Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction, and Sequencing
2.2. Sequence Analysis and Gene Annotation
2.3. Phylogenetic Analysis
3. Result
3.1. Genome Organization and Base Composition
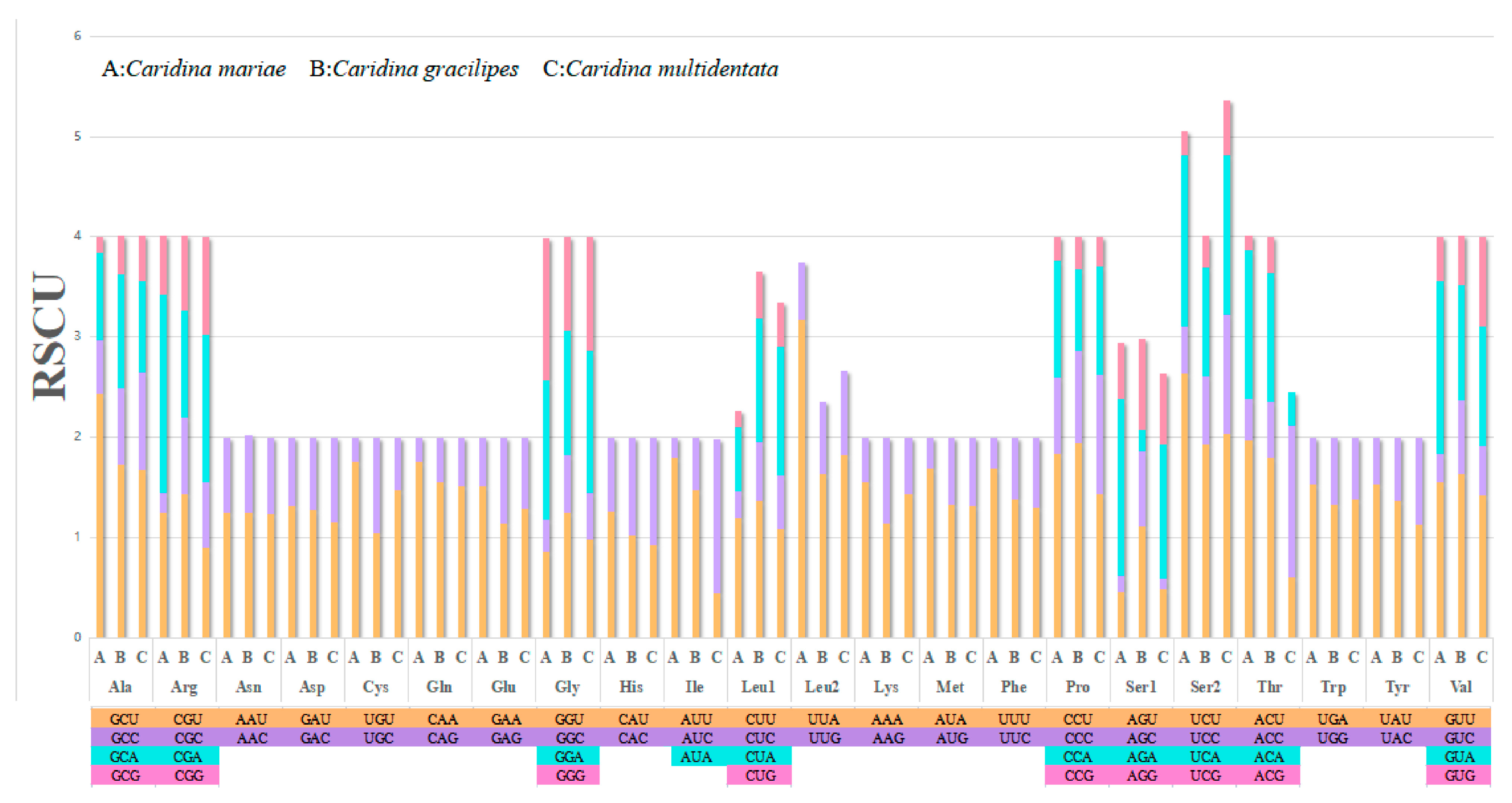
3.2. Protein-Coding Genes and Non-Coding Regions
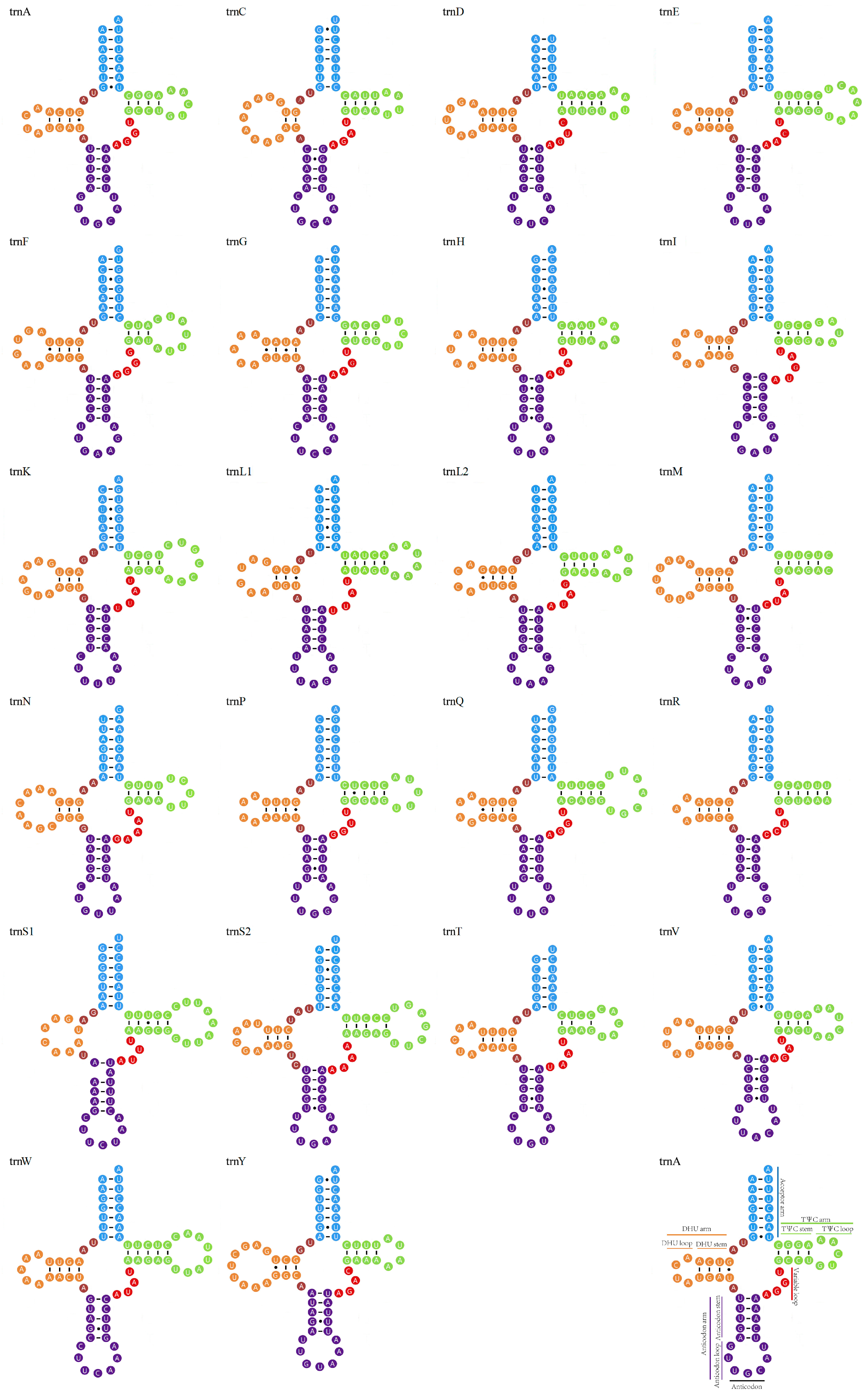
3.3. Transfer and Ribosomal RNA Genes
3.4. Phylogenetic Relationship
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grave, S.D.; Fransen, C.H. Carideorumcatalogus: The recent species of the dendrobranchiate, stenopodidean, procarididean and caridean shrimps (Crustacea: Decapoda). ZoologischeMededelingen 2011, 85, 195–589. [Google Scholar]
- Shen, H.; Braband, A.; Scholtz, G. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenetics Evol. 2013, 66, 776–789. [Google Scholar] [CrossRef] [PubMed]
- Nong, W.; Chai, Z.Y.; Jiang, X.; Qin, J.; Ma, K.Y.; Chan, K.M.; Chan, T.; Chow, B.K.; Kwan, H.S.; Wong, C.K.; et al. A crustacean annotated transcriptome (CAT) database. BMC Genom. 2020, 21, 32. [Google Scholar] [CrossRef] [PubMed]
- Ivey, J.L.; Santos, S.R. The complete mitochondrial genome of the Hawaiian anchialine shrimp Halocaridina rubra Holthuis, 1963 (Crustacea: Decapoda: Atyidae). Gene 2007, 394, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Grave, S.D.; Pentcheff, N.D.; Ahyong, S.T.; Chan, T.; Crandall, K.A.; Dworschak, P.C.; Felder, D.L.; Feldmann, R.M.; Fransen, C.H.; Goulding, L.Y.; et al. A Classification of Living and Fossil Genera of Decapod Crustaceans. Raffles Bull. Zool. 2009, 21, 1–109. [Google Scholar]
- Yi, Z.; Huang, Y.; Yi, H.Q.; Zhang, X.; Li, W. Biodiversity of macrozoobenthos in the Chebaling National Nature Reserve, Guangdong Province. Biodivers. Sci. 2021, 29, 680–687. [Google Scholar] [CrossRef]
- Zhu, L.Q.; Zhu, Z.H.; Zhu, L.Y.; Wang, D.Q.; Wang, J.X.; Lin, Q. Characteristics and phylogenetic analysis of mitogenome in the Atyidae. J. Dalian Ocean Univ. 2022, 37, 428–434. [Google Scholar]
- De Grave, S.; Smith, K.G.; Adeler, N.A.; Allen, D.J.; Alvarez, F.; Anker, A.; Cai, Y.; Carrizo, S.F.; Klotz, W.; Mantelatto, F.L.; et al. Dead shrimp blues: A global assessment of extinction risk in freshwater shrimps (Crustacea: Decapoda: Caridea). PLoS ONE 2015, 10, e0120198. [Google Scholar] [CrossRef]
- Tu, D.V.; Dong, D.; Rintelen, T.V. Description of one new species of freshwater shrimp of the genus Caridina (Crustacea: Decapoda: Atyidae) from two karst caves of Northern Vietnam. Zootaxa 2021, 4999, 228–242. [Google Scholar] [CrossRef]
- Mazancourt, V.D.; Klotz, W.; Marquet, G.; Mos, B.; Rogers, D.C.; Keith, P. New Insights on Biodiversity and Conservation of Amphidromous Shrimps of the Indo-Pacific islands (Decapoda: Atyidae: Caridina). In Recent Advances in Freshwater Crustacean Biodiversity and Conservation; CRC Press: Boca Raton, FL, USA, 2021; pp. 381–404. [Google Scholar]
- De Grave, S.; Cai, Y.; Anker, A. Global diversity of shrimps (Crustacea: Decapoda: Caridea) in freshwater. Hydrobiologia 2008, 595, 287–293. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, X. Caridina longiacuta, a new species of freshwater atyid shrimp (Decapoda, Atyidae) from Hunan Province, China. Zootaxa 2005, 1008, 13–20. [Google Scholar] [CrossRef]
- Liu, X.; Guo, Z.; Yu, H.J. Caridina xiangnanensis, a new freshwater atyid shrimp (Crustacea, Decapoda, Atyidae) from Hunan Province, China. Zootaxa 2006, 1153, 43–50. [Google Scholar] [CrossRef]
- Li, S.; Li, J. Description of Caridina alba, a new species of blind atyid shrimp from Tenglongdong cave, Hubei Province, China (Decapoda, Atyidae). Crustaceana 2010, 83, 17–27. [Google Scholar] [CrossRef]
- Cai, Y. Atyid shrimps of Hainan Island, southern China, with the description of a new species of Caridina (Crustacea, Decapoda, Atyidae). In Advances in Freshwater Decapod Systematics and Biology; Yeo, D.C.J., Cumberlidge, N., Klaus, S., Eds.; BRILL: Leiden, The Netherlands, 2014; Volume 19, pp. 207–231. [Google Scholar]
- Klotz, W.; Von Rintelen, T. To “bee” or not to be-on some ornamental shrimp from Guangdong Province, Southern China and Hong Kong SAR, with descriptions of three new species. Zootaxa 2014, 3889, 151–184. [Google Scholar] [CrossRef]
- Cai, Y.; Ng, P.K.L. Freshwater Shrimps from Karst Caves of Southern China, with Descriptions of Seven New Species and the Identity of Typhlocaridina linyunensis Li and Luo, 2001 (Crustacea: Decapoda: Caridea). Zool. Stud. 2018, 57, e27. [Google Scholar]
- Chen, Q.H.; Chen, W.J.; Zheng, X.Z.; Guo, Z.L. Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species. ZooKeys 2020, 923, 15–32. [Google Scholar] [CrossRef]
- Xu, D.; Li, D.; Zheng, X.; Guo, Z. Caridina sinanensis, a new species of stygobiotic atyid shrimp (Decapoda, Caridea, Atyidae) from a karst cave in the Guizhou Province, southwestern China. ZooKeys 2020, 1008, 17–35. [Google Scholar] [CrossRef]
- Feng, S.; Chen, Q.H.; Guo, Z.L. Integrative taxonomy uncovers a new stygobiotic Caridina species (Decapoda, Caridea, Atyidae) from Guizhou Province, China. ZooKeys 2021, 1028, 29–47. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, W.; Wong, K.C.; Huang, J. Atyid shrimps of the genus Caridina (Decapoda, Caridea, Atyidae) of Taipa-Coloane Island, Macau, China, with description of a new species. Crustaceana 2021, 94, 1103–1111. [Google Scholar] [CrossRef]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef]
- Kolesnikov, A.A.; Gerasimov, E.S. Diversity of mitochondrial genome organization. Biochemistry 2012, 77, 1424–1435. [Google Scholar] [CrossRef]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Havird, J.; Santos, S. Performance of Single and Concatenated Sets of Mitochondrial Genes at Inferring Metazoan Relationships Relative to Full Mitogenome Data. PLoS ONE 2014, 9, e84080. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Chai, X.Y.; Wang, Z.F.; Zhang, H.B.; Zhou, C.L.; Tang, B.P.; Liu, Q.N. Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): Gene rearrangements and higher-level phylogeny of the Brachyura. Sci. Rep. 2017, 7, 4128. [Google Scholar] [CrossRef]
- Wang, Z.F.; Shi, X.; Guo, H.; Tang, D.; Bai, Y.; Wang, Z. Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics 2020, 112, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Z.F.; Tang, D.; Xu, X.; Tao, Y.; Ji, C.; Wang, Z. Characterization and comparison of the mitochondrial genomes from two Alpheidae species and insights into the phylogeny of Caridea. Genomics 2020, 112, 65–70. [Google Scholar] [CrossRef]
- Wang, Z.F.; Wang, Z.; Shi, X.; Wu, Q.; Tao, Y.; Guo, H.; Ji, C.; Bai, Y. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): Gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int. J. Biol. Macromol 2018, 118 Pt A, 31–40. [Google Scholar] [CrossRef]
- Wang, Z.F.; Xu, X.Y.; Zheng, Y.Q.; Wang, J.; Yu, Q.; Liu, B. Taxonomic status and phylogenetic relationship of Anomura (Crustacea: Decapoda) based on mitochondrial sequences and gene order rearrangements. Gene 2023, 851, 147042. [Google Scholar] [CrossRef]
- Wang, Z.F.; Zheng, Y.Q.; Zhao, X.Y.; Xu, X.Y.; Xu, Z.W.; Cui, C. Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences. Biology 2023, 12, 974. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 31–319. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.Q.; Yang, W.J.; Yang, J.S. The complete mitogenome of the Chinese swamp shrimp Neocaridina denticulata sinensis Kemp 1918 (Crustacea: Decapoda: Atyidae). Mitochondrial DNA 2014, 25, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Chirico, N.; Vianelli, A.; Belshaw, R. Why genes overlap in viruses. Proc. Biol. Sci. 2010, 277, 3809–3817. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Yongsung, K.; Kwon, W.; Xi, H.; Park, J. The complete mitochondrial genome of Neocaridina heteropodakoreana Kubo, 1938 (Decapoda: Atyidae). Mitochondrial DNA Part B 2019, 4, 2332–2334. [Google Scholar] [CrossRef]
- Jain, K.; Panigrahi, M.; Nayak, S.S.; Rajawat, D.; Sharma, A.; Sahoo, S.P.; Bhushan, B.; Dutt, T. The evolution of contemporary livestock species: Insights from mitochondrial genome. Gene 2024, 927, 148728. [Google Scholar] [CrossRef]
- Nayak, S.S.; Rajawat, D.; Jain, K.; Sharma, A.; Gondro, C.; Tarafdar, A.; Dutt, T.; Panigrahi, M. A comprehensive review of livestock development: Insights into domestication, phylogenetics, diversity, and genomic advances. Mamm. Genome 2024, 35, 577–599. [Google Scholar] [CrossRef]
- Holthuis, L.B. FAO species catalogue. In Shrimps and Prawns of the World. An Annotated Catalogue of Species of Interest to Fisheries; FAO: Rome, Italy, 1980; Volume 1, pp. 1–271. [Google Scholar]
- De Grave, S.; Li, C.; Tsang, L.M.; Chu, K.H.; Chan, T. Unweaving hippolytoid systematics (Crustacea, Decapoda, Hippolytidae): Resurrection of several families. Zool. Scr. 2014, 43, 496–507. [Google Scholar] [CrossRef]
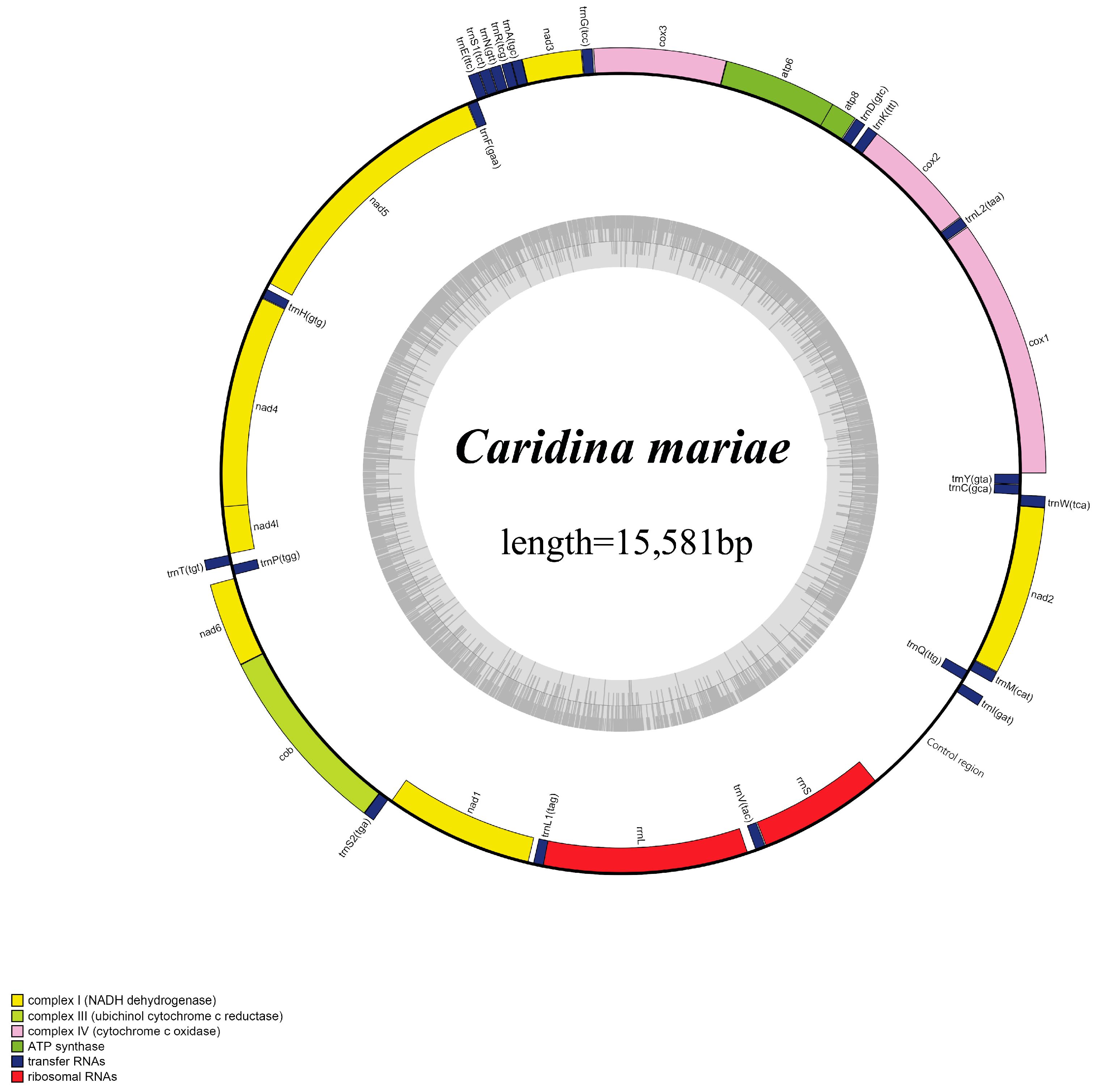

| Gene | Direction | Location | Size (bp) | Anticodon | Start Codon | Stop Codon | Intergenic Nucleotides |
|---|---|---|---|---|---|---|---|
| cox1 | + | 1–1533 | 1533 | CAA | TAA | 2 | |
| trnL2 | + | 1536–1599 | 64 | TAA | 1 | ||
| cox2 | + | 1601–2308 | 708 | ATG | TAA | −20 | |
| trnK | + | 2289–2355 | 67 | TTT | 22 | ||
| trnD | + | 2378–2442 | 65 | GTC | 2 | ||
| atp8 | + | 2445–2603 | 159 | ATT | TAA | −7 | |
| atp6 | + | 2597–3271 | 675 | ATG | TAA | −1 | |
| cox3 | + | 3271–4056 | 786 | ATG | TAA | 3 | |
| trnG | + | 4060–4124 | 65 | TCC | 0 | ||
| nad3 | + | 4125–4478 | 354 | ATT | TAA | −2 | |
| trnA | + | 4477–4540 | 64 | TGC | −1 | ||
| trnR | + | 4540–4601 | 62 | TCG | 3 | ||
| trnN | + | 4605–4671 | 67 | GTT | 0 | ||
| trnS1 | + | 4672–4738 | 67 | TCT | 0 | ||
| trnE | + | 4739–4806 | 68 | TTC | −2 | ||
| trnF | − | 4805–4870 | 66 | GAA | 0 | ||
| nad5 | − | 4871–6562 | 1692 | ATT | TAA | 36 | |
| trnH | − | 6599–6662 | 64 | GTG | 0 | ||
| nad4 | − | 6663–8001 | 1339 | ATG | T(AA) | −7 | |
| nad4L | − | 7995–8294 | 300 | ATG | TAA | 2 | |
| trnT | + | 8297–8361 | 65 | TGT | 0 | ||
| trnP | − | 8362–8427 | 66 | TGG | 17 | ||
| nad6 | + | 8445–8945 | 501 | ATA | TAA | −1 | |
| cob | + | 8945–10,081 | 1137 | ATG | TAG | −2 | |
| trnS2 | + | 10,080–10,149 | 70 | TGA | 18 | ||
| nad1 | − | 10,168–11,106 | 939 | ATA | TAG | 30 | |
| trnL1 | − | 11,137–11,203 | 67 | TAG | −40 | ||
| rrnL | − | 11,164–12,489 | 1326 | 50 | |||
| trnV | − | 12,540–12,606 | 67 | TAC | 2 | ||
| rrnS | − | 12,609–13,405 | 797 | 0 | |||
| CR | 13,406–14,151 | 746 | 0 | ||||
| trnI | + | 14,152–14,216 | 65 | GAT | 19 | ||
| trnQ | − | 14,236–14,303 | 68 | TTG | 6 | ||
| trnM | + | 14,310–14,377 | 68 | CAT | 0 | ||
| nad2 | + | 14,378–15,382 | 1005 | ATT | TAA | −2 | |
| trnW | + | 15,381–15,450 | 70 | TCA | −1 | ||
| trnC | − | 15,450–15,513 | 64 | GCA | 0 | ||
| trnY | − | 15,514–15,579 | 66 | GTA | 2 |
| C. mariae | Size (bp) | T(U) (%) | C (%) | A (%) | G (%) | A + T (%) | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|
| Mitogenome | 15,581 | 33.4 | 18.9 | 35.5 | 12.2 | 68.9 | 0.030 | −0.214 |
| PCGs | 11,128 | 39.6 | 16.3 | 27.4 | 16.6 | 67.1 | −0.182 | 0.007 |
| cox1 | 1533 | 35.5 | 18.3 | 28.0 | 18.2 | 63.5 | −0.118 | −0.004 |
| cox2 | 708 | 33.1 | 20.1 | 32.8 | 14.1 | 65.8 | −0.004 | −0.174 |
| atp8 | 159 | 39.6 | 20.1 | 33.3 | 6.9 | 73.0 | −0.086 | −0.488 |
| atp6 | 675 | 38.1 | 18.5 | 28.9 | 14.5 | 67.0 | −0.137 | −0.121 |
| cox3 | 786 | 35.9 | 19.2 | 29.1 | 15.8 | 65.0 | −0.104 | −0.098 |
| cob | 354 | 40.1 | 20.1 | 28.8 | 11.0 | 68.9 | −0.164 | −0.291 |
| nad5 | 1692 | 41.5 | 11.6 | 26.1 | 20.7 | 67.7 | −0.228 | 0.283 |
| nad4 | 1339 | 43.6 | 11.0 | 24.4 | 21.0 | 68.0 | −0.282 | 0.313 |
| nad4l | 300 | 44.3 | 8.7 | 26.3 | 20.7 | 70.7 | −0.255 | 0.409 |
| nad3 | 501 | 40.7 | 21.6 | 29.1 | 8.6 | 69.9 | −0.166 | −0.430 |
| nad1 | 1137 | 38.5 | 19.3 | 26.9 | 15.3 | 65.4 | −0.177 | −0.115 |
| nad6 | 939 | 44.9 | 11.2 | 25.1 | 18.7 | 70.1 | −0.283 | 0.253 |
| nad2 | 1005 | 40.4 | 21.4 | 27.7 | 10.5 | 68.1 | −0.187 | −0.340 |
| tRNAs | 1455 | 33.5 | 14.0 | 34.6 | 17.9 | 68.1 | 0.015 | 0.125 |
| rRNAs | 2123 | 39.0 | 9.2 | 33.9 | 17.9 | 72.9 | −0.070 | 0.323 |
| Control region | 746 | 39.7 | 7.0 | 47.1 | 6.3 | 86.7 | 0.085 | −0.051 |
| Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UUU(F) | 258 | 1.69 | UCU(S) | 115 | 2.63 | UAU(Y) | 104 | 1.53 | UGU(C) | 37 | 1.76 |
| UUC(F) | 47 | 0.31 | UCC(S) | 21 | 0.48 | UAC(Y) | 32 | 0.47 | UGC(C) | 5 | 0.24 |
| UUA(L) | 305 | 3.17 | UCA(S) | 75 | 1.71 | UAA(*) | 10 | 1.67 | UGA(W) | 75 | 1.53 |
| UUG(L) | 55 | 0.57 | UCG(S) | 10 | 0.23 | UAG(*) | 2 | 0.33 | UGG(W) | 23 | 0.47 |
| CUU(L) | 115 | 1.19 | CCU(P) | 70 | 1.84 | CAU(H) | 51 | 1.26 | CGU(R) | 19 | 1.25 |
| CUC(L) | 26 | 0.27 | CCC(P) | 29 | 0.76 | CAC(H) | 30 | 0.74 | CGC(R) | 3 | 0.2 |
| CUA(L) | 62 | 0.64 | CCA(P) | 44 | 1.16 | CAA(Q) | 71 | 1.75 | CGA(R) | 30 | 1.97 |
| CUG(L) | 15 | 0.16 | CCG(P) | 9 | 0.24 | CAG(Q) | 10 | 0.25 | CGG(R) | 9 | 0.59 |
| AUU(I) | 271 | 1.8 | ACU(T) | 102 | 1.97 | AAU(N) | 76 | 1.25 | AGU(S) | 20 | 0.46 |
| AUC(I) | 30 | 0.2 | ACC(T) | 21 | 0.41 | AAC(N) | 46 | 0.75 | AGC(S) | 7 | 0.16 |
| AUA(M) | 169 | 1.69 | ACA(T) | 77 | 1.49 | AAA(K) | 59 | 1.55 | AGA(S) | 77 | 1.76 |
| AUG(M) | 31 | 0.31 | ACG(T) | 7 | 0.14 | AAG(K) | 17 | 0.45 | AGG(S) | 25 | 0.57 |
| GUU(V) | 100 | 1.56 | GCU(A) | 152 | 2.44 | GAU(D) | 53 | 1.31 | GGU(G) | 54 | 0.86 |
| GUC(V) | 18 | 0.28 | GCC(A) | 33 | 0.53 | GAC(D) | 28 | 0.69 | GGC(G) | 20 | 0.32 |
| GUA(V) | 110 | 1.72 | GCA(A) | 54 | 0.87 | GAA(E) | 54 | 1.52 | GGA(G) | 87 | 1.39 |
| GUG(V) | 28 | 0.44 | GCG(A) | 10 | 0.16 | GAG(E) | 17 | 0.48 | GGG(G) | 89 | 1.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Jiang, W.; Ye, J.; Wu, H.; Wang, Y.; Xiong, F. Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps. Genes 2025, 16, 457. https://doi.org/10.3390/genes16040457
Wang Z, Jiang W, Ye J, Wu H, Wang Y, Xiong F. Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps. Genes. 2025; 16(4):457. https://doi.org/10.3390/genes16040457
Chicago/Turabian StyleWang, Zhengfei, Weijie Jiang, Jingxue Ye, Huiwen Wu, Yan Wang, and Fei Xiong. 2025. "Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps" Genes 16, no. 4: 457. https://doi.org/10.3390/genes16040457
APA StyleWang, Z., Jiang, W., Ye, J., Wu, H., Wang, Y., & Xiong, F. (2025). Decoding the Mitochondrial Genome of the Tiger Shrimp: Comparative Genomics and Phylogenetic Placement Within Caridean Shrimps. Genes, 16(4), 457. https://doi.org/10.3390/genes16040457






