GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Description of GC and ENC Histograms in Bees (Apis mellifera), Rice (Oryza sativa), and Yeast (Saccharomyces cerevisiae)
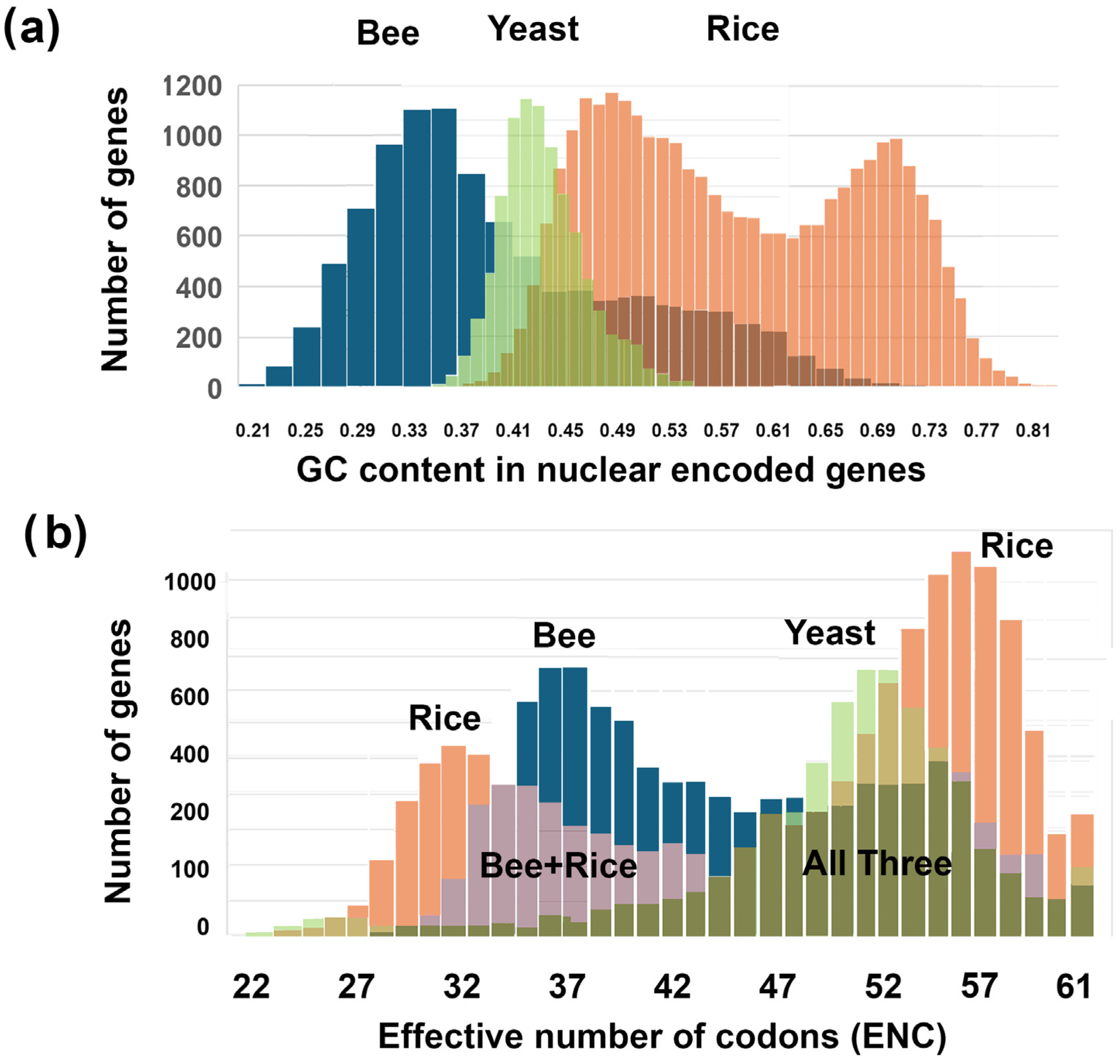
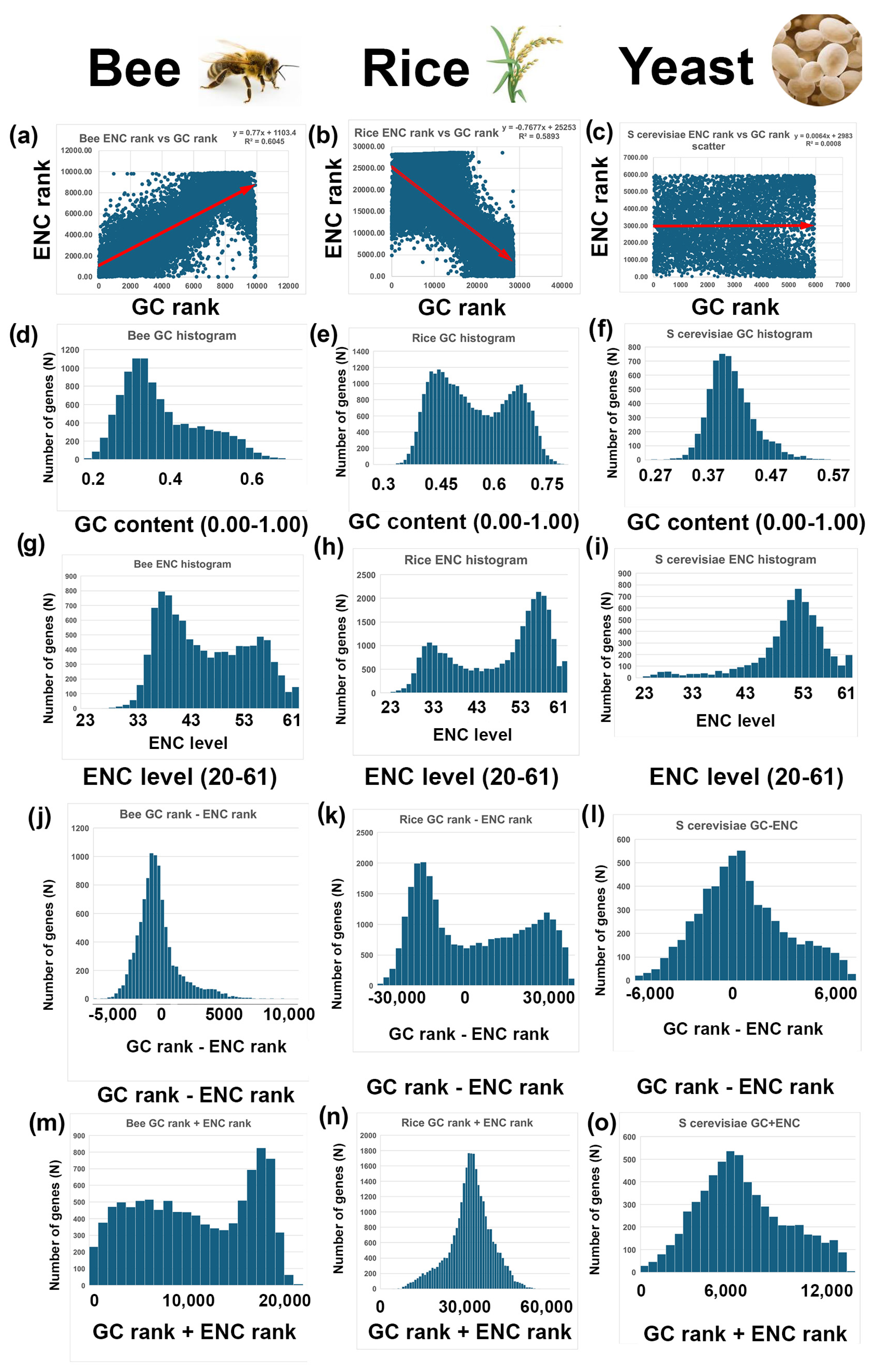
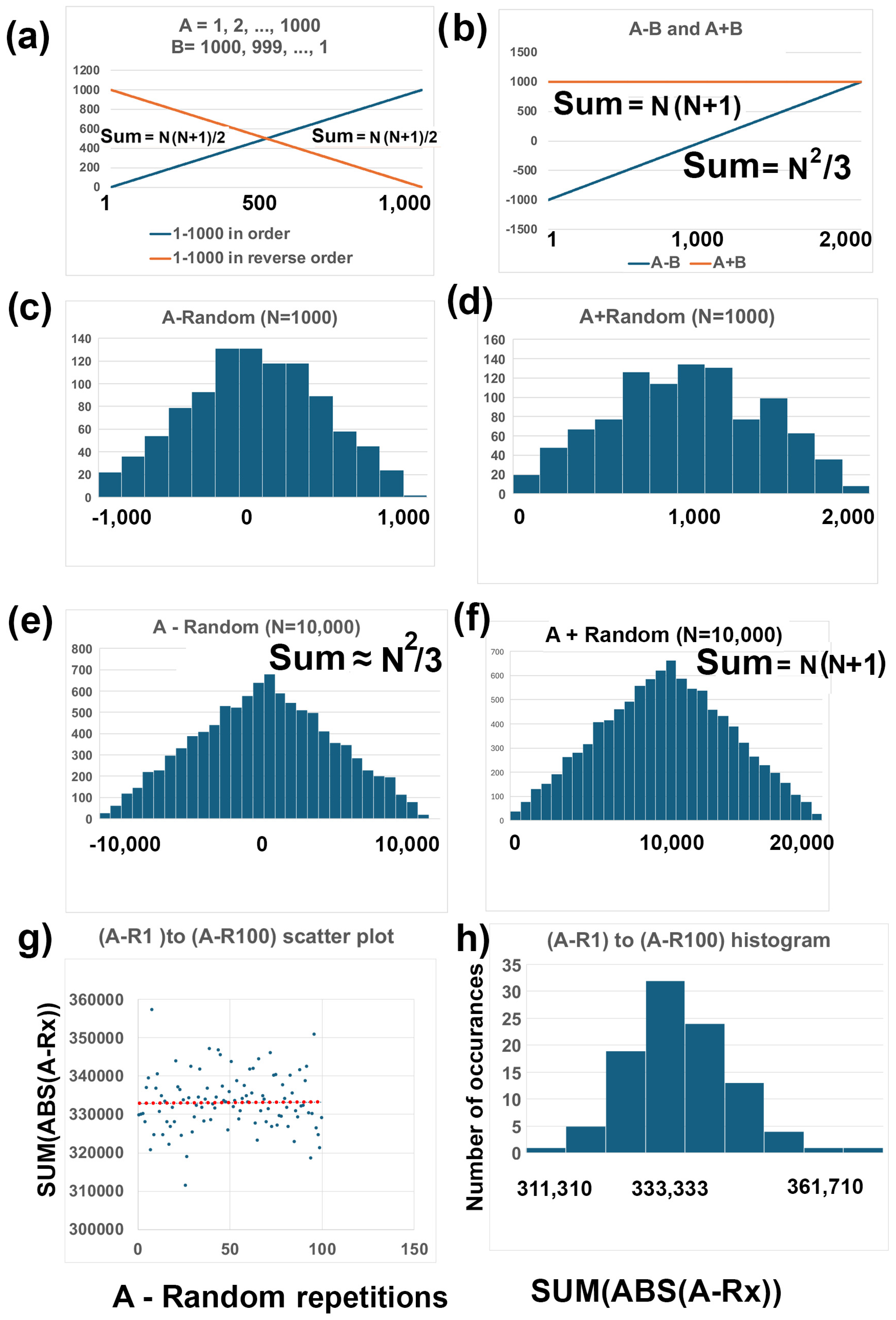
3.2. Description of Two-Rank Order Normalization (TRON) Mathematics
3.3. Comparisons Between GC and ENC Distributions
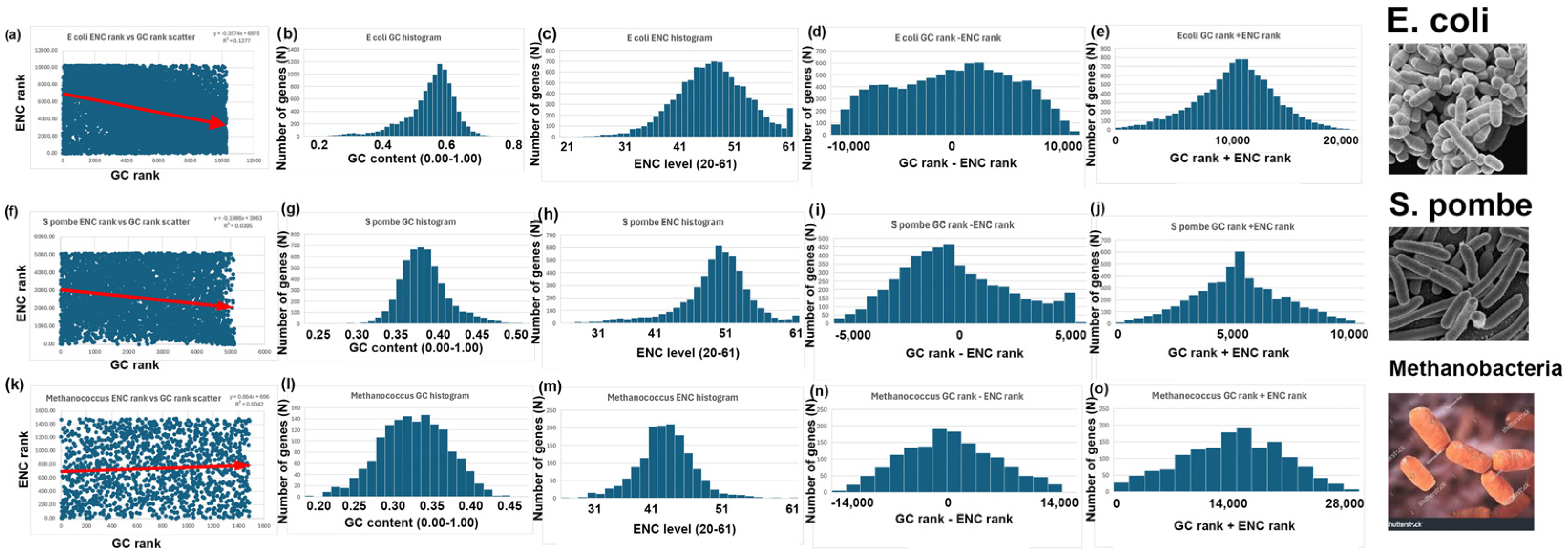
- GC and ENC distributions are positively correlated in bees (Apis mellifera) and negatively correlated in rice (Oryza sativa). There is little or no correlation between GC and ENC in yeast (Saccharomyces cerevisiae).
- 2.
- Rice (Oryza sativa) has bimodal GC and ENC distributions, GC-ENC is a bimodal distribution, and GC+ENC is a unimodal distribution; (2 − 2 = 2; 2 + 2 = 1).
- 3.
- The bee (Apis mellifera) has bimodal GC and ENC distributions, GC-ENC has a unimodal distribution, and GC+ENC forms a bimodal distribution (2 − 2 = 1; 2 + 2 = 2)
- 4.
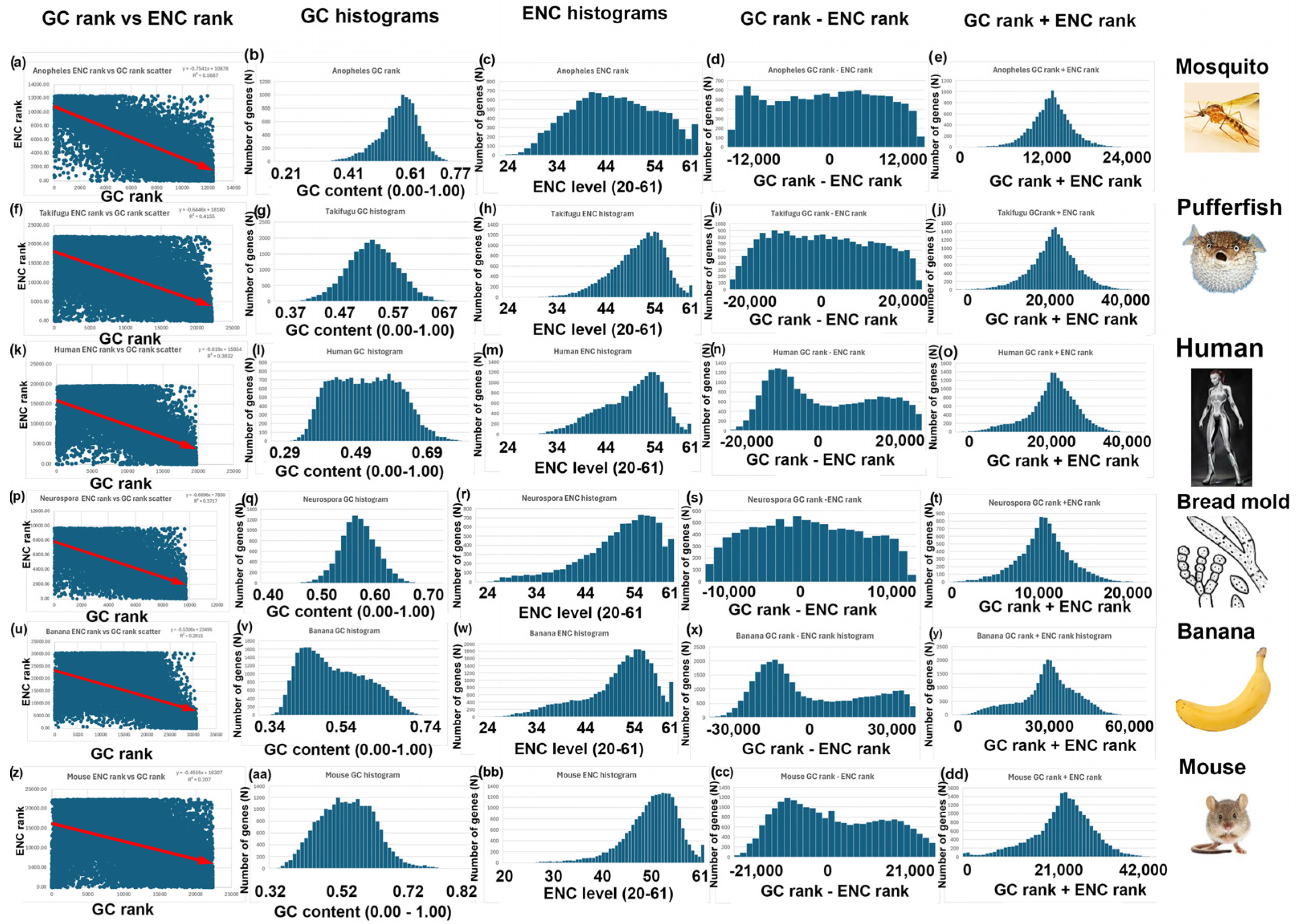
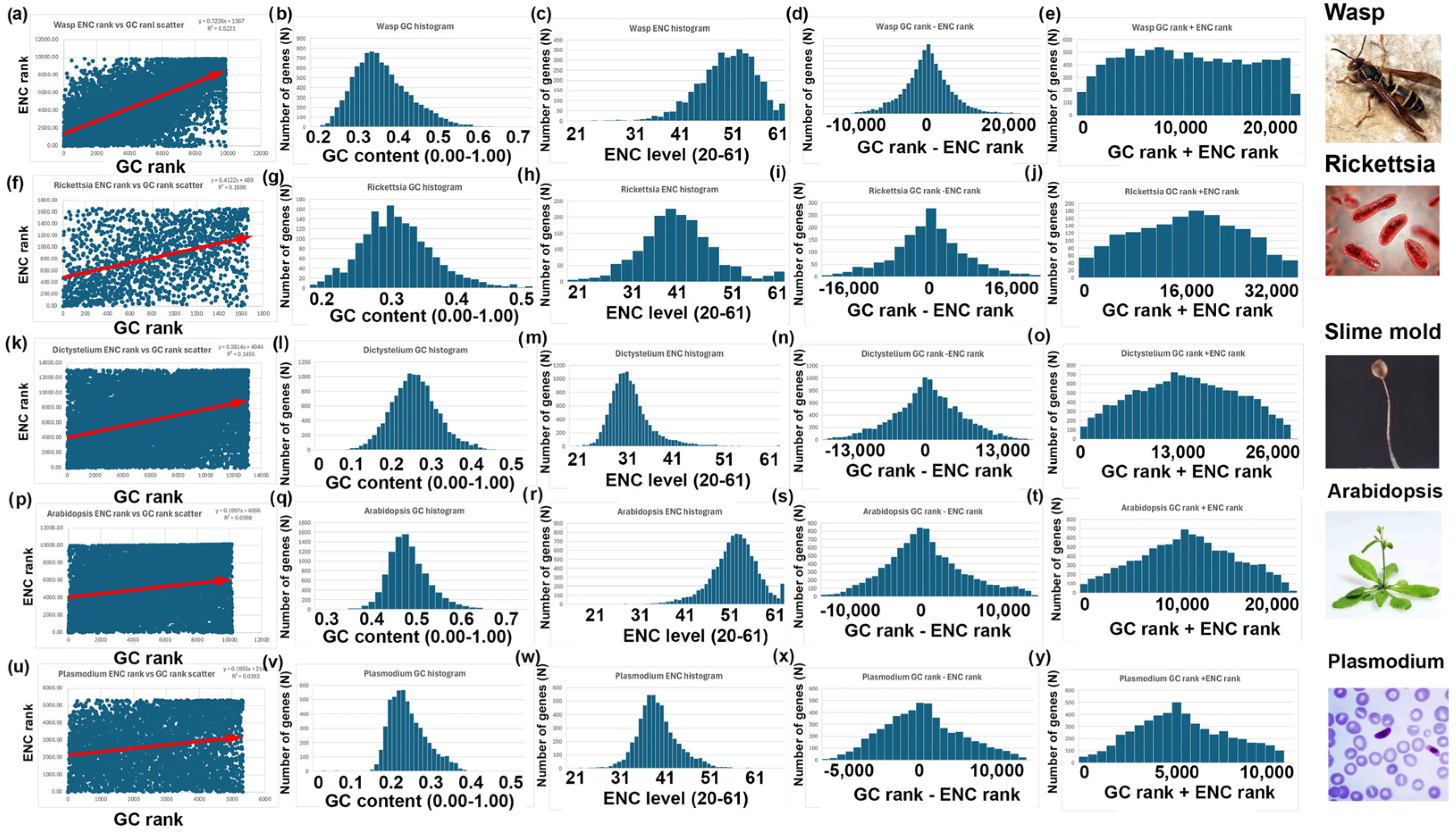
- I repeated the analyses described above for bees, rice, and yeast with 14 other species. Six additional species have a negative correlation between GC and ENC, as I found with bees (Figure 3). Five additional species have a positive correlation between GC and ENC, as I found with rice (Figure 4). Three additional species have little or no correlation between GC and ENC, as I found with yeast (Figure 5).
- 5.
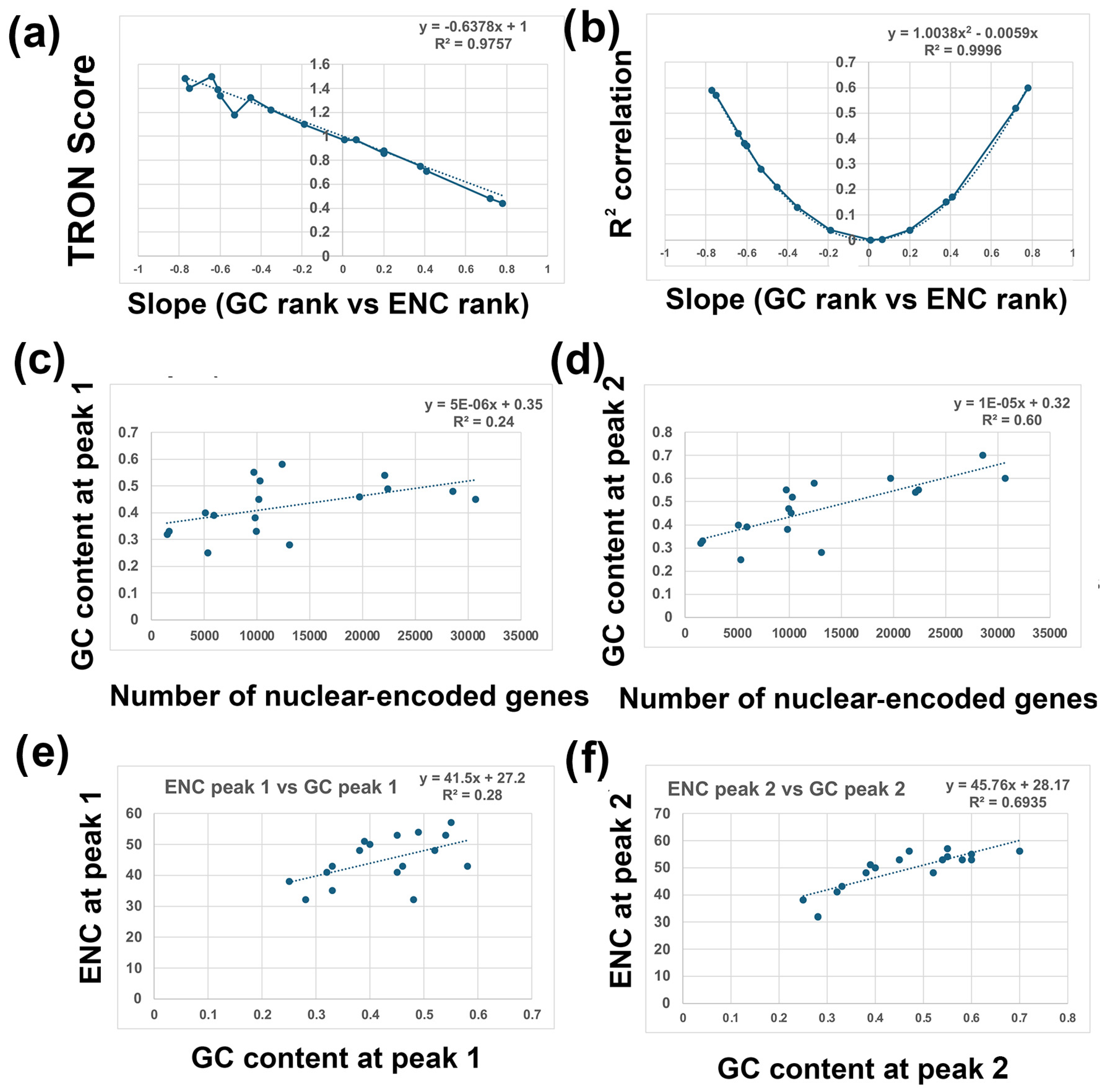
- Two-Rank Order Normalization (TRON) was plotted for the 17 species analyzed in this study (SUM(ABS(GC-ENC))/(N2/3)). Table 1 is a summary of several of the variables that were used to make Figure 7. I compared all the variables with each other and highlighted the strongest correlations in Figure 7. I found a strong inverse correlation between TRON in the 17 species with slope of the GC and ENC correlations in the 17 species (Figure 7a; R2 = 0.98). I also found a strong second-order parabolic (ax2-bx) correlation between R2 and the slope of the GC versus ENC correlations (Figure 7b: R2 = 0.99).
4. Discussion
4.1. Interpretation of GC and ENC Distributions Across Species
4.2. Correlation Between GC Content and ENC Across Species
4.3. Evolutionary and Functional Implications
4.4. Implications of GC-ENC and GC+ENC Transformations
4.5. Experimental Validation of the Possible Importance Between the Correlation Between GC Content and ENC Levels
4.6. Further Uses for the Two-Rank Order Normalization (TRON) Approach
4.7. Broader Implications and Future Directions
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GC | GC content for a nuclear-encoded gene (range 0.00 to 1.00) |
| ENC | Effective Number of codons (range 20–61). |
| GC-ENC | GC rank minus ENC rank |
| GC+ENC | GC rank plus ENC rank |
References
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Wright, F. The ’effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. A more accurate relationship between ’effective number of codons’ and GC3s under assumptions of no selection. Comput. Biol. Chem. 2013, 42, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallvé, S. E-CAI: A novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinform. 2008, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Zaytsev, K.; Bogatyreva, N.; Fedorov, A. Link Between Individual Codon Frequencies and Protein Expression: Going Beyond Codon Adaptation Index. Int. J. Mol. Sci. 2024, 25, 11622. [Google Scholar] [CrossRef]
- Gu, X.; Li, W.H. A model for the correlation of mutation rate with GC content and the origin of GC-rich isochores. J. Mol. Evol. 1994, 38, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D.; Williams, E.J. Covariation of GC content and the silent site substitution rate in rodents: Implications for methodology and for the evolution of isochores. Gene 2000, 261, 107–114. [Google Scholar] [CrossRef]
- Belle, E.M.; Duret, L.; Galtier, N.; Eyre-Walker, A. The decline of isochores in mammals: An assessment of the GC content variation along the mammalian phylogeny. J. Mol. Evol. 2004, 58, 653–660. [Google Scholar] [CrossRef]
- Huttener, R.; Thorrez, L.; In’t Veld, T.; Granvik, M.; Snoeck, L.; Van Lommel, L.; Schuit, F. GC content of vertebrate exome landscapes reveal areas of accelerated protein evolution. BMC Evol. Biol. 2019, 19, 144. [Google Scholar] [CrossRef]
- Bohlin, J. A simple stochastic model describing the evolution of genomic GC content in asexually reproducing organisms. Sci. Rep. 2022, 12, 18569. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Cao, X.; Khetani, R.S.; Chen, C.C.; Coon, M.; Sammak, A.A.; Bollig-Fischer, A.; Land, S.; Huang, Y.; Hudson, M.E.; et al. Intronic non-CG DNA hydroxymethylation and alternative mRNA splicing in honey bees. BMC Genom. 2013, 14, 666. [Google Scholar] [CrossRef]
- Deng, X.; Fan, G. Tuning up gene transcription via direct crosstalk of DNA and RNA methylation. Mol. Cell 2025, 85, 674–676. [Google Scholar] [CrossRef]
- Huang, K.Y.; Feng, Y.Y.; Du, H.; Ma, C.W.; Xie, D.; Wan, T.; Feng, X.Y.; Dai, X.G.; Yin, T.M.; Wang, X.Q.; et al. DNA methylation dynamics in gymnosperm duplicate genes: Implications for genome evolution and stress adaptation. Plant J. 2025, 121, e70006. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Li, D.; Zhao, X.; Wang, Y.; Wang, B. Genome-wide DNA methylation regulation analysis provides novel insights on post-radiation breast cancer. Sci. Rep. 2025, 15, 5641. [Google Scholar]
- Vollger, M.R.; Korlach, J.; Eldred, K.C.; Swanson, E.; Underwood, J.G.; Bohaczuk, S.C.; Mao, Y.; Cheng, Y.H.H.; Ranchalis, J.; Blue, E.E.; et al. Synchronized long-read genome, methylome, epigenome and transcriptome profiling resolve a Mendelian condition. Nat. Genet. 2025, 57, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Zhu, J.K. RNA Splicing Factors and RNA-Directed DNA Methylation. Biology 2014, 3, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, S.; Wang, T.; Tao, Z.; Huang, S.; Lin, N.; Zhao, Y.; Wang, C.; Li, P. Cooperative condensation of RNA-DIRECTED DNA METHYLATION 16 splicing isoforms enhances heat tolerance in Arabidopsis. Nat. Commun. 2025, 16, 433. [Google Scholar] [CrossRef]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef]
- Tatarinova, T.; Elhaik, E.; Pellegrini, M. Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol. Evol. 2013, 5, 1443–1456. [Google Scholar]
- Clement, Y.; Fustier, M.A.; Nabholz, B.; Glemin, S. The bimodal distribution of genic GC content is ancestral to monocot species. Genome Biol. Evol. 2014, 7, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Tang, H.; Burke, J.M.; Paterson, A.H. GC content of plant genes is linked to past gene duplications. PLoS ONE 2022, 17, e0261748. [Google Scholar] [CrossRef] [PubMed]
- Teng, W.; Liao, B.; Chen, M.; Shu, W. Genomic Legacies of Ancient Adaptation Illuminate GC-Content Evolution in Bacteria. Microbiol. Spectr. 2023, 11, e0214522. [Google Scholar]
- Mazumdar, P.; Binti Othman, R.; Mebus, K.; Ramakrishnan, N.; Ann Harikrishna, J. Codon usage and codon pair patterns in non-grass monocot genomes. Ann. Bot. 2017, 120, 893–909. [Google Scholar]
- Jørgensen, F.G.; Schierup, M.H.; Clark, A.G. Heterogeneity in regional GC content and differential usage of codons and amino acids in GC-poor and GC-rich regions of the genome of Apis mellifera. Mol. Biol. Evol. 2007, 24, 611–619. [Google Scholar] [CrossRef]
- Scapoli, C.; Bartolomei, E.; De Lorenzi, S.; Carrieri, A.; Salvatorelli, G.; Rodriguez-Larralde, A.; Barrai, I. Codon and aminoacid usage patterns in mycobacteria. J. Mol. Microbiol. Biotechnol. 2009, 17, 53–60. [Google Scholar] [CrossRef]
- Gaona-Mendoza, A.S.; Massange-Sánchez, J.A.; Barboza-Corona, J.E.; Abraham-Juárez, M.J.; Casados-Vázquez, L.E. Codon Optimization is Required to Express Fluorogenic Reporter Proteins in Lactococcus lactis. Mol. Biotechnol. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Steindorff, A.S.; Aguilar-Pontes, M.V.; Robinson, A.J.; Andreopoulos, B.; LaButti, K.; Kuo, A.; Mondo, S.; Riley, R.; Otillar, R.; Haridas, S.; et al. Comparative genomic analysis of thermophilic fungi reveals convergent evolutionary adaptations and gene losses. Commun. Biol. 2024, 7, 1124. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, K.L.; Schmitt, B.M.; Villar, D.; White, R.J.; Marioni, J.C.; Kutter, C.; Odom, D.T. Codon-Driven Translational Efficiency Is Stable across Diverse Mammalian Cell States. PLoS Genet. 2016, 12, e1006024. [Google Scholar] [CrossRef] [PubMed]
- López, J.L.; Lozano, M.J.; Fabre, M.L.; Lagares, A. Codon Usage Optimization in the Prokaryotic Tree of Life: How Synonymous Codons Are Differentially Selected in Sequence Domains with Different Expression Levels and Degrees of Conservation. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Payne, B.; Feyertag, F.; Alvarez-Ponce, D. The Codon Statistics Database: A Database of Codon Usage Bias. Mol. Biol. Evol. 2022, 39, msac157. [Google Scholar] [CrossRef] [PubMed]
- Sabi, R.; Tuller, T. Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res. 2014, 21, 511–526. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Y.; Chai, Y. Optimization of CRISPR/LbCas12a-mediated gene editing in Arabidopsis. PLoS ONE 2022, 17, e0265114. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, P.; Bhasin, M.; Varadarajan, R. Molecular bases for strong phenotypic effects of single synonymous codon substitutions in the E. coli ccdB toxin gene. BMC Genom. 2023, 24, 732. [Google Scholar] [CrossRef]
- Ando, D.; Rashad, S.; Begley, T.J.; Endo, H.; Aoki, M.; Dedon, P.C.; Niizuma, K. Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation. Int. J. Mol. Sci. 2025, 26, 706. [Google Scholar] [CrossRef]
- Ding, N.Q. Advanced Algebra; World Scientific: Hackensack, NJ, USA, 2025; Volume XVI, 495p. [Google Scholar]
- Jabbari, K.; Bernardi, G. An Isochore Framework Underlies Chromatin Architecture. PLoS ONE 2017, 12, e0168023. [Google Scholar] [CrossRef] [PubMed]
- Karro, J.E.; Peifer, M.; Hardison, R.C.; Kollmann, M.; Von Grünberg, H.H. Exponential decay of GC content detected by strand-symmetric substitution rates influences the evolution of isochore structure. Mol. Biol. Evol. 2008, 25, 362–374. [Google Scholar] [CrossRef]
- Matsuo, K.; Clay, O.; Takahashi, T.; Silke, J.; Schaffner, W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat. Cell Mol. Genet. 1993, 19, 543–555. [Google Scholar] [CrossRef]
- Schmegner, C.; Hoegel, J.; Vogel, W.; Assum, G. The rate, not the spectrum, of base pair substitutions changes at a GC-content transition in the human NF1 gene region: Implications for the evolution of the mammalian genome structure. Genetics 2007, 175, 421–428. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Wang, G.G. Wang, Polycomb Gene Silencing Mechanisms: PRC2 Chromatin Targeting, H3K27me3 ‘Readout’, and Phase Separation-Based Compaction. Trends Genet. 2021, 37, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2017, 8, 5629–5637. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.W.T.; Jenkins, L.J.; Chionh, F.; Mariadason, J.M. Aberrant DNA Methylation in Colorectal Cancer: What Should We Target? Trends Cancer 2017, 3, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Matassi, G.; Montero, L.M.; Salinas, J.; Bernardi, G. The isochore organization and the compositional distribution of homologous coding sequences in the nuclear genome of plants. Nucleic Acids Res. 1989, 17, 5273–5290. [Google Scholar] [CrossRef] [PubMed]
- Salinas, J.; Matassi, G.; Montero, L.M.; Bernardi, G. Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res. 1988, 16, 4269–4285. [Google Scholar] [CrossRef] [PubMed]
- Vogl, C.; Karapetiants, M.; Yıldırım, B.; Kjartansdóttir, H.; Kosiol, C.; Bergman, J.; Majka, M.; Mikula, L.C. Inference of genomic landscapes using ordered Hidden Markov Models with emission densities (oHMMed). BMC Bioinform. 2024, 25, 151. [Google Scholar] [CrossRef]
- Shi, C.; Xie, Y.; Guan, D.; Qin, G. Transcriptomic Analysis Reveals Adaptive Evolution and Conservation Implications for the Endangered Magnolia lotungensis. Genes 2024, 15, 787. [Google Scholar] [CrossRef] [PubMed]
- Kimsey, I.; Al-Hashimi, H.M. Increasing occurrences and functional roles for high energy purine-pyrimidine base-pairs in nucleic acids. Curr. Opin. Struct. Biol. 2014, 24, 72–80. [Google Scholar] [CrossRef]
- Woodside, M.T.; García-García, C.; Block, S.M. Folding and unfolding single RNA molecules under tension. Curr. Opin. Chem. Biol. 2008, 12, 640–646. [Google Scholar] [CrossRef]
- Aruda, J.; Grote, S.L.; Rouskin, S. Untangling the pseudoknots of SARS-CoV-2: Insights into structural heterogeneity and plasticity. Curr. Opin. Struct. Biol. 2024, 88, 102912. [Google Scholar] [CrossRef]
- Kiliushik, D.; Goenner, C.; Law, M.; Schroeder, G.M.; Srivastava, Y.; Jenkins, J.L.; Wedekind, J.E. Knotty is nice: Metabolite binding and RNA-mediated gene regulation by the preQ(1) riboswitch family. J. Biol. Chem. 2024, 300, 107951. [Google Scholar] [CrossRef]
- Chełkowska-Pauszek, A.; Kosiński, J.G.; Marciniak, K.; Wysocka, M.; Bąkowska-Żywicka, K.; Żywicki, M. The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. Int. J. Mol. Sci. 2021, 22, 7845. [Google Scholar] [CrossRef] [PubMed]
- Alghoul, F.; Eriani, G.; Martin, F. RNA Secondary Structure Study by Chemical Probing Methods Using DMS and CMCT. Methods Mol. Biol. 2021, 2300, 241–250. [Google Scholar] [PubMed]
- Zhou, Y.; Huang, Q.; Wu, C.; Xu, Y.; Guo, Y.; Yuan, X.; Xu, C.; Zhou, L. m(6)A-modified HOXC10 promotes HNSCC progression via co-activation of ADAM17/EGFR and Wnt/beta-catenin signaling. Int. J. Oncol. 2024, 64, 10. [Google Scholar] [PubMed]
| Common Name | Species | Genes (N) | GC Peak 1 | GC Peak 2 | ENC Peak 1 | ENC Peak 2 | Line Equation | Slope | R2 | (GC-ENC) /(N2/3) | (GC+ENC) /N(N+1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rice | Oryza sativa | 28,571 | 0.48 | 0.7 | 32 | 56 | y = −0.77x + 25253 | −0.77 | 0.59 | 1.48 | 1 |
| Mosquito | Anopheles gambiae | 12,402 | 0.58 | 0.58 | 43 | 53 | y = −0.75x + 10878 | −0.75 | 0.57 | 1.4 | 1 |
| Puffer fish | Takifugu rubripes | 22,107 | 0.54 | 0.54 | 53 | 53 | y = −0.64x + 18180 | −0.64 | 0.42 | 1.5 | 1 |
| Humans | Homo sapiens | 19,708 | 0.46 | 0.6 | 43 | 53 | y = −0.61x + 15954 | −0.61 | 0.38 | 1.39 | 1 |
| Bread mold | Neurospora crassa | 9728 | 0.55 | 0.55 | 57 | 57 | y = −0.60x + 7830 | −0.60 | 0.37 | 1.34 | 1 |
| Banana | Musa acuminata | 30,700 | 0.45 | 0.6 | 41 | 55 | y = −0.53x + 23495 | −0.53 | 0.28 | 1.18 | 1 |
| Mouse | Mus musculus | 22,405 | 0.49 | 0.55 | 54 | 54 | y = −0.45x + 16307 | −0.45 | 0.21 | 1.32 | 1 |
| E. coli bacteria | Escherichia coli | 10,276 | 0.52 | 0.52 | 48 | 48 | y = −0.35x + 6975 | −0.35 | 0.13 | 1.22 | 1 |
| Pombe yeast | pombe | 5110 | 0.4 | 0.4 | 50 | 50 | y = −0.19x + 3063 | −0.19 | 0.04 | 1.1 | 1 |
| Methanobacteria | Methanococcus aeolicus | 1485 | 0.32 | 0.32 | 41 | 41 | y = 0.064x + 696 | 0.064 | 0.004 | 0.97 | 1 |
| Bakers yeast | Saccharomyces | 5958 | 0.39 | 0.39 | 51 | 51 | y = 0.0064x + 2983 | 0.0064 | 0.0008 | 0.97 | 1 |
| Honey bee | Apis mellifera | 9918 | 0.33 | 0.47 | 35 | 56 | y = 0.78x + 1103 | 0.78 | 0.6 | 0.44 | 0.99 |
| Red paper wasp | Polistes canadensis | 9854 | 0.38 | 0.38 | 48 | 48 | y = 0.72x + 1367 | 0.72 | 0.52 | 0.48 | 0.99 |
| Spotted fever parasite | Rickettsia hoogstraalii | 1663 | 0.33 | 0.33 | 43 | 43 | y = 0.41x + 489 | 0.41 | 0.17 | 0.71 | 1 |
| Slime mold | Dictyostelium discoideum | 13,078 | 0.28 | 0.28 | 32 | 32 | y = 0.38x + 4044 | 0.38 | 0.15 | 0.75 | 1 |
| Mustard weed | Arabidopsis thaliana | 10,160 | 0.45 | 0.45 | 53 | 53 | y = 0.20x + 4066 | 0.20 | 0.04 | 0.86 | 1 |
| Malaria parasite | Plasmodium falciparum | 5321 | 0.25 | 0.25 | 38 | 38 | y = 0.20x + 2141 | 0.20 | 0.04 | 0.88 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruden, D.M. GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species. Genes 2025, 16, 432. https://doi.org/10.3390/genes16040432
Ruden DM. GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species. Genes. 2025; 16(4):432. https://doi.org/10.3390/genes16040432
Chicago/Turabian StyleRuden, Douglas M. 2025. "GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species" Genes 16, no. 4: 432. https://doi.org/10.3390/genes16040432
APA StyleRuden, D. M. (2025). GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species. Genes, 16(4), 432. https://doi.org/10.3390/genes16040432







