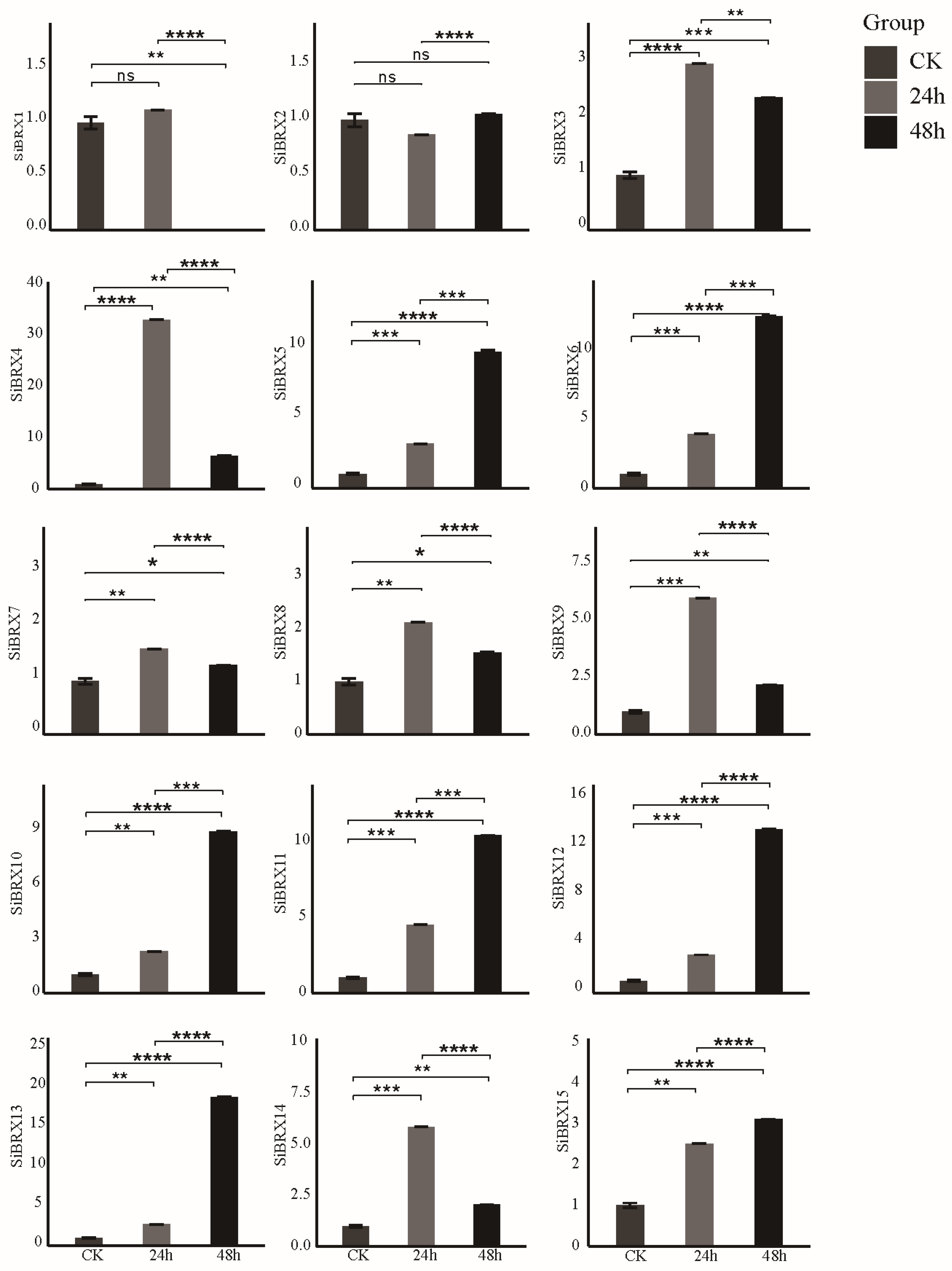
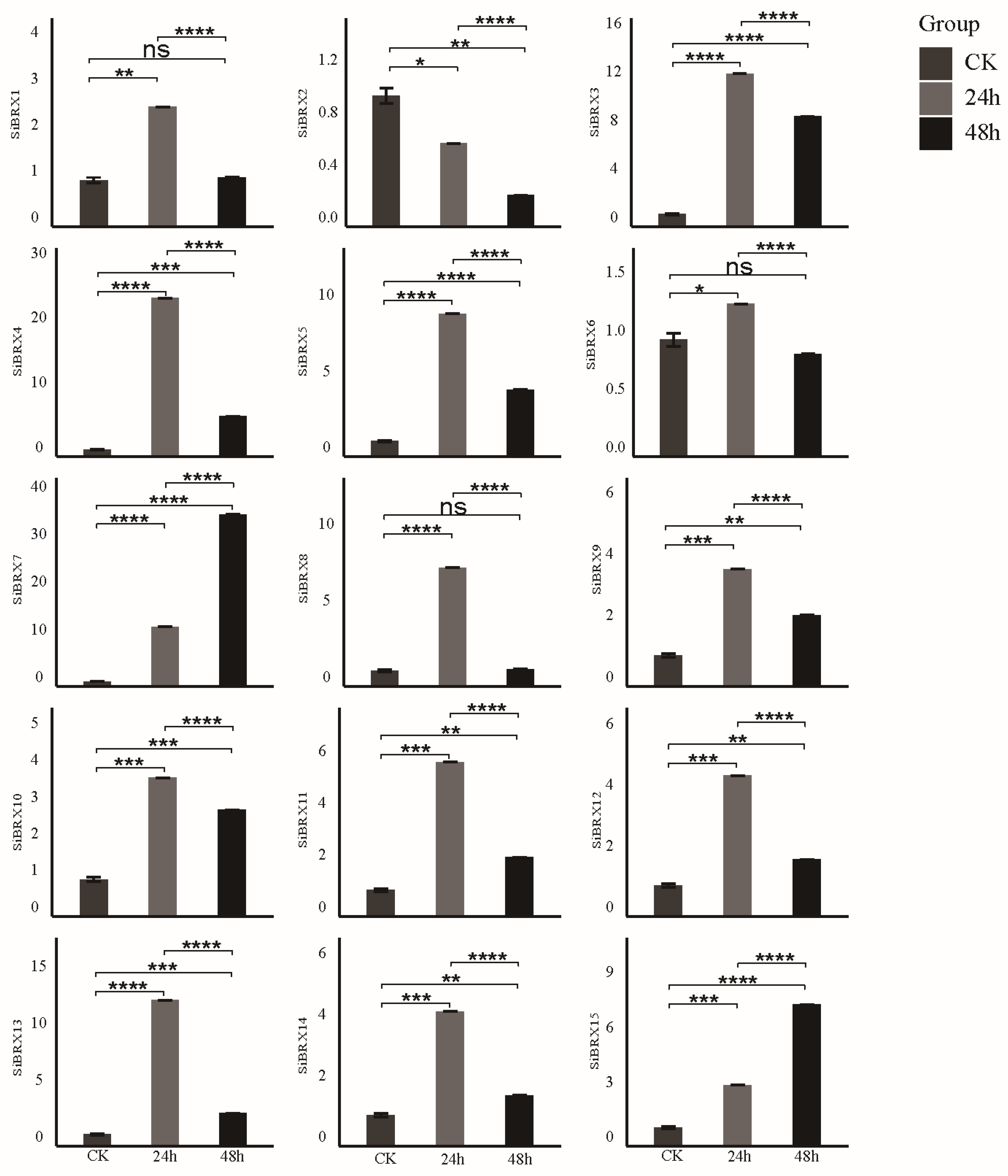
3.9. Analysis of SiBRX Expression Under Different Abiotic Stress Conditions
4. Discussion
4.1. Identification and Analysis of SiBRX Genes in Foxtail Millet
4.2. Evolution of SiBRX Genes
4.3. Spatio-Temporal Patterns of Expression for SiBRX Genes and Their Responses to Abiotic Stress
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Name | Sequence |
|---|---|
| SiActin-RTF | AACATTGTGCTCAGCGGTGG |
| SiActin-RTR | TGGAAGGTGCTAAGGGAGGC |
| Seita.2G222100-F | TTCGTCTCACTGGCGCAAGG |
| Seita.2G222100-R | GGGCTGTCTTCCTTGGAGCT |
| Seita.7G224300-F | TCTCACCTCACAGCTCAAGG |
| Seita.7G224300-R | TGAAGCTGGTGTCGTCGATG |
| Seita.1G286900-F | TACGGTTACATGAGGACGGC |
| Seita.1G286900-R | ATCCTCCTCCTCCTCGTCCT |
| Seita.6G175500-F | CCAAAGAGGATAACCCAGCT |
| Seita.6G175500-R | TGTCTTTGCTCCACTAATGC |
| Seita.9G007000-F | ATGACGTGTCCATCAGCAAC |
| Seita.9G007000-R | TTGTTCTCCTCCCACCACAG |
| Seita.5G455200-F | TAGGATAATTCCTGGGCAGC |
| Seita.5G455200-R | TTTGGTGGCTACGGGAAATC |
| Seita.3G069300-F | GAGCAAGCATTTCTAGTGCC |
| Seita.3G069300-R | GCTTGACCCCACAATCTACA |
| Seita.5G464200-F | GAGTTGGTTCTGACACGGTA |
| Seita.5G464200-R | GGATGAACAGAGTCTTCCCT |
| Seita.7G135400-F | ACTGCGACAAGTGCTGCTCT |
| Seita.7G135400-R | TCGACGATGCGGTCGCAGTT |
| Seita.3G006800-F | TGTCTAGGATTCTCTCAGGG |
| Seita.3G006800-R | GATACCAAGGCACTAAGACC |
| Seita.7G212700-F | CCATCGTTGCTCTGAAGAAG |
| Seita.7G212700-R | AGCTGACACAGAGCTTAAGC |
| Seita.3G160900-F | TATAGCCGGAAGGGAAAACC |
| Seita.3G160900-R | AGGTAGTCCTTCTCAGGGTG |
| Seita.4G097300-F | GGTCCCTTTGATAGGGATGT |
| Seita.4G097300-R | CAGGAAGATAGCCGTGCGTT |
| Seita.1G275300-F | CTACAACCAGAGGAGTTGAG |
| Seita.1G275300-R | CAAGAACTACACTGGACACA |
| Seita.5G285000-F | GCTTATCAAGTATAGCCGGA |
| Seita.5G285000-R | CCCGTTCTTGTATATGAGTG |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Seita.1G275300 | Sevir.1G280600 | 0.00164637 | 0.00275704 | 0.59715271 |
| Seita.1G286900 | Sevir.1G292100 | 0.00000000 | 0.00689264 | 0.00000000 |
| Seita.3G006800 | Sevir.3G008200 | 0.00000000 | 0.00521741 | 0.00000000 |
| Seita.3G069300 | Sevir.3G070700 | 0.00096254 | 0.00600454 | 0.16030237 |
| Seita.3G160900 | Sevir.3G164300 | 0.00169050 | 0.01541030 | 0.10969926 |
| Seita.4G097300 | Sevir.4G096600 | 0.00086549 | 0.00279135 | 0.31006098 |
| Seita.5G285000 | Sevir.5G287800 | 0.00082625 | 0.00818469 | 0.10095034 |
| Seita.5G455200 | Sevir.5G461800 | 0.00000000 | 0.00888335 | 0.00000000 |
| Seita.5G464200 | Sevir.5G470500 | 0.00095919 | 0.00000000 | NaN |
| Seita.6G175500 | Sevir.6G181700 | 0.00107239 | 0.00000000 | NaN |
| Seita.7G135400 | Sevir.7G143700 | 0.00000000 | 0.00458367 | 0.00000000 |
| Seita.7G212700 | Sevir.7G224500 | 0.00162031 | 0.00000000 | NaN |
| Seita.7G224300 | Sevir.7G235900 | 0.00000000 | 0.00000000 | NaN |
| Seita.9G007000 | Sevir.9G007200 | 0.00000000 | 0.01439756 | 0.00000000 |
References
- Li, X.; Gao, J.; Song, J.; Guo, K.; Hou, S.; Wang, X.; He, Q.; Zhang, Y.; Zhang, Y.; Yang, Y.; et al. Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects. Mol. Plant 2022, 15, 1367–1383. [Google Scholar] [PubMed]
- Saxena, R.; Vanga, S.K.; Wang, J.; Orsat, V.; Raghavan, V. Millets for Food Security in the Context of Climate Change: A Review. Sustainability 2018, 10, 2228. [Google Scholar] [CrossRef]
- Chanwala, J.; Khadanga, B.; Jha, D.K.; Sandeep, I.S.; Dey, N. MYB Transcription Factor Family in Pearl Millet: Genome-Wide Identification, Evolutionary Progression and Expression Analysis under Abiotic Stress and Phytohormone Treatments. Plants 2023, 12, 355. [Google Scholar] [CrossRef]
- Wasaya, A.; Hassan, J.; Yasir, T.A.; Ateeq, M.; Raza, M.A. Foliar Application of Silicon Improved Physiological Indicators, Yield Attributes, and Yield of Pearl Millet (Pennisetum glaucum L.) Under Terminal Drought Stress. J. Soil Sci. Plant Nutr. 2022, 22, 4458–4472. [Google Scholar]
- Zhang, J.; Wu, J.; Guo, M.; Aslam, M.; Wang, Q.; Ma, H.; Li, S.; Zhang, X.; Cao, S. Genome-wide characterization and expression profiling of Eucalyptus grandis HD-Zip gene family in response to salt and temperature stress. BMC Plant Biol. 2020, 20, 451. [Google Scholar]
- Lai, D.; Yao, X.; Yan, J.; Gao, A.; Yang, H.; Xiang, D.; Ruan, J.; Fan, Y.; Cheng, J. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 549. [Google Scholar]
- Zhang, X.; Zhou, Y.; Ding, L.; Wu, Z.; Liu, R.; Meyerowitz, E.M. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 2013, 25, 83–101. [Google Scholar]
- Santos, L.A.; de Souza, S.R.; Fernandes, M.S. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol. Rep. 2012, 6, 327–337. [Google Scholar]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 2011, 6, e17458. [Google Scholar]
- Yang, B.; Jiang, Y.; Rahman, M.H.; Deyholos, M.K.; Kav, N.N. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, V.; Coego, A.; López, A.; Agorio, A.; Flors, V.; Vera, P. Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant J. 2009, 58, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Briggs, G.C.; Mouchel, C.F.; Hardtke, C.S. Characterization of the Plant-Specific BREVIS RADIX Gene Family Reveals Limited Genetic Redundancy Despite High Sequence Conservation. Plant Physiol. 2006, 140, 1306–1316. [Google Scholar] [CrossRef]
- Mouchel, C.F.; Osmont, K.S.; Hardtke, C.S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 2006, 443, 458–461. [Google Scholar]
- Tiwari, S.; Muthusamy, S.K.; Roy, P.; Dalal, M. Genome wide analysis of BREVIS RADIX gene family from wheat (Triticum aestivum): A conserved gene family differentially regulated by hormones and abiotic stresses. Int. J. Biol. Macromol. 2023, 232, 123081. [Google Scholar]
- Liu, L.; Jose, S.; Campoli, C.; Bayer, M.M.; Sánchez-Diaz, M.A.; McAllister, T.; Zhou, Y.; Eskan, M.; Milne, L.; Schreiber, M.; et al. Conserved signalling components coordinate epidermal patterning and cuticle deposition in barley. Nat. Commun. 2022, 13, 6050. [Google Scholar] [CrossRef] [PubMed]
- Yoshiaki, I.; Masaichi, M.; Yasuo, N.; Hidemi, K.; Akira, Y. Characterization of Rice Mutants Deficient in the Formation of Crown Roots. Breed. Sci. 2001, 51, 123–129. [Google Scholar]
- Kocsy, G.; Tóth, B.; Berzy, T.; Szalai, G.; Jednákovits, A.; Galiba, G. Glutathione reductase activity and chilling tolerance are induced by a hydroxylamine derivative BRX-156 in maize and soybean. Plant Sci. 2001, 160, 943–950. [Google Scholar]
- Zhang, Y.; Liang, J.; Cai, X.; Chen, H.; Wu, J.; Lin, R.; Cheng, F.; Wang, X. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 2021, 8, 68. [Google Scholar] [CrossRef]
- Shindo, C.; Bernasconi, G.; Hardtke, C.S. Intraspecific competition reveals conditional fitness effects of single gene polymorphism at the Arabidopsis root growth regulator BRX. New Phytol. 2008, 180, 71–80. [Google Scholar]
- Beuchet, J.; Scacchi, E.; Tarkowska, E.; Ragn, L.; Stmad, M.; Hardtke, C.S. BRX promotes Arabidopsis shoot growth. New Phytol. 2010, 1, 23–29. [Google Scholar]
- Liu, J.; Liang, D.; Song, Y.; Xiong, L. Systematic identification and expression analysis of BREVIS RADIX-like homologous genes in rice. Plant Sci. 2010, 178, 183–191. [Google Scholar]
- He, Q.; Wang, C.; Zhang, J.; Liang, H.; Lu, Z.; Xie, K.; Tang, S.; Zhou, Y.; Liu, B.; Zhi, H.; et al. A complete reference genome assembly for foxtail millet and Setaria-db, a comprehensive database for Setaria. Mol. Plant 2023, 17, 219–222. [Google Scholar] [PubMed]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Alam, O.; Li, H.; Zhang, H.; Xing, L.; et al. A graph-based genome and pan-genome variation of the model plant Setaria. Nat. Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef]
- Tang, S.; Shahriari, M.; Xiang, J.; Pasternak, T.; Igolkina, A.; Aminizade, S.; Zhi, H.; Gao, Y.; Roodbarkelari, F.; Sui, Y.; et al. The role of AUX1 during lateral root development in the domestication of the model C4 grass Setaria italica. J. Exp. Bot. 2021, 73, 2021–2034. [Google Scholar]
- Liu, D.; Cui, Y.; Zhao, Z.; Li, S.; Liang, D.; Wang, C.; Feng, G.; Wang, J.; Liu, Z. Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genom. 2021, 22, 682. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Mouchel, C.F.; Briggs, G.C.; Hardtke, C.S. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 2004, 18, 700–714. [Google Scholar]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar]
- Larkin, M. Clustal W and Clustal X v. 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [PubMed]
- Wang, J.; Farideh, C.; Myra, K.D.; Noreen, R.G.; Marc, G.; Lu, G.; Gabriele, H.M.; Song, J.; Narmada, T.; Roxanne, A.Y.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2022, 51, D384–D388. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar]
- Lin, M.; Dong, Z.; Zhou, H.; Wu, G.; Xu, L.; Ying, S.; Chen, M. Genome-Wide Identification and Transcriptional Analysis of the MYB Gene Family in Pearl Millet (Pennisetum glaucum). Int. J. Mol. Sci. 2023, 24, 2484. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and SwissPdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef] [PubMed]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from Standard Atomic Volumes as a Quality Measure for Protein Crystal Structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]


| Name | Primary Transcript ID | Amino Acid (aa) | MW (kDa) | Theoretical pI | Grand Average of Hydropathicity | Instability Index | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| SiBRX1 | Seita.1G275300.1 | 1053 | 116,648.95 | 9.07 | −0.449 | 37.32 | 77.59 | outer membrane |
| SiBRX2 | Seita.1G286900.1 | 433 | 47,506.54 | 5.93 | −0.576 | 51.7 | 62.51 | extracellular |
| SiBRX3 | Seita.2G222100.1 | 410 | 44,938.51 | 5.99 | −0.77 | 52.24 | 54.87 | extracellular |
| SiBRX4 | Seita.3G006800.1 | 1064 | 111,694.82 | 8.55 | −0.0469 | 40.09 | 73.75 | outer membrane |
| SiBRX5 | Seita.3G069300.1 | 910 | 31,415.71 | 8.57 | −0.406 | 40.39 | 77.19 | outer membrane |
| SiBRX6 | Seita.3G160900.1 | 1030 | 111,684.82 | 8.6 | −0.429 | 42.31 | 71.41 | extracellular |
| SiBRX7 | Seita.4G097300.1 | 1010 | 109,925.71 | 7.19 | −0.353 | 41.15 | 78.42 | extracellular |
| SiBRX8 | Seita.5G285000.1 | 1053 | 115,184.1 | 8.56 | −0.402 | 39.01 | 75.12 | extracellular |
| SiBRX9 | Seita.5G455200.1 | 1092 | 99,108.17 | 8.94 | −0.464 | 44.68 | 71.05 | outer membrane |
| SiBRX10 | Seita.5G464200.1 | 915 | 116,388.59 | 8.73 | −0.44 | 44.69 | 75.88 | outer membrane |
| SiBRX11 | Seita.6G175500.1 | 411 | 44,966.19 | 5.67 | −0.845 | 51.21 | 54.96 | outer membrane |
| SiBRX12 | Seita.7G135400.1 | 300 | 117,900.21 | 4.91 | −0.859 | 55.38 | 48.3 | extracellular |
| SiBRX13 | Seita.7G212700.1 | 1069 | 109,925.71 | 9.18 | −0.437 | 43.7 | 76.11 | outer membrane |
| SiBRX14 | Seita.7G224300.1 | 413 | 44,061.81 | 6.42 | −0.549 | 52.05 | 57.31 | extracellular |
| SiBRX15 | Seita.9G007000.1 | 376 | 118,008.43 | 8.64 | −0.817 | 55.64 | 54.87 | outer membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Bai, X.; Yu, J.; Jia, Z.; Wang, C. Genome-Wide Identification of the BREVIS RADIX Gene Family in Foxtail Millet: Function, Evolution, and Expression. Genes 2025, 16, 374. https://doi.org/10.3390/genes16040374
Yuan X, Bai X, Yu J, Jia Z, Wang C. Genome-Wide Identification of the BREVIS RADIX Gene Family in Foxtail Millet: Function, Evolution, and Expression. Genes. 2025; 16(4):374. https://doi.org/10.3390/genes16040374
Chicago/Turabian StyleYuan, Xiaorui, Xionghui Bai, Jin Yu, Zhijie Jia, and Chenyu Wang. 2025. "Genome-Wide Identification of the BREVIS RADIX Gene Family in Foxtail Millet: Function, Evolution, and Expression" Genes 16, no. 4: 374. https://doi.org/10.3390/genes16040374
APA StyleYuan, X., Bai, X., Yu, J., Jia, Z., & Wang, C. (2025). Genome-Wide Identification of the BREVIS RADIX Gene Family in Foxtail Millet: Function, Evolution, and Expression. Genes, 16(4), 374. https://doi.org/10.3390/genes16040374





