A Comprehensive Evolutionary Analysis of the Dihydroflavonol 4-Reductase (DFR) Gene Family in Plants: Insights from 237 Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Construction of Species Phylogenetic Tree
2.3. Identification of DFR Gene Family Members
2.4. Construction of DFR Gene Family Phylogenetic Tree
2.5. Conserved Motif Analysis of Gene Family Sequences
2.6. Analysis of the Duplication Type for DFR Family Genes
2.7. Positive Selection
2.8. Gene Expression Analysis
3. Results
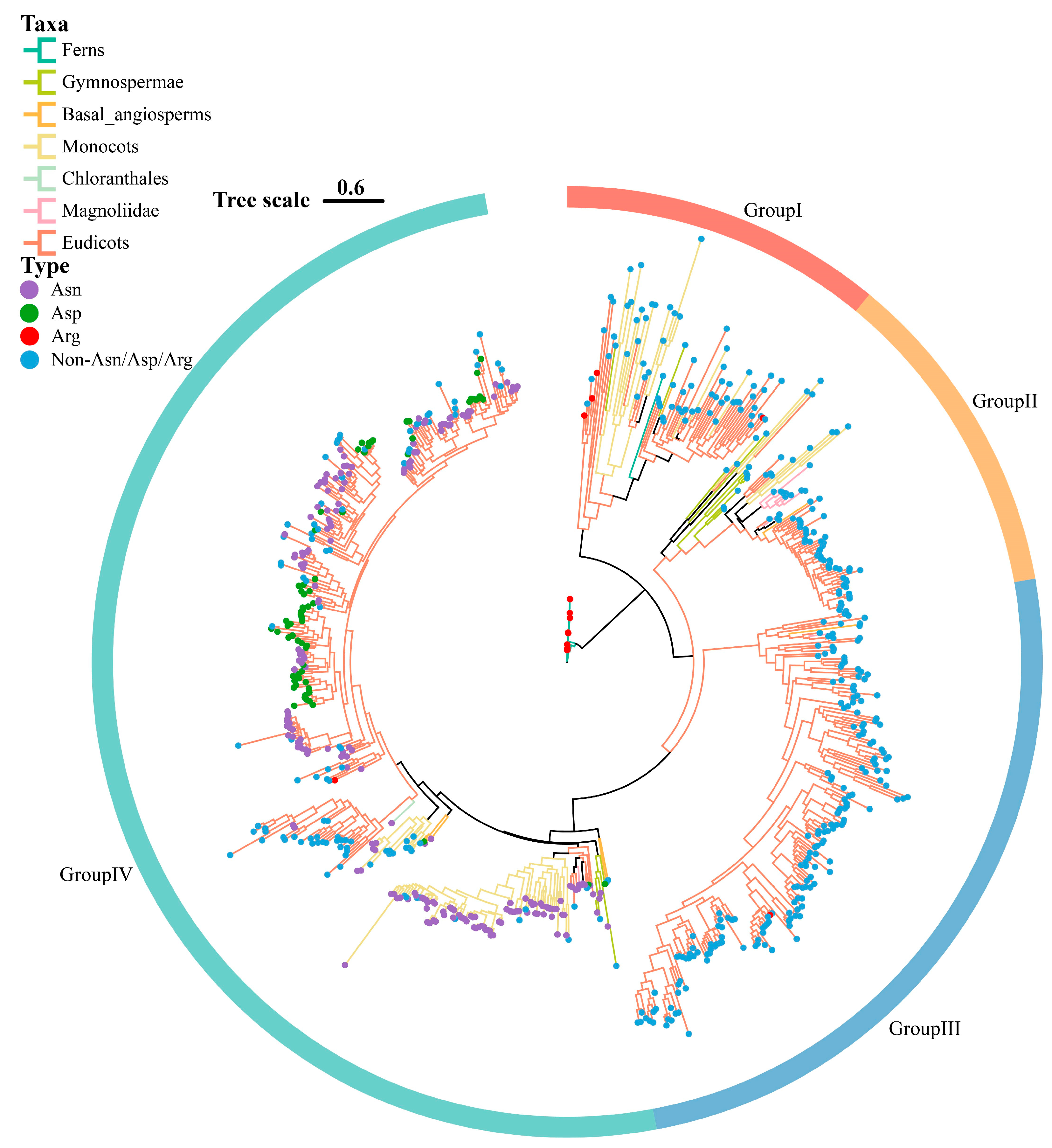
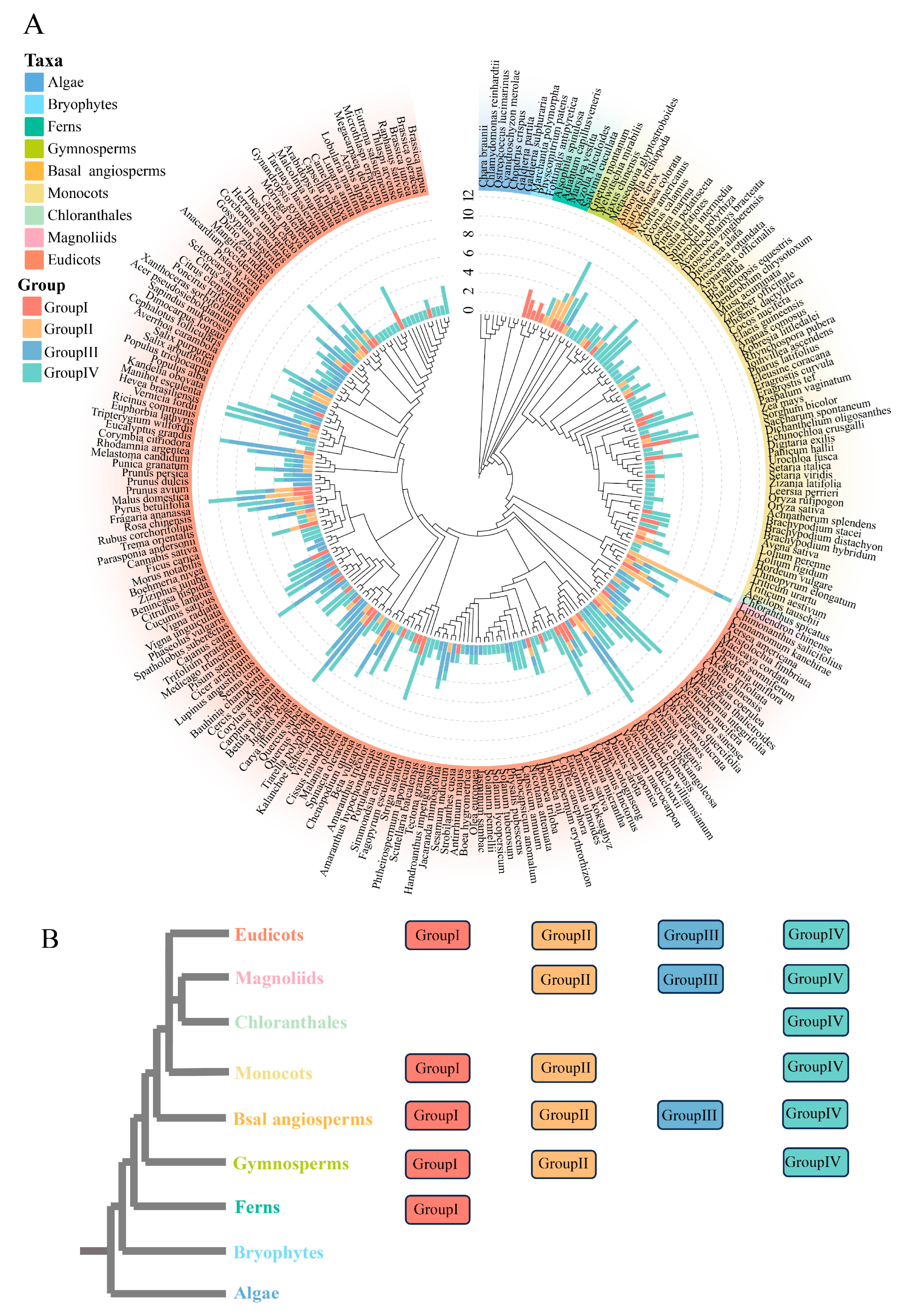
3.1. Identification and Classification of DFR Gene Family
3.2. Phylogenetic Analysis of DFR Gene Family
3.3. Member Number and Distribution of the DFR Gene Family Across Different Plant Taxa
3.4. Conserved Motif Analysis of Representative Species
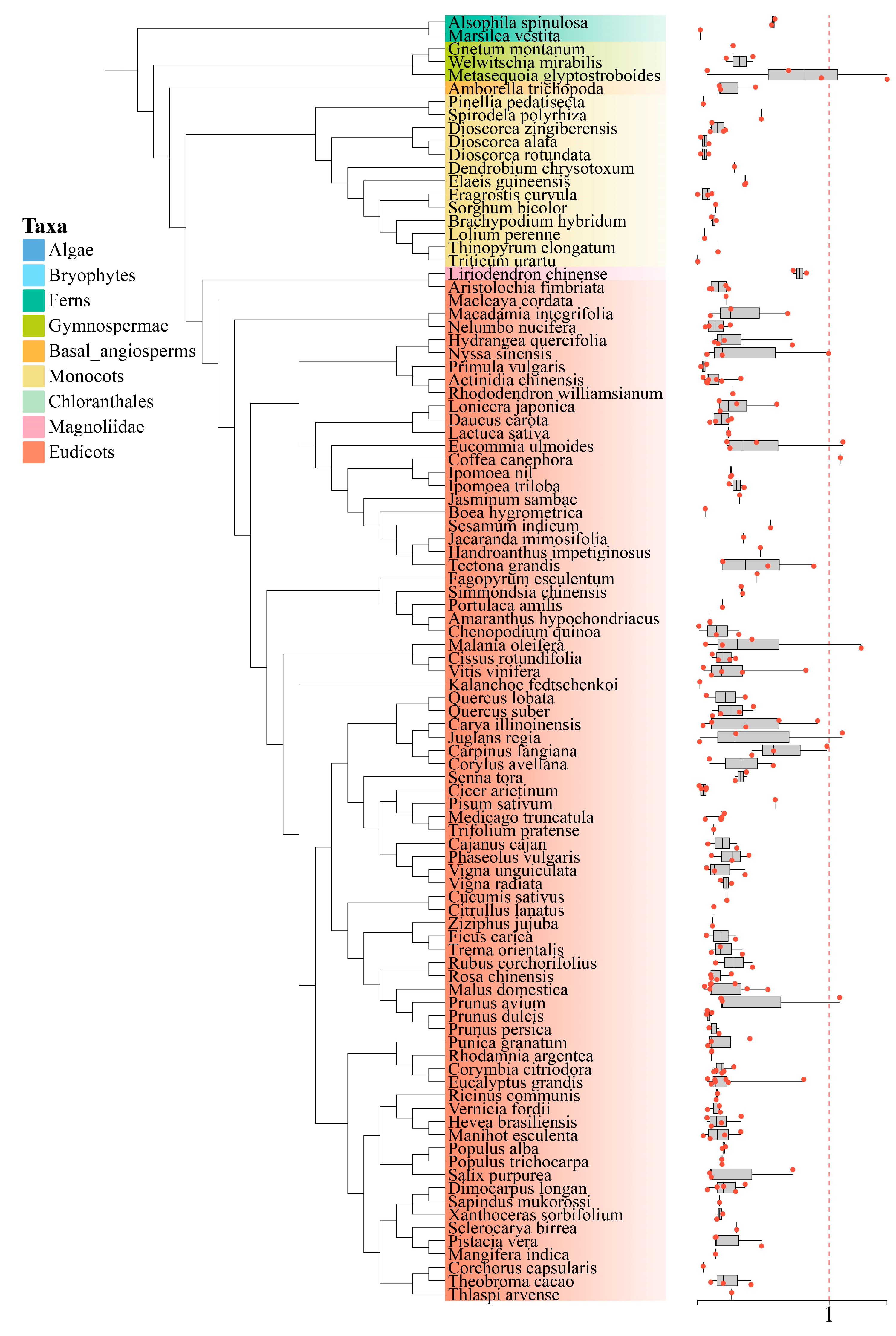
3.5. Identification of Gene Duplication Types
3.6. Ka/Ks Calculation
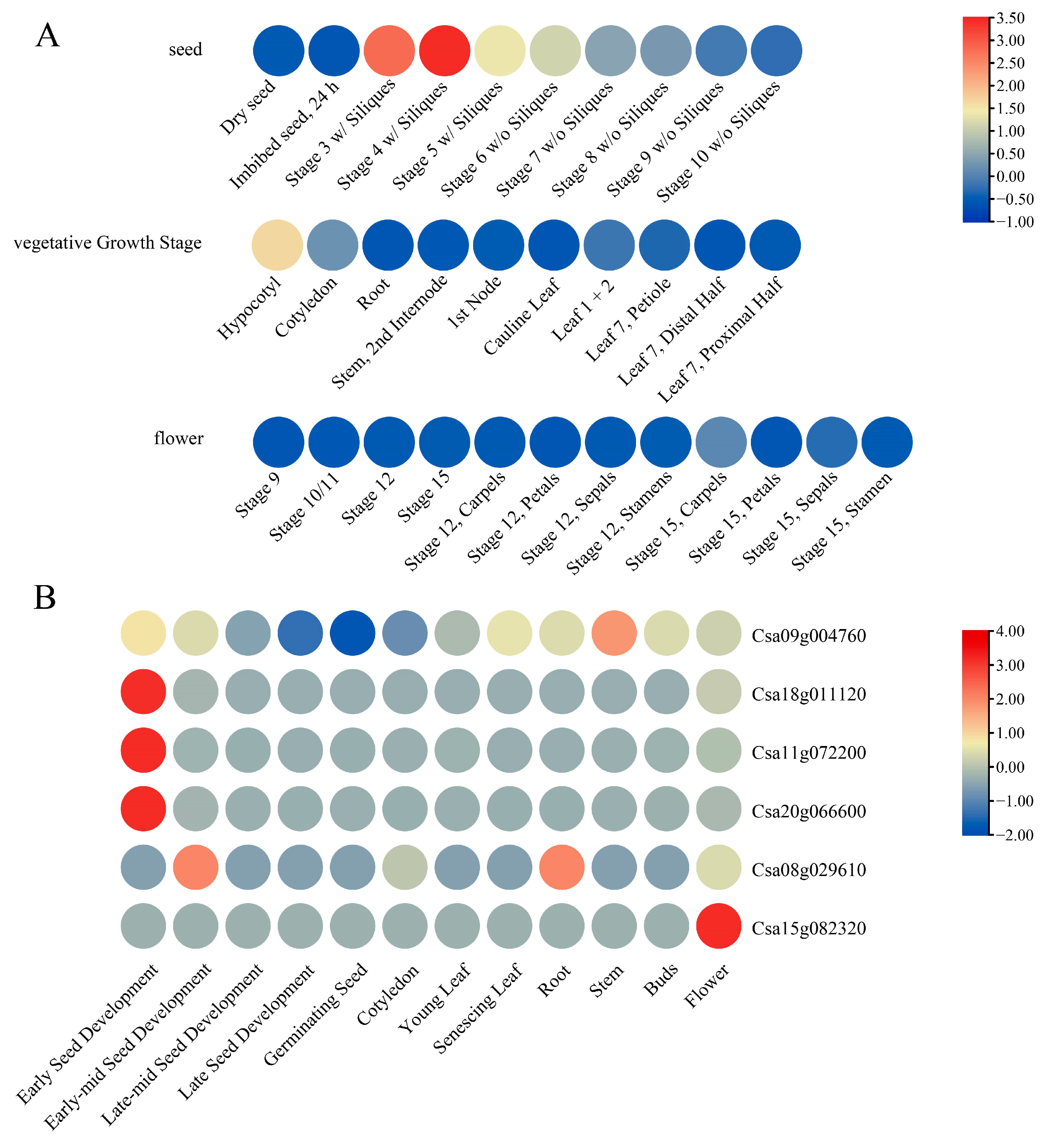
3.7. Expression Analysis of DFR Genes Under Different Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wen, W.; Alseekh, S.; Fernie, A.R. Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol. 2020, 55, 100–108. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Weng, J.-K.; Philippe, R.N.; Noel, J.P. The rise of chemodiversity in plants. Science 2012, 336, 1667–1670. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Li, Z.; Ahammed, G.J. Plant stress response and adaptation via anthocyanins: A review. Plant Stress 2023, 10, 100230. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Saewan, N.; Jimtaisong, A. Photoprotection of natural flavonoids. J. Appl. Pharm. Sci. 2013, 3, 129–141. [Google Scholar] [CrossRef]
- Li, S.-J.; Bai, Y.-C.; Li, C.-L.; Yao, H.-P.; Chen, H.; Zhao, H.-X.; Wu, Q. Anthocyanins accumulate in tartary buckwheat (Fagopyrum tataricum) sprout in response to cold stress. Acta Physiol. Plant 2015, 37, 159. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Yang, Y. Anthocyanin-mediated arsenic tolerance in plants. Environ. Pollut. 2022, 292, 118475. [Google Scholar] [CrossRef]
- Wrolstad, R. Anthocyanin pigments—Bioactivity and coloring properties. J. Food Sci. 2004, 69, C419–C425. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Diharce, J.; Bignon, E.; Fiorucci, S.; Antonczak, S. Exploring Dihydroflavonol-4-Reductase Reactivity and Selectivity by QM/MM-MD Simulations. ChemBioChem 2022, 23, e202100553. [Google Scholar] [CrossRef]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Lewis, D.H.; Bloor, S.J.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Davies, K.; Winefield, C.; Lewis, D.; Nielsen, K.; Bradley, M.; Schwinn, K.; Deroles, S.; Manson, D.; Jordan, B. Research into control of flower colour and flowering time in Eustoma grandiflorum (Lisianthus). Flower. Newsl. 1997, 23, 24–32. [Google Scholar]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef]
- Nielsen, K.; Deroles, S.C.; Markham, K.R.; Bradley, M.J.; Podivinsky, E.; Manson, D. Antisense flavonol synthase alters copigmentation and flower color in lisianthus. Mol. Breed. 2002, 9, 217–229. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Johnson, E.T.; Yi, H.; Shin, B.; Oh, B.J.; Cheong, H.; Choi, G. Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 1999, 19, 81–85. [Google Scholar] [CrossRef]
- Helariutta, Y.; Elomaa, P.; Kotilainen, M.; Seppänen, P.; Teeri, T.H. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 1993, 22, 183–193. [Google Scholar] [CrossRef]
- Forkmann, G.; Ruhnau, B. Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of Petunia hybrida. Z. Naturforsch. C 1987, 42, 1146–1148. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Huang, X.; Fu, H.; Wang, Q.; Wang, Y.; Cao, J. Arg-type dihydroflavonol 4-reductase genes from the fern Dryopteris erythrosora play important roles in the biosynthesis of anthocyanins. PLoS ONE 2020, 15, e0232090. [Google Scholar] [CrossRef]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Inagaki, Y.; Nagata, H.; Miykzaki, C.; Iida, S. Molecular characterization of the gene for dihydroflavonol 4-reductase of japonica rice varieties. Plant Biotechnol. 1998, 15, 221–225. [Google Scholar] [CrossRef]
- Ruan, H.; Shi, X.; Gao, L.; Rashid, A.; Li, Y.; Lei, T.; Dai, X.; Xia, T.; Wang, Y. Functional analysis of the dihydroflavonol 4-reductase family of Camellia sinensis: Exploiting key amino acids to reconstruct reduction activity. Hortic. Res. 2022, 9, uhac098. [Google Scholar] [CrossRef]
- Qian, X.; Zheng, W.; Hu, J.; Ma, J.; Sun, M.; Li, Y.; Liu, N.; Chen, T.; Wang, M.; Wang, L. Identification and Expression Analysis of DFR Gene Family in Brassica napus L. Plants 2023, 12, 2583. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Liu, H.; Wang, Q.; Feng, X.; Wang, C.; Sun, Y.; Zhang, X.; Zhu, S. Genome-wide identification of putative dihydroflavonol 4-reductase (DFR) gene family in eight Solanaceae species and expression analysis in Solanum lycopersicum. PeerJ 2023, 11, e16124. [Google Scholar] [CrossRef]
- Li, W.-F.; Gao, J.; Ma, Z.-H.; Hou, Y.-J.; Li, X.; Mao, J.; Chen, B.-H. Molecular evolution and expression assessment of DFR s in apple. Chem. Biol. Technol. Agric. 2023, 10, 98. [Google Scholar] [CrossRef]
- Chen, L.-Z.; Tian, X.-C.; Feng, Y.-Q.; Qiao, H.-L.; Wu, A.-Y.; Li, X.; Hou, Y.-J.; Ma, Z.-H. The Genome-Wide Identification of the Dihydroflavonol 4-Reductase (DFR) Gene Family and Its Expression Analysis in Different Fruit Coloring Stages of Strawberry. Int. J. Mol. Sci. 2024, 25, 9911. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Rabiee, M.; Sayyari, E.; Mirarab, S. Multi-allele species reconstruction using ASTRAL. Mol. Phylogenet. Evol. 2019, 130, 286–296. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Yu, T.; Bai, Y.; Liu, Z.; Wang, Z.; Yang, Q.; Wu, T.; Feng, S.; Zhang, Y.; Shen, S.; Li, Q. Large-scale analyses of heat shock transcription factors and database construction based on whole-genome genes in horticultural and representative plants. Hortic. Res. 2022, 9, uhac035. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Li, N.; Chen, H.; Liu, Z.; Li, C.; Zhou, R.; Zhang, Y.; Cao, R.; Ma, X.; Song, X. Large-scale analysis of the ARF and Aux/IAA gene families in 406 horticultural and other plants. Mol. Hortic. 2024, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Skolnick, J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003, 333, 863–882. [Google Scholar] [CrossRef]
- Katsu, K.; Suzuki, R.; Tsuchiya, W.; Inagaki, N.; Yamazaki, T.; Hisano, T.; Yasui, Y.; Komori, T.; Koshio, M.; Kubota, S. A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 2017, 17, 239. [Google Scholar] [CrossRef]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.-W.; Lee, H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Zhen, X.; Zhang, J.; Zhang, Q.; Liu, Z.; Hou, S.; Han, Y.; Zhang, B. Uncovering the mechanism of anthocyanin accumulation in a purple-leaved variety of foxtail millet (Setaria italica) by transcriptome analysis. PeerJ 2022, 10, e14099. [Google Scholar] [CrossRef]
- Fürst-Jansen, J.M.; de Vries, S.; de Vries, J. Evo-physio: On stress responses and the earliest land plants. J. Exp. Bot. 2020, 71, 3254–3269. [Google Scholar] [CrossRef]
- Davies, K.M.; Jibran, R.; Zhou, Y.; Albert, N.W.; Brummell, D.A.; Jordan, B.R.; Bowman, J.L.; Schwinn, K.E. The evolution of flavonoid biosynthesis: A bryophyte perspective. Front. Plant Sci. 2020, 11, 7. [Google Scholar] [CrossRef]
- Valenta, K.; Kalbitzer, U.; Razafimandimby, D.; Omeja, P.; Ayasse, M.; Chapman, C.A.; Nevo, O. The evolution of fruit colour: Phylogeny, abiotic factors and the role of mutualists. Sci. Rep. 2018, 8, 14302. [Google Scholar] [CrossRef] [PubMed]
- Nyegaard Kristiansen, K.; Rohde, W. Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant 18 mutants blocked in flavonoid synthesis. Mol. Gen. Genet. 1991, 230, 49–59. [Google Scholar] [CrossRef]
- Hirner, A.A.; Veit, S.; Seitz, H.U. Regulation of anthocyanin biosynthesis in UV-A-irradiated cell cultures of carrot and in organs of intact carrot plants. Plant Sci. 2001, 161, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 2004, 134, 979–994. [Google Scholar] [CrossRef] [PubMed]
- De Jong, W.; De Jong, D.; De Jong, H.; Kalazich, J.; Bodis, M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor. Appl. Genet. 2003, 107, 1375–1383. [Google Scholar] [CrossRef]
- Cheng, H.; Li, L.; Cheng, S.; Cao, F.; Xu, F.; Yuan, H.; Wu, C. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS ONE 2013, 8, e72017. [Google Scholar] [CrossRef]
- Copley, S.D. Evolution of new enzymes by gene duplication and divergence. FEBS J. 2020, 287, 1262–1283. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef]
- Yang, F.-S.; Nie, S.; Liu, H.; Shi, T.-L.; Tian, X.-C.; Zhou, S.-S.; Bao, Y.-T.; Jia, K.-H.; Guo, J.-F.; Zhao, W. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 2020, 11, 5269. [Google Scholar] [CrossRef]
- Bennetzen, J.L. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 2002, 115, 29–36. [Google Scholar] [CrossRef]
- Lawton-Rauh, A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 2003, 29, 396–409. [Google Scholar] [CrossRef]
- Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Mol. Biol. 2000, 7, 786–791. [Google Scholar] [CrossRef]
- Ferrer, J.-L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Mol. Biol. 1999, 6, 775–784. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, S.; Chu, D.; Wang, X. Exploring the evolution of CHS gene family in plants. Front. Genet. 2024, 15, 1368358. [Google Scholar] [CrossRef]
- Shimada, N.; Aoki, T.; Sato, S.; Nakamura, Y.; Tabata, S.; Ayabe, S.-I. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy (iso) flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Ralston, L.; Subramanian, S.; Matsuno, M.; Yu, O. Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol. 2005, 137, 1375–1388. [Google Scholar] [CrossRef]
- Cheng, A.X.; Zhang, X.; Han, X.J.; Zhang, Y.Y.; Gao, S.; Liu, C.J.; Lou, H.X. Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for synthesis of flavonoids. New Phytol. 2018, 217, 909–924. [Google Scholar] [CrossRef]
- Ngaki, M.N.; Louie, G.V.; Philippe, R.N.; Manning, G.; Pojer, F.; Bowman, M.E.; Li, L.; Larsen, E.; Wurtele, E.S.; Noel, J.P. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 2012, 485, 530–533. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Wang, S.; Yang, L.; Luo, K.; Cheng, J.; Ning, Y.; Dong, Y.; Wang, W. A Comprehensive Evolutionary Analysis of the Dihydroflavonol 4-Reductase (DFR) Gene Family in Plants: Insights from 237 Species. Genes 2025, 16, 396. https://doi.org/10.3390/genes16040396
Luo S, Wang S, Yang L, Luo K, Cheng J, Ning Y, Dong Y, Wang W. A Comprehensive Evolutionary Analysis of the Dihydroflavonol 4-Reductase (DFR) Gene Family in Plants: Insights from 237 Species. Genes. 2025; 16(4):396. https://doi.org/10.3390/genes16040396
Chicago/Turabian StyleLuo, Senlin, Shiping Wang, Ling Yang, Kaiyong Luo, Jia Cheng, Ya Ning, Yang Dong, and Weibin Wang. 2025. "A Comprehensive Evolutionary Analysis of the Dihydroflavonol 4-Reductase (DFR) Gene Family in Plants: Insights from 237 Species" Genes 16, no. 4: 396. https://doi.org/10.3390/genes16040396
APA StyleLuo, S., Wang, S., Yang, L., Luo, K., Cheng, J., Ning, Y., Dong, Y., & Wang, W. (2025). A Comprehensive Evolutionary Analysis of the Dihydroflavonol 4-Reductase (DFR) Gene Family in Plants: Insights from 237 Species. Genes, 16(4), 396. https://doi.org/10.3390/genes16040396





