New Insights into Genetic Diversity and Differentiation of 11 Buffalo Populations Using Validated SNPs for Dairy Improvement
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and DNA Isolation
2.2. PCR Amplification and Genotyping
2.3. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DBI | Davies−Bouldin index |
| MAF | minor allele frequency |
| PCA | principal component analysis |
| PCR | polymerase chain reaction |
| QTL | quantitative trait locus |
| RFLP | restriction fragment length polymorphism |
| ROH | runs of homozygosity |
| SNP | single nucleotide polymorphism |
| SS | silhouette score |
| UPGMA | unweighted pair group method with the arithmetic mean |
References
- Borghese, A. Buffalo Production and Research. REU Technical Series; Food and Agriculture Organization of the United Nations: Rome, Italy, 2005; pp. 1–315. [Google Scholar]
- Zicarelli, L. Buffalo milk: Its properties, dairy yield and mozzarella production. Vet. Res. Commun. 2004, 28, 127. [Google Scholar] [PubMed]
- Di Stasio, L.; Brugiapaglia, A. Current knowledge on river buffalo meat: A critical analysis. Animals 2021, 11, 2111. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.S.F.; Moore, S.; Hetzel, D.; Evans, D.; Byrne, K.; Tan, S. Genetic diversity of Asian water buffalo (Bubalus bubalis): Microsatellite variation and a comparison with protein-coding loci. Anim. Genet. 1997, 28, 103–115. [Google Scholar] [CrossRef]
- Kumar, S.; Nagarajan, M.; Sandhu, J.; Kumar, N.; Behl, V.; Nishanth, G. Mitochondrial DNA analyses of Indian water buffalo support a distinct genetic origin of river and swamp buffalo. Anim. Genet. 2007, 38, 227–232. [Google Scholar] [PubMed]
- Kumar, S.; Nagarajan, M.; Sandhu, J.S.; Kumar, N.; Behl, V. Phylogeography and domestication of Indian river buffalo. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar]
- Moioli, B.; Georgoudis, A.; Napolitano, F.; Catillo, G.; Lucioli, S.; Ligda, C.; Boyazoglu, J. Genetic diversity between Italian and Greek buffalo populations. Anim. Genet. Resour. Resour. Génétiques Anim. Recur. Genéticos Anim. 2001, 29, 31–40. [Google Scholar]
- Yindee, M.; Vlamings, B.; Wajjwalku, W.; Techakumphu, M.; Lohachit, C.; Sirivaidyapong, S.; Thitaram, C.; Amarasinghe, A.; Alexander, P.; Colenbrander, B. Y-chromosomal variation confirms independent domestications of swamp and river buffalo. Anim. Genet. 2010, 41, 433–435. [Google Scholar]
- Zhang, Y.; Colli, L.; Barker, J.S.F. Asian water buffalo: Domestication, history and genetics. Anim. Genet. 2020, 51, 177–191. [Google Scholar]
- Zhang, Y.; Lu, Y.; Yindee, M.; Li, K.Y.; Kuo, H.Y.; Ju, Y.T.; Ye, S.; Faruque, M.O.; Li, Q.; Wang, Y. Strong and stable geographic differentiation of swamp buffalo maternal and paternal lineages indicates domestication in the China/Indochina border region. Mol. Ecol. 2016, 25, 1530–1550. [Google Scholar] [CrossRef]
- Zhang, Y.; Vankan, D.; Zhang, Y.; Barker, J.S. Genetic differentiation of water buffalo (Bubalus bubalis) populations in China, Nepal and south-east Asia: Inferences on the region of domestication of the swamp buffalo. Anim. Genet. 2011, 42, 366–377. [Google Scholar]
- Colli, L.; Milanesi, M.; Vajana, E.; Iamartino, D.; Bomba, L.; Puglisi, F.; Del Corvo, M.; Nicolazzi, E.L.; Ahmed, S.S.; Herrera, J.R. New insights on water buffalo genomic diversity and post-domestication migration routes from medium density SNP chip data. Front. Genet. 2018, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Noce, A.; Qanbari, S.; González-Prendes, R.; Brenmoehl, J.; Luigi-Sierra, M.G.; Theerkorn, M.; Fiege, M.-A.; Pilz, H.; Bota, A.; Vidu, L. Genetic diversity of Bubalus bubalis in Germany and global relations of its genetic background. Front. Genet. 2021, 11, 610353. [Google Scholar]
- Dhakal, A.; Si, J.; Sapkota, S.; Pauciullo, A.; Han, J.; Gorkhali, N.A.; Zhao, X.; Zhang, Y. Whole-genome sequencing reveals genetic structure and adaptive genes in Nepalese buffalo breeds. BMC Genom. 2024, 25, 1082. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Dai, D.; Gorkhali, N.A.; Wang, M.; Wang, S.; Sapkota, S.; Kadel, R.C.; Sadaula, A.; Dhakal, A.; Faruque, M.O. Complete Genomic Landscape Reveals Hidden Evolutionary History and Selection Signature in Asian Water Buffaloes (Bubalus bubalis). Adv. Sci. 2025, 12, 2407615. [Google Scholar] [CrossRef]
- Wang, S.; Chen, N.; Capodiferro, M.R.; Zhang, T.; Lancioni, H.; Zhang, H.; Miao, Y.; Chanthakhoun, V.; Wanapat, M.; Yindee, M. Whole mitogenomes reveal the history of swamp buffalo: Initially shaped by glacial periods and eventually modelled by domestication. Sci. Rep. 2017, 7, 4708. [Google Scholar] [CrossRef]
- Bonfatti, V.; Giantin, M.; Gervaso, M.; Coletta, A.; Dacasto, M.; Carnier, P. Effect of CSN1S1-CSN3 (αS1-κ-casein) composite genotype on milk production traits and milk coagulation properties in Mediterranean water buffalo. J. Dairy Sci. 2012, 95, 3435–3443. [Google Scholar] [CrossRef]
- Pauciullo, A.; Gaspa, G.; Zhang, Y.; Liu, Q.; Cosenza, G. CSN1S1, CSN3 and LPL: Three validated gene polymorphisms useful for more sustainable dairy production in the Mediterranean river buffalo. Animals 2024, 14, 1414. [Google Scholar] [CrossRef]
- Pauciullo, A.; Martorello, S.; Carku, K.; Versace, C.; Coletta, A.; Cosenza, G. A novel duplex ACRS-PCR for composite CSN1S1–CSN3 genotype discrimination in domestic buffalo. Ital. J. Anim. Sci. 2021, 20, 1264–1269. [Google Scholar] [CrossRef]
- Zicarelli, L.; Di Palo, R.; Napolano, R.; Tonhati, H.; De Carlo, E.; Gagliardi, R.; Di Luccia, A.; la Gatta, B. Influence of αS1-casein and κ-casein polymorphism on the curd yield of Italian Mediterranean buffalo (Bubalus bubalis L.) milk. Int. Dairy J. 2020, 100, 104559. [Google Scholar] [CrossRef]
- Cosenza, G.; Pauciullo, A.; Macciotta, N.; Apicella, E.; Steri, R.; La Battaglia, A.; Jemma, L.; Coletta, A.; Di Berardino, D.; Ramunno, L. Mediterranean river buffalo CSN1S1 gene: Search for polymorphisms and association studies. Anim. Prod. Sci. 2015, 55, 654–660. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Li, J.; Li, H.; Yang, L. Genome-wide identification of Diacylglycerol Acyltransferases (DGAT) family genes influencing Milk production in Buffalo. BMC Genet. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schennink, A.; Stoop, W.M.; Visker, M.W.; Heck, J.M.; Bovenhuis, H.; Van Der Poel, J.J.; Van Valenberg, H.J.; Van Arendonk, J.A. DGAT1 underlies large genetic variation in milk-fat composition of dairy cows. Anim. Genet. 2007, 38, 467–473. [Google Scholar] [CrossRef]
- Thaller, G.; Kühn, C.; Winter, A.; Ewald, G.; Bellmann, O.; Wegner, J.; Zühlke, H.; Fries, R. DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Anim. Genet. 2003, 34, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Krämer, W.; Werner, F.A.; Kollers, S.; Kata, S.; Durstewitz, G.; Buitkamp, J.; Womack, J.E.; Thaller, G.; Fries, R. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA: Diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 2002, 99, 9300–9305. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Iannaccone, M.; Macciotta, N.; Guo, Y.; Di Stasio, L.; Pauciullo, A. The single nucleotide polymorphism g. 133A> C in the stearoyl CoA desaturase gene (SCD) promoter affects gene expression and quali-quantitative properties of river buffalo milk. J. Dairy Sci. 2019, 102, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Schennink, A.; Heck, J.M.; Bovenhuis, H.; Visker, M.H.; van Valenberg, H.J.; van Arendonk, J.A. Milk fatty acid unsaturation: Genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA: Diacylglycerol acyltransferase 1 (DGAT1). J. Dairy Sci. 2008, 91, 2135–2143. [Google Scholar] [CrossRef]
- Guo, Y.; Wei, Z.; Zhang, Y.; Cao, J. Research progress on the mechanism of milk fat synthesis in cows and the effect of conjugated linoleic acid on milk fat metabolism and its underlying mechanism: A Review. Animals 2024, 14, 204. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Gaspa, G.; Iannaccone, M.; Macciotta, N.; Chemello, G.; Di Stasio, L.; Pauciullo, A. Sequencing of lipoprotein lipase gene in the Mediterranean river buffalo identified novel variants affecting gene expression. J. Dairy Sci. 2020, 103, 6374–6382. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Nicolae, I.; Bota, A.; Guo, Y.; Di Stasio, L.; Pauciullo, A. Transcript analysis at DGAT1 reveals different mRNA profiles in river buffaloes with extreme phenotypes for milk fat. J. Dairy Sci. 2017, 100, 8265–8276. [Google Scholar] [CrossRef]
- Everitt, B.S.; Landau, S.; Leese, M. Cluster Analysis; Wiley: Hoboken, NJ, USA, 2001. [Google Scholar]
- Sarataphan, N.; Narongwanichgarn, W.; Maneerat, S. Phylogenetic analysis of a Thai wild water buffalo (Bubalus arnee) through mitochondrial control region. Int. J. Conserv. Sci. 2017, 8, 105. [Google Scholar]
- Flamand, J.; Vankan, D.; Gairhe, K.; Duong, H.; Barker, J. Genetic identification of wild Asian water buffalo in Nepal. In Animal Conservation Forum; Cambridge University Press: New York, NY, USA, 2003; pp. 265–270. [Google Scholar]
- Sun, T.; Shen, J.; Achilli, A.; Chen, N.; Chen, Q.; Dang, R.; Zheng, Z.; Zhang, H.; Zhang, X.; Wang, S. Genomic analyses reveal distinct genetic architectures and selective pressures in buffaloes. Gigascience 2020, 9, giz166. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Ma, Y.; Ma, J.; Xiao, J.; Liu, Y.; Liu, S.; Khan, A.; Khan, I.M.; Cao, Z. Association of DGAT1 with cattle, buffalo, goat, and sheep milk and meat production traits. Front. Vet. Sci. 2021, 8, 712470. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Hofer, T.; Foll, M. Detecting loci under selection in a hierarchically structured population. Heredity 2009, 103, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Moioli, B.; Borghese, A. Buffalo breeds and management systems. Buffalo Prod. Res. 2005, 51–76. [Google Scholar]
- Borghese, A.; Chiariotti, A.; Barile, V.L. Buffalo in the world: Situation and perspectives. In Biotechnological Applications in Buffalo Research; Springer: New York, NY, USA, 2022; pp. 3–31. [Google Scholar]
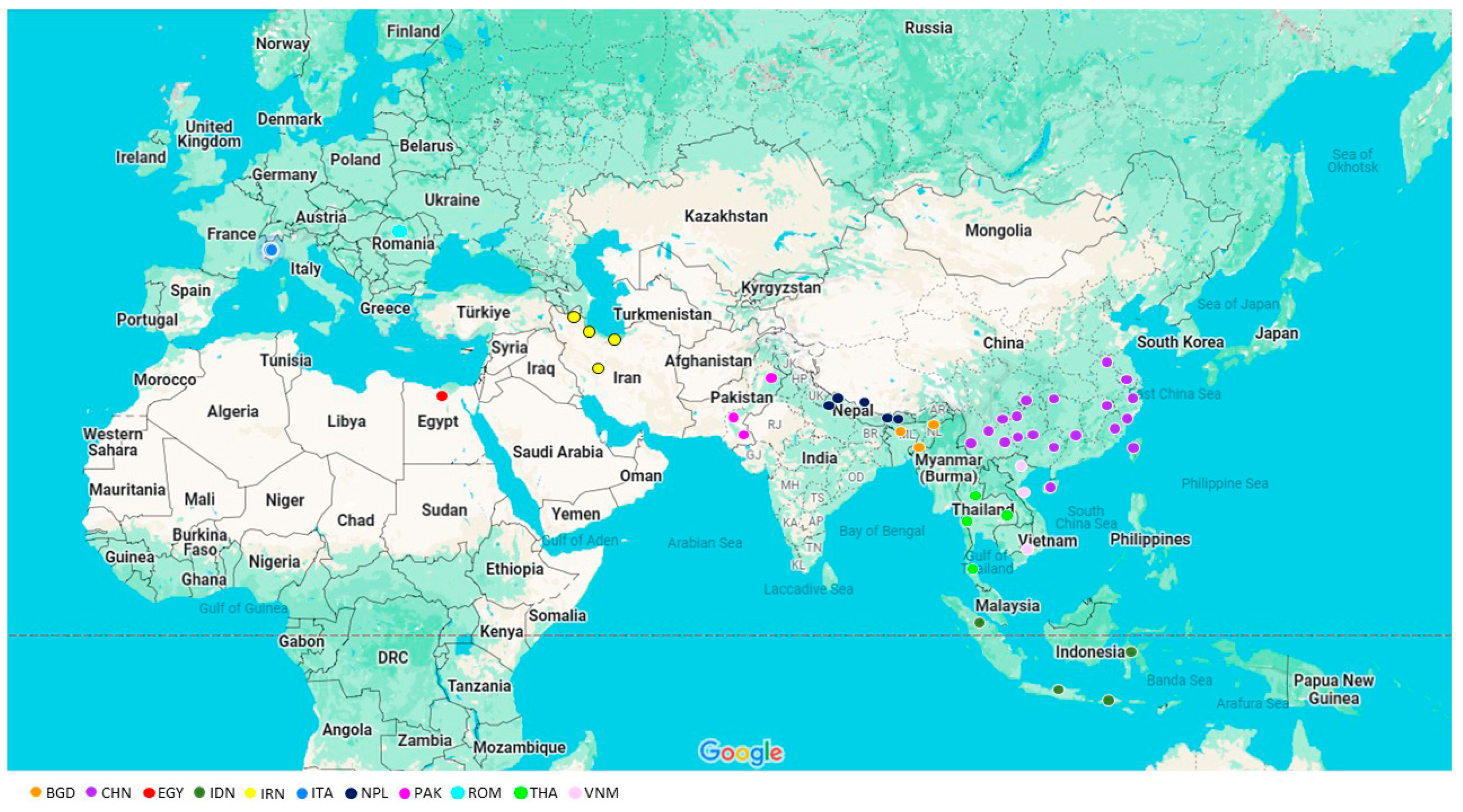
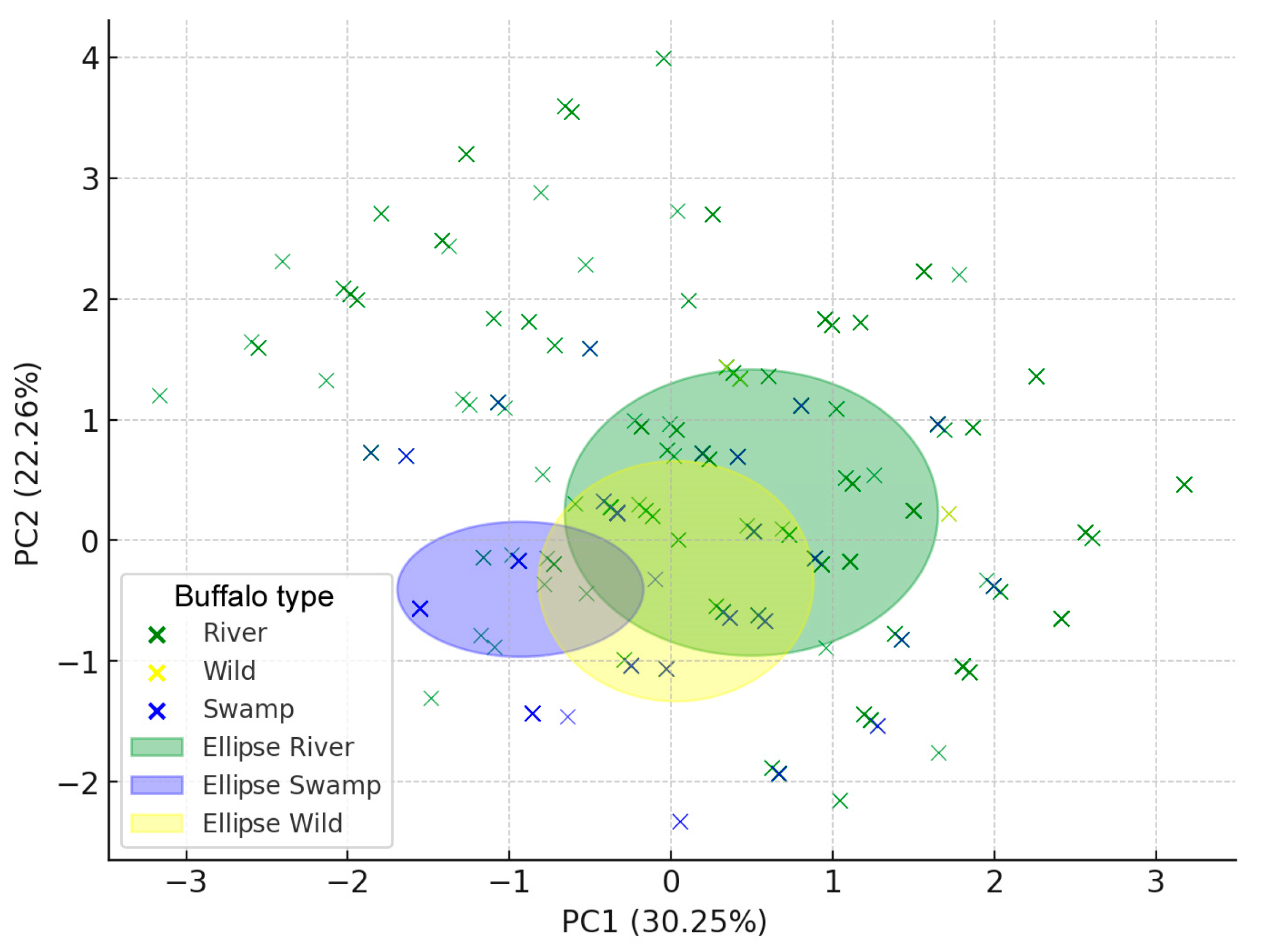

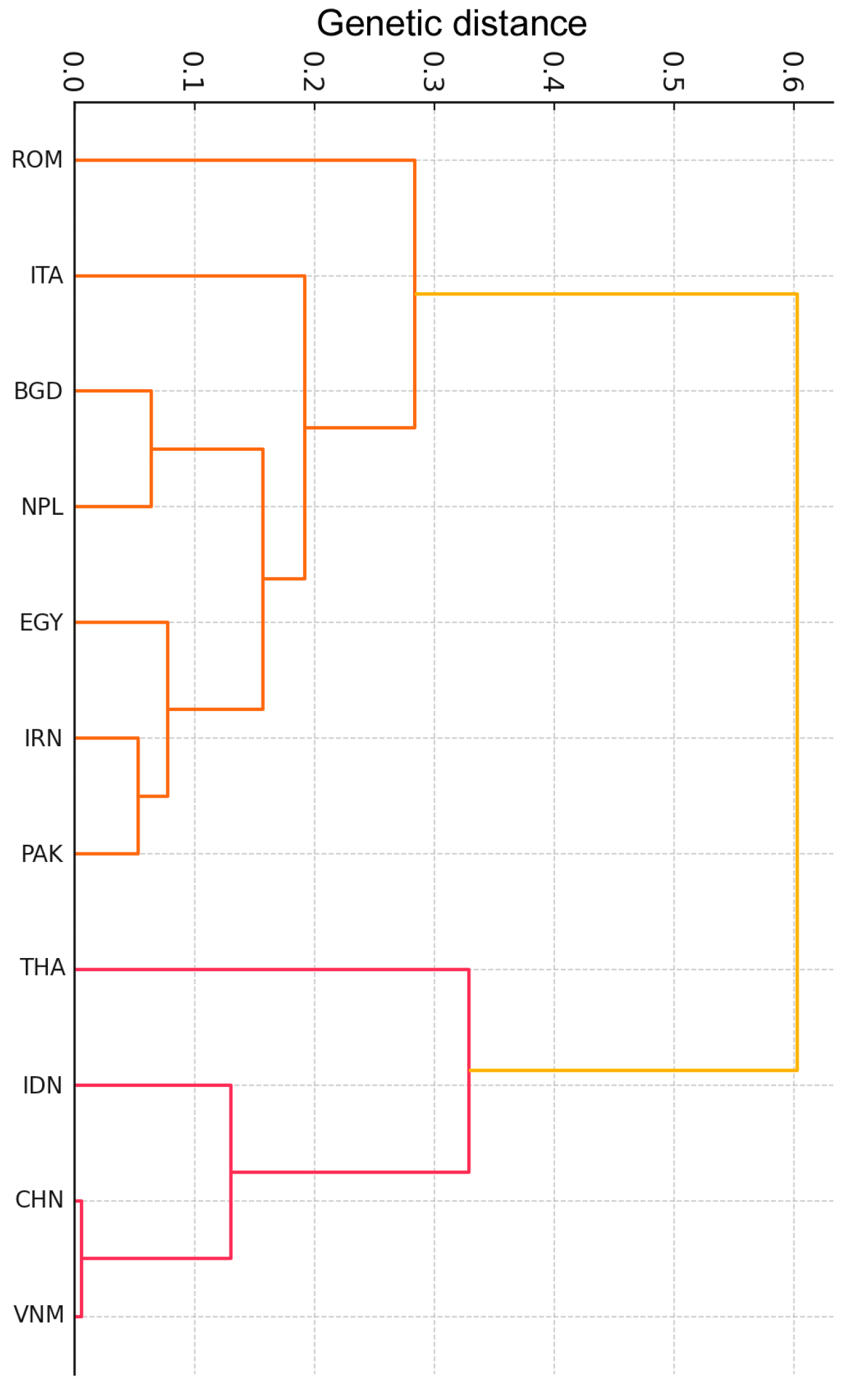
| Chr. | Genome Position | Gene | Location | Reference Allele 5′→3′ | Mutation | Genotyping Method | Endonuclease | Reference |
|---|---|---|---|---|---|---|---|---|
| 3 | 129,635,007 | LPL | Exon 1 | G | A | RFLP | DdeI | Gu et al. [29] |
| 7 | 32,148,856 | CSN1S1 | Exon 17 | A | G | ACRS | Mbo I | Pauciullo et al. [19] |
| 7 | 31,917,000 | CSN3 | Exon 4 | A | G | ACRS | Hinf I | Pauciullo et al. [19] |
| 15 | 81,685,203 | DGAT1 | Exon 13 | G | A | RFLP | DdeI | Gu et al. [30] |
| 23 | 21,066,603 | SCD | Promoter | C | A | RFLP | TaqI | Gu et al. [26] |
| SNP | Allele | Population | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BGD | CHN | EGY | IDN | IRN | ITA | NPL | PAK | ROM | THA | VNM | All | ||
| (45) | (149) | (19) | (56) | (132) | (232) | (53) | (27) | (98) | (91) | (15) | (918) | ||
| LPL g.129,635,007G>A | G | 0.822 | 0.225 | 0.947 | 0.096 | 0.943 | 0.567 | 0.868 | 0.944 | 0.954 | 0.423 | 0.233 | 0.607 |
| A | 0.178 | 0.775 | 0.053 | 0.904 | 0.057 | 0.433 | 0.132 | 0.056 | 0.046 | 0.577 | 0.767 | 0.393 | |
| χ2 | 3.080 | 0.537 | 0.028 | 10.061 | 0.445 | 2.095 | 1.933 | 0.022 | 0.200 | 14.404 | 0.168 | 152.165 | |
| PHW | 0.07925 | 0.46333 | 0.86577 | 0.00151 | 0.50448 | 0.14779 | 0.16441 | 0.88149 | 0.65408 | 0.00015 | 0.68132 | 0.00000 | |
| CSN1S1 g.32,148,856A>G | A | 0.211 | 0.000 | 0.474 | 0.000 | 0.333 | 0.306 | 0.208 | 0.222 | 0.194 | 0.115 | 0.000 | 0.196 |
| G | 0.789 | 1.000 | 0.526 | 1.000 | 0.667 | 0.694 | 0.792 | 0.778 | 0.806 | 0.885 | 1.000 | 0.804 | |
| χ2 | 1.042 | - | 0.634 | - | 0.030 | 2.768 | 0.455 | 0.003 | 2.426 | 0.028 | - | 30.015 | |
| PHW | 0.30739 | - | 0.42587 | - | 0.86154 | 0.09615 | 0.49966 | 0.95699 | 0.11932 | 0.86715 | - | 0.00000 | |
| CSN3 g.31,917,000A>G | A | 0.178 | 0.000 | 0.211 | 0.000 | 0.356 | 0.422 | 0.283 | 0.259 | 0.597 | 0.324 | 0.000 | 0.291 |
| G | 0.822 | 1.000 | 0.789 | 1.000 | 0.644 | 0.578 | 0.717 | 0.741 | 0.403 | 0.676 | 1.000 | 0.709 | |
| χ2 | 0.111 | - | 0.119 | - | 2.031 | 0.038 | 0.009 | 0.199 | 13.661 | 2.738 | - | 14.925 | |
| PHW | 0.73939 | - | 0.72935 | - | 0.15405 | 0.84515 | 0.92261 | 0.65497 | 0.00022 | 0.09795 | - | 0.00011 | |
| DGAT1 g.81,685,203G>A | G | 0.711 | 0.990 | 0.711 | 0.991 | 0.708 | 0.433 | 0.726 | 0.667 | 0.515 | 0.912 | 1.000 | 0.705 |
| A | 0.289 | 0.010 | 0.289 | 0.009 | 0.292 | 0.567 | 0.274 | 0.333 | 0.485 | 0.088 | 0.000 | 0.295 | |
| χ2 | 3.961 | 0.010 | 0.336 | 0.004 | 0.902 | 0.833 | 2.196 | 0.170 | 0.589 | 19.978 | - | 58.428 | |
| PHW | 0.04656 | 0.91940 | 0.56180 | 1.00000 | 0.34215 | 0.36148 | 0.13834 | 0.67974 | 0.44283 | 0.00004 | - | 0.00000 | |
| SCD g.21,066,603C>A | C | 0.044 | 0.000 | 0.079 | 0.000 | 0.140 | 0.246 | 0.123 | 0.167 | 0.107 | 0.033 | 0.000 | 0.113 |
| A | 0.956 | 1.000 | 0.921 | 1.000 | 0.860 | 0.754 | 0.877 | 0.833 | 0.893 | 0.967 | 1.000 | 0.887 | |
| χ2 | 0.070 | - | 0.091 | - | 1.245 | 1.067 | 0.948 | 0.825 | 1.338 | 0.088 | - | 158.077 | |
| PHW | 0.79056 | - | 0.76322 | - | 0.26443 | 0.30162 | 0.33020 | 0.36353 | 0.24739 | 0.76717 | - | 0.00000 | |
| Average heterozygosity | |||||||||||
| BGD | CHN | EGY | IDN | IRN | ITA | NPL | PAK | ROM | THA | VNM | |
| Observed | 0.244 | 0.070 | 0.274 | 0.021 | 0.320 | 0.464 | 0.294 | 0.341 | 0.355 | 0.235 | 0.067 |
| Expected | 0.286 | 0.074 | 0.306 | 0.046 | 0.334 | 0.454 | 0.318 | 0.317 | 0.316 | 0.272 | 0.074 |
| Expected heterozygosity per locus and population | |||||||||||
| BGD | CHN | EGY | IDN | IRN | ITA | NPL | PAK | ROM | THA | VNM | |
| LPL g.129,635,007G>A | 0.296 | 0.350 | 0.102 | 0.177 | 0.108 | 0.492 | 0.232 | 0.107 | 0.088 | 0.492 | 0.371 |
| CSN1S1 g.32,148,856A>G | 0.337 | 0.000 | 0.515 | 0.000 | 0.446 | 0.426 | 0.332 | 0.352 | 0.314 | 0.205 | 0.000 |
| CSN3 g.31,917,000A>G | 0.295 | 0.000 | 0.342 | 0.000 | 0.461 | 0.489 | 0.410 | 0.390 | 0.483 | 0.440 | 0.000 |
| DGAT1 g.81,685,203G>A | 0.417 | 0.020 | 0.424 | 0.052 | 0.415 | 0.492 | 0.402 | 0.453 | 0.502 | 0.162 | 0.000 |
| SCD g.21,066,603C>A | 0.086 | 0.000 | 0.149 | 0.000 | 0.242 | 0.371 | 0.217 | 0.282 | 0.192 | 0.064 | 0.000 |
| Wright’s fixation index (Fis) as a measure of heterozygote deficiency or excess | |||||||||||
| BGD | CHN | EGY | IDN | IRN | ITA | NPL | PAK | ROM | THA | VNM | |
| LPL g.129,635,007G>A | 0.254 | 0.060 | −0.029 | 0.403 | −0.056 | −0.095 | 0.186 | −0.040 | −0.043 | 0.397 | 0.103 |
| CSN1S1 g.32,148,856A>G | 0.148 | - | 0.182 | - | 0.015 | 0.109 | 0.092 | −0.053 | 0.156 | −0.017 | - |
| CSN3 g.31,917,000A>G | −0.048 | - | 0.077 | - | 0.124 | 0.013 | −0.013 | −0.139 | −0.374 | −0.173 | - |
| DGAT1 g.81,685,203G>A | 0.291 | −0.007 | 0.131 | 0.000 | 0.083 | −0.060 | 0.202 | 0.019 | −0.078 | 0.456 | - |
| SCD g.21,066,603C>A | −0.034 | - | −0.059 | - | −0.096 | −0.068 | −0.130 | −0.182 | −0.115 | −0.029 | - |
| BGD | CHN | EGY | IDN | IRN | ITA | NPL | PAK | ROM | THA | VNM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BGD | *** | NS | *** | *** | *** | NS | NS | *** | *** | *** | |
| CHN | 0.4562 | *** | NS | *** | *** | *** | *** | *** | *** | NS | |
| EGY | 0.0382 | 0.6351 | *** | NS | *** | NS | NS | *** | *** | *** | |
| IDN | 0.4912 | 0.0562 | 0.6830 | *** | *** | *** | *** | *** | *** | NS | |
| IRN | 0.0350 | 0.4640 | 0.0104 | 0.4669 | *** | NS | NS | *** | *** | *** | |
| ITA | 0.0992 | 0.3308 | 0.1163 | 0.3183 | 0.0983 | *** | *** | *** | *** | *** | |
| NPL | 0.0025 | 0.4722 | 0.0331 | 0.4879 | 0.0095 | 0.0886 | NS | *** | *** | *** | |
| PAK | 0.0095 | 0.5786 | 0.0251 | 0.6122 | 0.0024 | 0.0890 | 0.0067 | * | *** | *** | |
| ROM | 0.1259 | 0.5771 | 0.1345 | 0.5654 | 0.0619 | 0.0930 | 0.0813 | 0.0732 | *** | *** | |
| THA | 0.1410 | 0.1777 | 0.2376 | 0.2059 | 0.1918 | 0.1420 | 0.1415 | 0.1971 | 0.2598 | ** | |
| VNM | 0.2941 | 0.0194 | 0.4495 | 0.0785 | 0.3462 | 0.2521 | 0.3057 | 0.3882 | 0.4440 | 0.0999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauciullo, A.; Gaspa, G.; Versace, C.; Cosenza, G.; Piscopo, N.; Gu, M.; Coletta, A.; Hussain, T.; Seidavi, A.; Nicolae, I.; et al. New Insights into Genetic Diversity and Differentiation of 11 Buffalo Populations Using Validated SNPs for Dairy Improvement. Genes 2025, 16, 400. https://doi.org/10.3390/genes16040400
Pauciullo A, Gaspa G, Versace C, Cosenza G, Piscopo N, Gu M, Coletta A, Hussain T, Seidavi A, Nicolae I, et al. New Insights into Genetic Diversity and Differentiation of 11 Buffalo Populations Using Validated SNPs for Dairy Improvement. Genes. 2025; 16(4):400. https://doi.org/10.3390/genes16040400
Chicago/Turabian StylePauciullo, Alfredo, Giustino Gaspa, Carmine Versace, Gianfranco Cosenza, Nadia Piscopo, Meichao Gu, Angelo Coletta, Tanveer Hussain, Alireza Seidavi, Ioana Nicolae, and et al. 2025. "New Insights into Genetic Diversity and Differentiation of 11 Buffalo Populations Using Validated SNPs for Dairy Improvement" Genes 16, no. 4: 400. https://doi.org/10.3390/genes16040400
APA StylePauciullo, A., Gaspa, G., Versace, C., Cosenza, G., Piscopo, N., Gu, M., Coletta, A., Hussain, T., Seidavi, A., Nicolae, I., Kovitvadhi, A., Liu, Q., Shang, J., Si, J., Dai, D., & Zhang, Y. (2025). New Insights into Genetic Diversity and Differentiation of 11 Buffalo Populations Using Validated SNPs for Dairy Improvement. Genes, 16(4), 400. https://doi.org/10.3390/genes16040400










