Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethic Statement
2.2. Animals and Study Design
2.3. Acquisition of Biological Samples and RNA Seq Pre-Processing
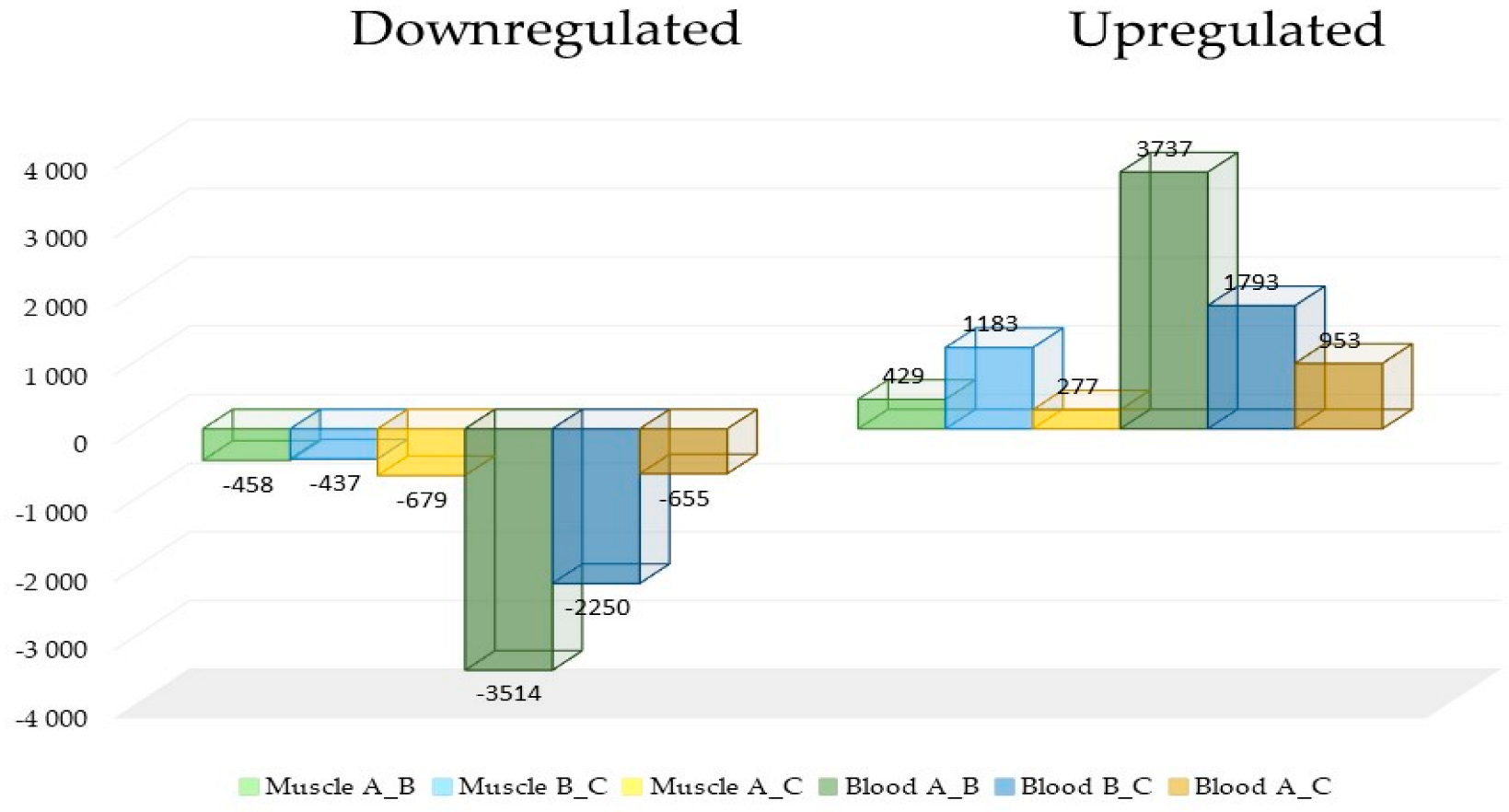
3. Results
Identification and Analysis of DEGs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Librado, P.; Khan, N.; Fages, A.; Kusliy, M.A.; Suchan, T.; Tonasso-Calvière, L.; Schiavinato, S.; Alioglu, D.; Fromentier, A.; Perdereau, A.; et al. The Origins and Spread of Domestic Horses from the Western Eurasian Steppes. Nature 2021, 598, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Anthony, D.W.; Brown, D.R. The Secondary Products Revolution, Horse-Riding, and Mounted Warfare. J. World Prehistory 2011, 24, 131–160. [Google Scholar] [CrossRef]
- McGivney, B.A.; McGettigan, P.A.; Browne, J.A.; Evans, A.C.O.; Fonseca, R.G.; Loftus, B.J.; Lohan, A.; MacHugh, D.E.; Murphy, B.A.; Katz, L.M.; et al. Characterization of the Equine Skeletal Muscle Transcriptome Identifies Novel Functional Responses to Exercise Training. BMC Genom. 2010, 11, 398. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Musiał, A.D.; Velie, B.D. The Genetics of Racing Performance in Arabian Horses. Int. J. Genom. 2019, 2019, 9013239. [Google Scholar] [CrossRef]
- Silva, A.P.A.; Curi, R.A.; Langlois, B.; Silva, J.A.I.V. Genetic Parameters for Earnings in Quarter Horse. Genet. Mol. Res. 2014, 13, 5840–5848. [Google Scholar] [CrossRef]
- Velie, B.D.; Jäderkvist Fegraeus, K.; Ihler, C.F.; Lindgren, G.; Strand, E. Competition Lifespan Survival Analysis in the Norwegian-Swedish Coldblooded Trotter Racehorse. Equine Vet. J. 2019, 51, 206–211. [Google Scholar] [CrossRef]
- Hill, E.W.; Gu, J.; Eivers, S.S.; Fonseca, R.G.; Mcgivney, B.A. A Sequence Polymorphism in MSTN Predicts Sprinting Ability and Racing Stamina in Thoroughbred Horses. PLoS ONE 2010, 5, 8645. [Google Scholar] [CrossRef]
- Gu, J.; Orr, N.; Park, S.D.; Katz, L.M.; Sulimova, G.; MacHugh, D.E.; Hill, E.W. A Genome Scan for Positive Selection in Thoroughbred Horses. PLoS ONE 2009, 4, 5767. [Google Scholar] [CrossRef]
- Capomaccio, S.; Vitulo, N.; Verini-Supplizi, A.; Barcaccia, G.; Albiero, A.; D’Angelo, M.; Campagna, D.; Valle, G.; Felicetti, M.; Silvestrelli, M.; et al. RNA Sequencing of the Exercise Transcriptome in Equine Athletes. PLoS ONE 2013, 8, e83504. [Google Scholar] [CrossRef]
- Mach, N.; Plancade, S.; Pacholewska, A.; Lecardonnel, J.; Rivière, J.; Moroldo, M.; Vaiman, A.; Morgenthaler, C.; Beinat, M.; Nevot, A.; et al. Integrated MRNA and MiRNA Expression Profiling in Blood Reveals Candidate Biomarkers Associated with Endurance Exercise in the Horse. Sci. Rep. 2016, 6, 22932. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Zukowski, K.; Piórkowska, K.; Gurgul, A.; Bugno-Poniewierska, M. Transcriptome Profiling of Arabian Horse Blood during Training Regimens. BMC Genet. 2017, 18, 31. [Google Scholar] [CrossRef]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Spinsanti, G.; Silvestrelli, M.; Supplizi, A.V. Exercise Induced Stress in Horses: Selection of the Most Stable Reference Genes for Quantitative RT-PCR Normalization. BMC Mol. Biol. 2008, 9, 49. [Google Scholar] [CrossRef]
- Ricard, A.; Robert, C.; Blouin, C.; Baste, F.; Torquet, G.; Morgenthaler, C.; Rivière, J.; Mach, N.; Mata, X.; Schibler, L.; et al. Endurance Exercise Ability in the Horse: A Trait with Complex Polygenic Determinism. Front. Genet. 2017, 8, 89. [Google Scholar] [CrossRef]
- Gurgul, A.; Jasielczuk, I.; Semik-Gurgul, E.; Pawlina-Tyszkoid, K.; Stefaniuk-Szmukierid, M.; Szmatoła, T.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. A Genome-Wide Scan for Diversifying Selection Signatures in Selected Horse Breeds. PLoS ONE 2019, 14, e0210751. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Żukowski, K.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-Induced Modification of the Skeletal Muscle Transcriptome in Arabian Horses. Physiol. Genom. 2017, 49, 318–326. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Piórkowska, K.; Szmatoła, T.; Bugno-Poniewierska, M. Molecular Characterization of the Apoptosis-Related SH3RF1 and SH3RF2 Genes and Their Association with Exercise Performance in Arabian Horses. BMC Vet. Res. 2018, 14, 237. [Google Scholar] [CrossRef]
- Mach, N.; Ramayo-Caldas, Y.; Clark, A.; Moroldo, M.; Robert, C.; Barrey, E.; López, J.M.; Le Moyec, L. Understanding the Response to Endurance Exercise Using a Systems Biology Approach: Combining Blood Metabolomics, Transcriptomics and MiRNomics in Horses. BMC Genom. 2017, 18, 187. [Google Scholar] [CrossRef]
- Librado, P.; Fages, A.; Gaunitz, C.; Leonardi, M.; Wagner, S.; Khan, N.; Hanghøj, K.; Alquraishi, S.A.; Alfarhan, A.H.; Al-Rasheid, K.A.; et al. The Evolutionary Origin and Genetic Makeup of Domestic Horses. Genetics 2016, 204, 423–434. [Google Scholar] [CrossRef]
- Klecel, W.; Drobik-Czwarno, W.; Martyniuk, E. Judging the Arabian Beauty: What Are the Relationships Between Different Scoring Categories? J. Equine Vet. Sci. 2023, 123, 104247. [Google Scholar] [CrossRef]
- Prince, A.; Geor, R.; Harris, P.; Hoekstra, K.; Gardner, S.; Hudson, C.; Pagan, J. Comparison of the Metabolic Responses of Trained Arabians and Thoroughbreds during High- and Low-Intensity Exercise. Equine Vet. J. 2002, 34 (Suppl. S34), 95–99. [Google Scholar] [CrossRef]
- Stefaniuk-Szmukier, M.; Ropka-Molik, K.; Piórkowska, K.; Żukowski, K.; Bugno-Poniewierska, M. Transcriptomic Hallmarks of Bone Remodelling Revealed by RNA-Seq Profiling in Blood of Arabian Horses during Racing Training Regime. Gene 2018, 676, 256–262. [Google Scholar] [CrossRef]
- Chomczynski, P. A Reagent for the Single-Step Simultaneous Isolation of RNA, DNA and Proteins from Cell and Tissue Samples. Biotechniques 1993, 15, 532–537. [Google Scholar] [PubMed]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR-Flexible Barcode and Adapter Processing for next-Generation Sequencing Platforms. Biology 2012, 1, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 002832. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cosgrove, E.J.; Sadeghi, R.; Schlamp, F.; Holl, H.M.; Moradi-Shahrbabak, M.; Miraei-Ashtiani, S.R.; Abdalla, S.; Shykind, B.; Troedsson, M.; Stefaniuk-Szmukier, M.; et al. Genome Diversity and the Origin of the Arabian Horse. Sci. Rep. 2020, 10, 9702. [Google Scholar] [CrossRef]
- Rivero, J.L.L.; Serrano, A.L.; Henckel, P.; Aguera, E. Muscle Fiber Type Composition and Fiber Size in Successfully and Unsuccessfully Endurance-Raced Horses. J. Appl. Physiol. 1993, 75, 1758–1766. [Google Scholar] [CrossRef]
- Bean, C.; Verma, N.K.; Yamamoto, D.L.; Chemello, F.; Cenni, V.; Filomena, M.C.; Chen, J.; Bang, M.L.; Lanfranchi, G. Ankrd2 Is a Modulator of NF-ΚB-Mediated Inflammatory Responses during Muscle Differentiation. Cell Death Dis. 2014, 5, e1002. [Google Scholar] [CrossRef]
- Cenni, V.; Kojic, S.; Capanni, C.; Faulkner, G.; Lattanzi, G. Ankrd2 in Mechanotransduction and Oxidative Stress Response in Skeletal Muscle: New Cues for the Pathogenesis of Muscular Laminopathies. Oxidative Med. Cell. Longev. 2019, 2019, 7318796. [Google Scholar]
- Lord, S.O.; Lai, Y.C. Exercise Mediates Ubiquitin Signalling in Human Skeletal Muscle. FASEB BioAdv. 2022, 4, 402–407. [Google Scholar] [CrossRef]
- Carraro, F.; Stuart, C.A.; Hartl, W.H.; Rosenblatt, J.; Wolfe, R.R. Effect of Exercise and Recovery on Muscle Protein Synthesis in Human Subjects. Am. J. Physiol.-Endocrinol. Metab. 1990, 259, E470–E476. [Google Scholar] [CrossRef]
- Ramirez-Martinez, A.; Cenik, B.K.; Bezprozvannaya, S.; Chen, B.; Bassel-Duby, R.; Liu, N.; Olson, E.N. KLHL41 Stabilizes Skeletal Muscle Sarcomeres by Nonproteolytic Ubiquitination. Elife 2017, 6, e26439. [Google Scholar] [CrossRef]
- Yi, J.S.; Park, J.S.; Ham, Y.M.; Nguyen, N.; Lee, N.R.; Hong, J.; Kim, B.W.; Lee, H.; Lee, C.S.; Jeong, B.C.; et al. MG53-Induced IRS-1 Ubiquitination Negatively Regulates Skeletal Myogenesis and Insulin Signaling. Nat. Commun. 2013, 4, 2354. [Google Scholar] [CrossRef]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 Controls the Levels of P53 by Regulating the Deubiquitination Activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, G.; Zhang, W.; Qin, B.; Ye, Z.; Shi, H.; Zhao, X.; Chen, Y.; Song, B.; Mei, Z.; et al. USP13: Multiple Functions and Target Inhibition. Front. Cell Dev. Biol. 2022, 10, 875124. [Google Scholar] [CrossRef]
- Halaby, M.J.; Hakem, R.; Hakem, A. Pirh2: An E3 Ligase with Central Roles in the Regulation of Cell Cycle, DNA Damage Response, and Differentiation. Cell Cycle 2013, 12, 2733. [Google Scholar] [CrossRef] [PubMed]
- Kakihara, Y.; Houry, W.A. The R2TP Complex: Discovery and Functions. Biochim. Biophys. Acta-Mol. Cell Res. 2012, 1823, 101–107. [Google Scholar] [CrossRef]
- Inoue, M.; Saeki, M.; Egusa, H.; Niwa, H.; Kamisaki, Y. PIH1D1, a Subunit of R2TP Complex, Inhibits Doxorubicin-Induced Apoptosis. Biochem. Biophys. Res. Commun. 2010, 403, 340–344. [Google Scholar] [CrossRef]
- Kamano, Y.; Saeki, M.; Egusa, H.; Kakihara, Y.; Houry, W.A.; Yatani, H.; Kamisaki, Y. PIH1D1 Interacts with MTOR Complex 1 and Enhances Ribosome RNA Transcription. FEBS Lett. 2013, 587, 3303–3308. [Google Scholar] [CrossRef]
- Layman, D.K. Role of Leucine in Protein Metabolism During Exercise and Recovery. Artic. Can. J. Appl. Physiol. 2003, 27, 646–662. [Google Scholar] [CrossRef]
- Garlick, P.J. The Role of Leucine in the Regulation of Protein Metabolism. J. Nutr. 2005, 135, 1553S–1556S. [Google Scholar] [CrossRef] [PubMed]
- Holecek, M. Branched-Chain Amino Acids and Ammonia Metabolism in Liver Disease: Therapeutic Implications. Nutrition 2013, 29, 1186–1191. [Google Scholar] [CrossRef]
- Mann, G.; Mora, S.; Madu, G.; Adegoke, O.A.J. Branched-Chain Amino Acids: Catabolism in Skeletal Muscle and Implications for Muscle and Whole-Body Metabolism. Front. Physiol. 2021, 12, 702826. [Google Scholar] [CrossRef]
- Myćka, G.; Ropka-Molik, K.; Cywińska, A.; Szmatoła, T.; Stefaniuk-Szmukier, M. Molecular Insights into the Lipid-Carbohydrates Metabolism Switch under the Endurance Effort in Arabian Horses. Equine Vet. J. 2024, 56, 586–597. [Google Scholar] [CrossRef]
- Lee, S.M.; Lee, S.H.; Jung, Y.; Lee, Y.; Yoon, J.H.; Choi, J.Y.; Hwang, C.Y.; Son, Y.H.; Park, S.S.; Hwang, G.S.; et al. FABP3-Mediated Membrane Lipid Saturation Alters Fluidity and Induces ER Stress in Skeletal Muscle with Aging. Nat. Commun. 2020, 11, 5661. [Google Scholar] [CrossRef]
- Kusudo, T.; Kontani, Y.; Kataoka, N.; Ando, F.; Shimokata, H.; Yamashita, H. Fatty Acid-Binding Protein 3 Stimulates Glucose Uptake by Facilitating AS160 Phosphorylation in Mouse Muscle Cells. Genes Cells 2011, 16, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, B.; Flück, M.; Décombaz, J.; Kreis, R.; Boesch, C.; Wittwer, M.; Graber, F.; Vogt, M.; Howald, H.; Hoppeler, H. Transcriptional Adaptations of Lipid Metabolism in Tibialis Anterior Muscle of Endurance-Trained Athletes. Physiol. Genom. 2004, 15, 148–157. [Google Scholar] [CrossRef]
- Sorichter, S. Early Assesment of Exercise Induced Skeletal Muscle Injury Using Plasma Fatty Acid Binding Protein. Br. J. Sports Med. 1998, 32, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Neppl, R.L.; Kataok, M.; Wang, D.Z. Crystallin-AB Regulates Skeletal Muscle Homeostasis via Modulation of Argonaute2 Activity. J. Biol. Chem. 2014, 289, 17240–17248. [Google Scholar] [CrossRef]
- Jacko, D.; Bersiner, K.; Schulz, O.; Przyklenk, A.; Spahiu, F.; Höhfeld, J.; Bloch, W.; Gehlert, S. Coordinated Alpha-Crystallin B Phosphorylation and Desmin Expression Indicate Adaptation and Deadaptation to Resistance Exercise-Induced Loading in Human Skeletal Muscle. Am. J. Physiol. Cell Physiol. 2020, 319, C300–C312. [Google Scholar] [CrossRef]
- Jacko, D.; Bersiner, K.; Hebchen, J.; De Marées, M.; Bloch, W.; Gehlert, S. Phosphorylation of AB-Crystallin and Its Cytoskeleton Association Differs in Skeletal Myofiber Types Depending on Resistance Exercise Intensity and Volume. J. Appl. Physiol. 2019, 126, 1607–1618. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefaniuk-Szmukier, M.; Szmatoła, T.; Ropka-Molik, K. Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle. Genes 2025, 16, 431. https://doi.org/10.3390/genes16040431
Stefaniuk-Szmukier M, Szmatoła T, Ropka-Molik K. Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle. Genes. 2025; 16(4):431. https://doi.org/10.3390/genes16040431
Chicago/Turabian StyleStefaniuk-Szmukier, Monika, Tomasz Szmatoła, and Katarzyna Ropka-Molik. 2025. "Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle" Genes 16, no. 4: 431. https://doi.org/10.3390/genes16040431
APA StyleStefaniuk-Szmukier, M., Szmatoła, T., & Ropka-Molik, K. (2025). Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle. Genes, 16(4), 431. https://doi.org/10.3390/genes16040431




