Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns
Abstract
1. Introduction
2. ASD Genetics
Syndromic and Non-Syndromic ASD
3. ASD Inheritance Pattern
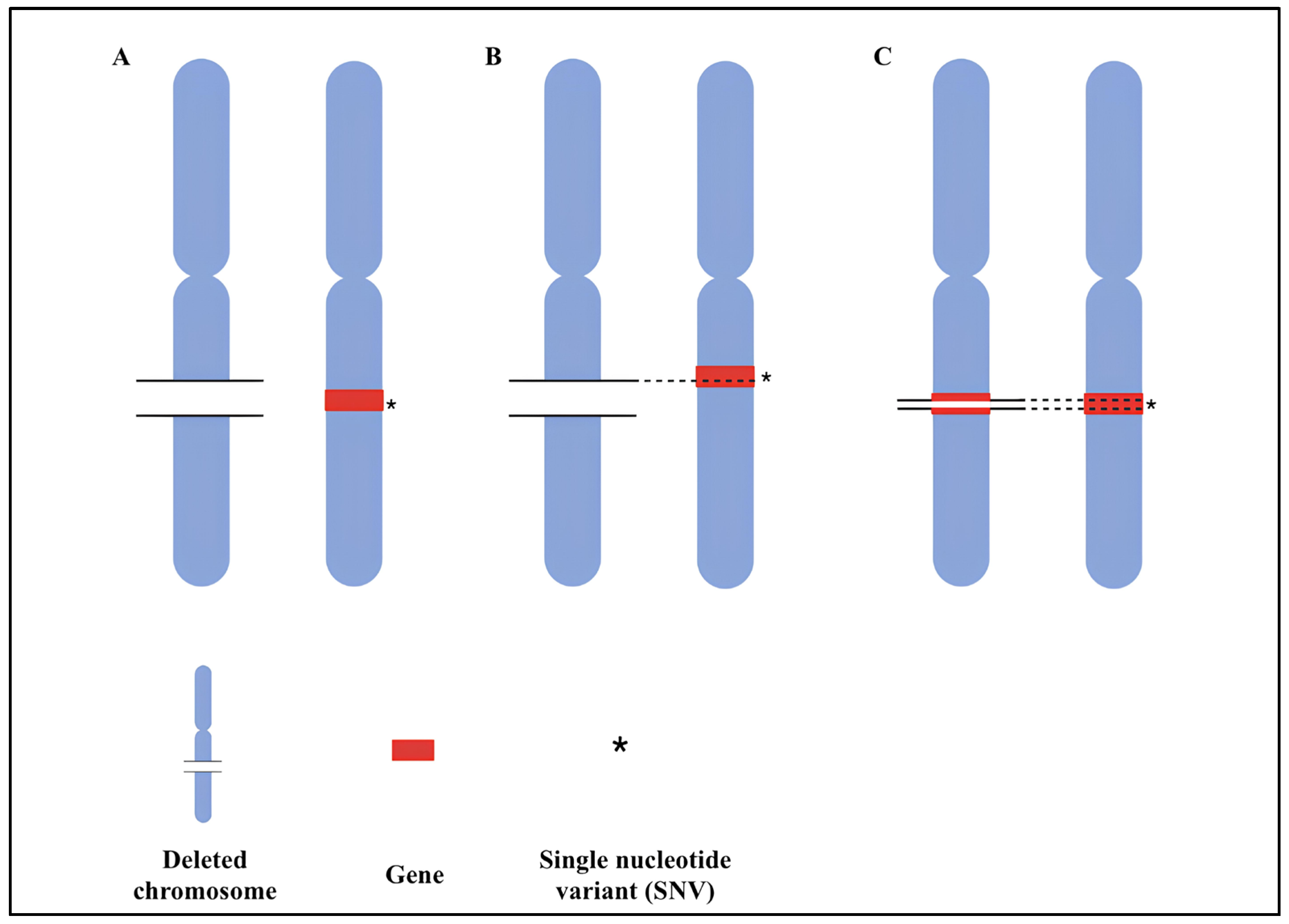
3.1. Double Hit Model
3.2. Modifier Genes
3.3. Incomplete Penetrance Model
3.4. Compound Heterozygosity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Masini, E.; Loi, E.; Vega-Benedetti, A.F.; Carta, M.; Doneddu, G.; Fadda, R.; Zavattari, P. An Overview of the Main Genetic, Epigenetic and Environmental Factors Involved in Autism Spectrum Disorder Focusing on Synaptic Activity. Int. J. Mol. Sci. 2020, 21, 8290. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 1994. [Google Scholar]
- Uljarević, M.; Frazier, T.W.; Chetcuti, L. Asperger Syndrome and Clinical Heterogeneity: Reflections on the Past, Present, and Future. Dev. Med. Child. Neurol. 2023, 65, 1414–1415. [Google Scholar] [CrossRef] [PubMed]
- Motlani, V.; Motlani, G.; Thool, A. Asperger Syndrome (AS): A Review Article. Cureus 2022, 14, e31395. [Google Scholar] [CrossRef]
- Newson, E. Pathological Demand Avoidance Syndrome: A Necessary Distinction within the Pervasive Developmental Disorders. Arch. Dis. Child. 2003, 88, 595–600. [Google Scholar] [CrossRef]
- Carigi, T.; Muratori, F.; Termine, C.; Veggiotti, P.; Derhemi, L.; Nardo, R.; Rossi, G.; Balottin, U. Diagnostic Boundaries of Autism Disorder Vs Pervasive Developmental Disorder Nos Comparative Observational Study and Literature Review. Curr. Clin. Pharmacol. 2014, 9, 377–386. [Google Scholar] [CrossRef][Green Version]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5), 5th ed.; American Psychiatric Association: Washington, DC, USA, 2014. [Google Scholar]
- Lyall, K.; Croen, L.; Daniels, J.; Fallin, M.D.; Ladd-Acosta, C.; Lee, B.K.; Park, B.Y.; Snyder, N.W.; Schendel, D.; Volk, H.; et al. The Changing Epidemiology of Autism Spectrum Disorders. Annu. Rev. Public Health 2017, 38, 81–102. [Google Scholar] [CrossRef]
- Paglia, L. CChildren Diagnosed with ASD Are First of All Children. Eur. J. Paediatr. Dent. 2020, 21, 8. [Google Scholar] [CrossRef]
- Elsabbagh, M.; Divan, G.; Koh, Y.-J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global Prevalence of Autism and Other Pervasive Developmental Disorders. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef]
- Hossain, M.M.; Khan, N.; Sultana, A.; Ma, P.; McKyer, E.L.J.; Ahmed, H.U.; Purohit, N. Prevalence of Comorbid Psychiatric Disorders among People with Autism Spectrum Disorder: An Umbrella Review of Systematic Reviews and Meta-Analyses. Psychiatry Res. 2020, 287, 112922. [Google Scholar] [CrossRef]
- Bosch, R.; Pagerols, M.; Rivas, C.; Sixto, L.; Bricollé, L.; Español-Martín, G.; Prat, R.; Ramos-Quiroga, J.A.; Casas, M. Neurodevelopmental Disorders among Spanish School-Age Children: Prevalence and Sociodemographic Correlates. Psychol. Med. 2022, 52, 3062–3072. [Google Scholar] [CrossRef]
- Dockrell, J.E.; Hurry, J. The Identification of Speech and Language Problems in Elementary School: Diagnosis and Co-Occurring Needs. Res. Dev. Disabil. 2018, 81, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Wiśniowiecka-Kowalnik, B.; Nowakowska, B.A. Genetics and Epigenetics of Autism Spectrum Disorder—Current Evidence in the Field. J. Appl. Genet. 2019, 60, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism Spectrum Disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Salari, N.; Rasoulpoor, S.; Rasoulpoor, S.; Shohaimi, S.; Jafarpour, S.; Abdoli, N.; Khaledi-Paveh, B.; Mohammadi, M. The Global Prevalence of Autism Spectrum Disorder: A Comprehensive Systematic Review and Meta-Analysis. Ital. J. Pediatr. 2022, 48, 112. [Google Scholar] [CrossRef]
- Werling, D.M. The Role of Sex-Differential Biology in Risk for Autism Spectrum Disorder. Biol. Sex. Differ. 2016, 7, 58. [Google Scholar] [CrossRef]
- Badcock, C.; Crespi, B. Imbalanced Genomic Imprinting in Brain Development: An Evolutionary Basis for the Aetiology of Autism. J. Evol. Biol. 2006, 19, 1007–1032. [Google Scholar] [CrossRef]
- Baron-Cohen, S.; Knickmeyer, R.C.; Belmonte, M.K. Sex Differences in the Brain: Implications for Explaining Autism. Science 2005, 310, 819–823. [Google Scholar] [CrossRef]
- Allen, N.D.; Logan, K.; Lally, G.; Drage, D.J.; Norris, M.L.; Keverne, E.B. Distribution of Parthenogenetic Cells in the Mouse Brain and Their Influence on Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 1995, 92, 10782–10786. [Google Scholar] [CrossRef]
- Halladay, A.K.; Bishop, S.; Constantino, J.N.; Daniels, A.M.; Koenig, K.; Palmer, K.; Messinger, D.; Pelphrey, K.; Sanders, S.J.; Singer, A.T.; et al. Sex and Gender Differences in Autism Spectrum Disorder: Summarizing Evidence Gaps and Identifying Emerging Areas of Priority. Mol. Autism 2015, 6, 36. [Google Scholar] [CrossRef]
- Lichtenstein, P.; Carlström, E.; Råstam, M.; Gillberg, C.; Anckarsäter, H. The Genetics of Autism Spectrum Disorders and Related Neuropsychiatric Disorders in Childhood. Am. J. Psychiatry 2010, 167, 1357–1363. [Google Scholar] [CrossRef]
- Newschaffer, C.J.; Croen, L.A.; Daniels, J.; Giarelli, E.; Grether, J.K.; Levy, S.E.; Mandell, D.S.; Miller, L.A.; Pinto-Martin, J.; Reaven, J.; et al. The Epidemiology of Autism Spectrum Disorders. Annu. Rev. Public Health 2007, 28, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Mbadiwe, T.; Millis, R.M. Epigenetics and Autism. Autism Res. Treat. 2013, 2013, 826156. [Google Scholar] [CrossRef] [PubMed]
- Losh, M.; Sullivan, P.F.; Trembath, D.; Piven, J. Current Developments in the Genetics of Autism: From Phenome to Genome. J. Neuropathol. Exp. Neurol. 2008, 67, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Freitag, C.M. Genetics of Autism. J. Intellect. Disabil. Res. 2008, 52, 817. [Google Scholar] [CrossRef]
- Constantino, J.N.; Todorov, A.; Hilton, C.; Law, P.; Zhang, Y.; Molloy, E.; Fitzgerald, R.; Geschwind, D. Autism Recurrence in Half Siblings: Strong Support for Genetic Mechanisms of Transmission in ASD. Mol. Psychiatry 2013, 18, 137–138. [Google Scholar] [CrossRef]
- Colvert, E.; Tick, B.; McEwen, F.; Stewart, C.; Curran, S.R.; Woodhouse, E.; Gillan, N.; Hallett, V.; Lietz, S.; Garnett, T.; et al. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry 2015, 72, 415. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The Familial Risk of Autism. JAMA 2014, 311, 1770. [Google Scholar] [CrossRef]
- Sebat, J.; Lakshmi, B.; Malhotra, D.; Troge, J.; Lese-Martin, C.; Walsh, T.; Yamrom, B.; Yoon, S.; Krasnitz, A.; Kendall, J.; et al. Strong Association of De Novo Copy Number Mutations with Autism. Science 2007, 316, 445–449. [Google Scholar] [CrossRef]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural Variation of Chromosomes in Autism Spectrum Disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef]
- Constantino, J.N.; Zhang, Y.; Frazier, T.; Abbacchi, A.M.; Law, P. Sibling Recurrence and the Genetic Epidemiology of Autism. Am. J. Psychiatry 2010, 167, 1349–1356. [Google Scholar] [CrossRef]
- Virkud, Y.V.; Todd, R.D.; Abbacchi, A.M.; Zhang, Y.; Constantino, J.N. Familial Aggregation of Quantitative Autistic Traits in Multiplex versus Simplex Autism. Am. J. Med. Genet. Part. B Neuropsychiatr. Genet. 2009, 150, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Chaste, P.; Leboyer, M. Autism Risk Factors: Genes, Environment, and Gene-Environment Interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Hultman, C.; Larsson, H.; Reichenberg, A. The Heritability of Autism Spectrum Disorder. JAMA 2017, 318, 1182. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Yip, B.H.K.; Windham, G.C.; Sourander, A.; Francis, R.; Yoffe, R.; Glasson, E.; Mahjani, B.; Suominen, A.; Leonard, H.; et al. Association of Genetic and Environmental Factors with Autism in a 5-Country Cohort. JAMA Psychiatry 2019, 76, 1035. [Google Scholar] [CrossRef]
- Thapar, A.; Rutter, M. Genetic Advances in Autism. J. Autism Dev. Disord. 2021, 51, 4321–4332. [Google Scholar] [CrossRef]
- Kainer, D.; Templeton, A.R.; Prates, E.T.; Jacboson, D.; Allan, E.R.O.; Climer, S.; Garvin, M.R. Structural Variants Identified Using Non-Mendelian Inheritance Patterns Advance the Mechanistic Understanding of Autism Spectrum Disorder. Hum. Genet. Genom. Adv. 2023, 4, 100150. [Google Scholar] [CrossRef]
- Lombardo, B.; Ceglia, C.; Tarsitano, M.; Pierucci, I.; Salvatore, F.; Pastore, L. Identification of a Deletion in the NDUFS4 Gene Using Array-Comparative Genomic Hybridization in a Patient with Suspected Mitochondrial Respiratory Disease. Gene 2014, 535, 376–379. [Google Scholar] [CrossRef]
- Iossa, S.; Costa, V.; Corvino, V.; Auletta, G.; Barruffo, L.; Cappellani, S.; Ceglia, C.; Cennamo, G.; D’Adamo, A.P.; D’Amico, A.; et al. Phenotypic and Genetic Characterization of a Family Carrying Two Xq21.1-21.3 Interstitial Deletions Associated with Syndromic Hearing Loss. Mol. Cytogenet. 2015, 8, 18. [Google Scholar] [CrossRef][Green Version]
- Sanna, V.; Ceglia, C.; Tarsitano, M.; Lombardo, B.; Coppola, A.; Zarrilli, F.; Castaldo, G.; Di Minno, G. Aberrant F8 Gene Intron 1 Inversion with Concomitant Duplication and Deletion in a Severe Hemophilia A Patient from Southern Italy. J. Thromb. Haemost. 2013, 11, 195–197. [Google Scholar] [CrossRef]
- Schaefer, G.B.; Mendelsohn, N.J. Clinical Genetics Evaluation in Identifying the Etiology of Autism Spectrum Disorders: 2013 Guideline Revisions. Genet. Med. 2013, 15, 399–407. [Google Scholar] [CrossRef]
- Ranieri, A.; La Monica, I.; Di Iorio, M.R.; Lombardo, B.; Pastore, L. Genetic Alterations in a Large Population of Italian Patients Affected by Neurodevelopmental Disorders. Genes 2024, 15, 427. [Google Scholar] [CrossRef] [PubMed]
- Falcone, N.; Ranieri, A.; Vitale, A.; Pastore, L.; Lombardo, B. Identification of a De Novo Deletion by Using A-CGH Involving PLNAX2: An Interesting Candidate Gene in Psychomotor Developmental Delay. Medicina 2022, 58, 524. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, B.; Esposito, D.; Iossa, S.; Vitale, A.; Verdesca, F.; Perrotta, C.; Di Leo, L.; Costa, V.; Pastore, L.; Franzé, A. Intragenic Deletion in MACROD2: A Family with Complex Phenotypes Including Microcephaly, Intellectual Disability, Polydactyly, Renal and Pancreatic Malformations. Cytogenet. Genome Res. 2019, 158, 25–31. [Google Scholar] [CrossRef]
- Genovese, A.; Butler, M.G. The Autism Spectrum: Behavioral, Psychiatric and Genetic Associations. Genes 2023, 14, 677. [Google Scholar] [CrossRef]
- Ranieri, A.; Veneruso, I.; La Monica, I.; Pascale, M.; Pastore, L.; D’Argenio, V.; Lombardo, B. Combined ACGH and Exome Sequencing Analysis Improves Autism Spectrum Disorders Diagnosis: A Case Report. Medicina 2022, 58, 522. [Google Scholar] [CrossRef]
- Veneruso, I.; Ranieri, A.; Falcone, N.; Tripodi, L.; Scarano, C.; La Monica, I.; Pastore, L.; Lombardo, B.; D’Argenio, V. The Potential Usefulness of the Expanded Carrier Screening to Identify Hereditary Genetic Diseases: A Case Report from Real-World Data. Genes 2023, 14, 1651. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of Common Genetic Risk Variants for Autism Spectrum Disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef]
- Butler, M.; Rafi, S.; Manzardo, A. High-Resolution Chromosome Ideogram Representation of Currently Recognized Genes for Autism Spectrum Disorders. Int. J. Mol. Sci. 2015, 16, 6464–6495. [Google Scholar] [CrossRef]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism Spectrum Disorder: Neuropathology and Animal Models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef]
- Kotulska, K.; Jóźwiak, S. Autism in Monogenic Disorders. Eur. J. Paediatr. Neurol. 2011, 15, 177–180. [Google Scholar] [CrossRef]
- Acero-Garcés, D.O.; Saldarriaga, W.; Cabal-Herrera, A.M.; Rojas, C.A.; Hagerman, R.J. Fragile X Syndrome in Children. Colomb. Med. 2023, 54, e4005089. [Google Scholar] [CrossRef] [PubMed]
- Ziats, C.A.; Patterson, W.G.; Friez, M. Syndromic Autism Revisited: Review of the Literature and Lessons Learned. Pediatr. Neurol. 2021, 114, 21–25. [Google Scholar] [CrossRef]
- Fernandez, B.A.; Scherer, S.W. Syndromic Autism Spectrum Disorders: Moving from a Clinically Defined to a Molecularly Defined Approach. Dialogues Clin. Neurosci. 2017, 19, 353–371. [Google Scholar] [CrossRef]
- Gao, S.; Shan, C.; Zhang, R.; Wang, T. Genetic Advances in Neurodevelopmental Disorders. Med. Rev. 2025, 5, 139–151. [Google Scholar] [CrossRef]
- Lombardo, B.; Pagani, M.; De Rosa, A.; Nunziato, M.; Migliarini, S.; Garofalo, M.; Terrile, M.; D’Argenio, V.; Galbusera, A.; Nuzzo, T.; et al. D-Aspartate Oxidase Gene Duplication Induces Social Recognition Memory Deficit in Mice and Intellectual Disabilities in Humans. Transl. Psychiatry 2022, 12, 305. [Google Scholar] [CrossRef]
- Schaefer, G.B.; Lutz, R.E. Diagnostic Yield in the Clinical Genetic Evaluation of Autism Spectrum Disorders. Genet. Med. 2006, 8, 549–556. [Google Scholar] [CrossRef]
- Miles, J.H.; Takahashi, T.N.; Hong, J.; Munden, N.; Flournoy, N.; Braddock, S.R.; Martin, R.A.; Spence, M.A.; Hillman, R.E.; Farmer, J.E. Development and Validation of a Measure of Dysmorphology: Useful for Autism Subgroup Classification. Am. J. Med. Genet. A 2008, 146, 1101–1116. [Google Scholar] [CrossRef]
- Wier, M.L.; Yoshida, C.K.; Odouli, R.; Grether, J.K.; Croen, L.A. Congenital Anomalies Associated with Autism Spectrum Disorders. Dev. Med. Child. Neurol. 2006, 48, 500. [Google Scholar] [CrossRef]
- Schaaf, C.P.; Sabo, A.; Sakai, Y.; Crosby, J.; Muzny, D.; Hawes, A.; Lewis, L.; Akbar, H.; Varghese, R.; Boerwinkle, E.; et al. Oligogenic Heterozygosity in Individuals with High-Functioning Autism Spectrum Disorders. Hum. Mol. Genet. 2011, 20, 3366–3375. [Google Scholar] [CrossRef]
- Griesi-Oliveira, K.; Acab, A.; Gupta, A.R.; Sunaga, D.Y.; Chailangkarn, T.; Nicol, X.; Nunez, Y.; Walker, M.F.; Murdoch, J.D.; Sanders, S.J.; et al. Modeling Non-Syndromic Autism and the Impact of TRPC6 Disruption in Human Neurons. Mol. Psychiatry 2015, 20, 1350–1365. [Google Scholar] [CrossRef]
- Vorstman, J.A.S.; Scherer, S.W. Contemplating Syndromic Autism. Genet. Med. 2023, 25, 100919. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, C.P.; Zoghbi, H.Y. Solving the Autism Puzzle a Few Pieces at a Time. Neuron 2011, 70, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, P.J. The Fragile X Prevalence Paradox. J. Med. Genet. 2008, 45, 498–499. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.E.; Weisenfeld, L.A.H.; Hatton, D.D.; Heath, M.; Kaufmann, W.E. Social Approach and Autistic Behavior in Children with Fragile X Syndrome. J. Autism Dev. Disord. 2007, 37, 1748–1760. [Google Scholar] [CrossRef]
- Laurvick, C.L.; de Klerk, N.; Bower, C.; Christodoulou, J.; Ravine, D.; Ellaway, C.; Williamson, S.; Leonard, H. Rett Syndrome in Australia: A Review of the Epidemiology. J. Pediatr. 2006, 148, 347–352. [Google Scholar] [CrossRef]
- Reed, M.L.; Leff, S.E. Maternal Imprinting of Human SNRPN, a Gene Deleted in Prader–Willi Syndrome. Nat. Genet. 1994, 6, 163–167. [Google Scholar] [CrossRef]
- Dimitropoulos, A.; Schultz, R.T. Autistic-like Symptomatology in Prader-Willi Syndrome: A Review of Recent Findings. Curr. Psychiatry Rep. 2007, 9, 159–164. [Google Scholar] [CrossRef]
- Veltman, M.W.M.; Craig, E.E.; Bolton, P.F. Autism Spectrum Disorders in Prader-Willi and Angelman Syndromes: A Systematic Review. Psychiatr. Genet. 2005, 15, 243–254. [Google Scholar] [CrossRef]
- Tassabehji, M. Williams-Beuren Syndrome: A Challenge for Genotype-Phenotype Correlations. Hum. Mol. Genet. 2003, 12, R229–R237. [Google Scholar] [CrossRef]
- Caglayan, A.O. Genetic Causes of Syndromic and Non-syndromic Autism. Dev. Med. Child. Neurol. 2010, 52, 130–138. [Google Scholar] [CrossRef]
- Moreno-De-Luca, D.; Sanders, S.J.; Willsey, A.J.; Mulle, J.G.; Lowe, J.K.; Geschwind, D.H.; State, M.W.; Martin, C.L.; Ledbetter, D.H. Using Large Clinical Data Sets to Infer Pathogenicity for Rare Copy Number Variants in Autism Cohorts. Mol. Psychiatry 2013, 18, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Tropeano, M.; Ahn, J.W.; Dobson, R.J.B.; Breen, G.; Rucker, J.; Dixit, A.; Pal, D.K.; McGuffin, P.; Farmer, A.; White, P.S.; et al. Male-Biased Autosomal Effect of 16p13.11 Copy Number Variation in Neurodevelopmental Disorders. PLoS ONE 2013, 8, e61365. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.A.; Shen, Y.; Korn, J.M.; Arking, D.E.; Miller, D.T.; Fossdal, R.; Saemundsen, E.; Stefansson, H.; Ferreira, M.A.R.; Green, T.; et al. Association between Microdeletion and Microduplication at 16p11.2 and Autism. N. Engl. J. Med. 2008, 358, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, B.A.; Roberts, W.; Chung, B.; Weksberg, R.; Meyn, S.; Szatmari, P.; Joseph-George, A.M.; MacKay, S.; Whitten, K.; Noble, B.; et al. Phenotypic Spectrum Associated with de Novo and Inherited Deletions and Duplications at 16p11.2 in Individuals Ascertained for Diagnosis of Autism Spectrum Disorder. J. Med. Genet. 2010, 47, 195–203. [Google Scholar] [CrossRef]
- Sanders, S.J.; He, X.; Willsey, A.J.; Ercan-Sencicek, A.G.; Samocha, K.E.; Cicek, A.E.; Murtha, M.T.; Bal, V.H.; Bishop, S.L.; Dong, S.; et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015, 87, 1215–1233. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Muotri, A.R.; Sebat, J. Getting to the Cores of Autism. Cell 2019, 178, 1287–1298. [Google Scholar] [CrossRef]
- Doelken, S.C.; Köhler, S.; Mungall, C.J.; Gkoutos, G.V.; Ruef, B.J.; Smith, C.; Smedley, D.; Bauer, S.; Klopocki, E.; Schofield, P.N.; et al. Phenotypic Overlap in the Contribution of Individual Genes to CNV Pathogenicity Revealed by Cross-Species Computational Analysis of Single-Gene Mutations in Humans, Mice and Zebrafish. Dis. Model. Mech. 2013, 6, 358–372. [Google Scholar] [CrossRef]
- Velinov, M. Genomic Copy Number Variations in the Autism Clinic—Work in Progress. Front. Cell. Neurosci. 2019, 13, 57. [Google Scholar] [CrossRef]
- Girirajan, S.; Rosenfeld, J.A.; Cooper, G.M.; Antonacci, F.; Siswara, P.; Itsara, A.; Vives, L.; Walsh, T.; McCarthy, S.E.; Baker, C.; et al. A Recurrent 16p12.1 Microdeletion Supports a Two-Hit Model for Severe Developmental Delay. Nat. Genet. 2010, 42, 203–209. [Google Scholar] [CrossRef]
- Duyzend, M.H.; Nuttle, X.; Coe, B.P.; Baker, C.; Nickerson, D.A.; Bernier, R.; Eichler, E.E. Maternal Modifiers and Parent-of-Origin Bias of the Autism-Associated 16p11.2 CNV. Am. J. Hum. Genet. 2016, 98, 45–57. [Google Scholar] [CrossRef]
- Pizzo, L.; Jensen, M.; Polyak, A.; Rosenfeld, J.A.; Mannik, K.; Krishnan, A.; McCready, E.; Pichon, O.; Le Caignec, C.; Van Dijck, A.; et al. Rare Variants in the Genetic Background Modulate Cognitive and Developmental Phenotypes in Individuals Carrying Disease-Associated Variants. Genet. Med. 2019, 21, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Veltman, J.A.; Brunner, H.G. Understanding Variable Expressivity in Microdeletion Syndromes. Nat. Genet. 2010, 42, 192–193. [Google Scholar] [CrossRef] [PubMed]
- Bateson, W.; Mendel, G.; Leighton, A.G. Mendel’s Principles of Heredity, by W. Bateson; University Press: Cambridge, UK, 1909. [Google Scholar]
- Fisher, R.A. XV—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Trans. R. Soc. Edinb. 1919, 52, 399–433. [Google Scholar] [CrossRef]
- Webber, C. Epistasis in Neuropsychiatric Disorders. Trends Genet. 2017, 33, 256–265. [Google Scholar] [CrossRef]
- Girirajan, S.; Rosenfeld, J.A.; Coe, B.P.; Parikh, S.; Friedman, N.; Goldstein, A.; Filipink, R.A.; McConnell, J.S.; Angle, B.; Meschino, W.S.; et al. Phenotypic Heterogeneity of Genomic Disorders and Rare Copy-Number Variants. N. Engl. J. Med. 2012, 367, 1321–1331. [Google Scholar] [CrossRef]
- Servetti, M.; Pisciotta, L.; Tassano, E.; Cerminara, M.; Nobili, L.; Boeri, S.; Rosti, G.; Lerone, M.; Divizia, M.T.; Ronchetto, P.; et al. Neurodevelopmental Disorders in Patients with Complex Phenotypes and Potential Complex Genetic Basis Involving Non-Coding Genes, and Double CNVs. Front. Genet. 2021, 12, 732002. [Google Scholar] [CrossRef]
- Um, J.W.; Ko, J. Neural Glycosylphosphatidylinositol-Anchored Proteins in Synaptic Specification. Trends Cell Biol. 2017, 27, 931–945. [Google Scholar] [CrossRef]
- Helbig, K.L.; Mroske, C.; Moorthy, D.; Sajan, S.A.; Velinov, M. Biallelic Loss-of-function Variants in DOCK3 Cause Muscle Hypotonia, Ataxia, and Intellectual Disability. Clin. Genet. 2017, 92, 430–433. [Google Scholar] [CrossRef]
- Demily, C.; Lesca, G.; Poisson, A.; Till, M.; Barcia, G.; Chatron, N.; Sanlaville, D.; Munnich, A. Additive Effect of Variably Penetrant 22q11.2 Duplication and Pathogenic Mutations in Autism Spectrum Disorder: To Which Extent Does the Tree Hide the Forest? J. Autism Dev. Disord. 2018, 48, 2886–2889. [Google Scholar] [CrossRef]
- Guo, H.; Wang, T.; Wu, H.; Long, M.; Coe, B.P.; Li, H.; Xun, G.; Ou, J.; Chen, B.; Duan, G.; et al. Inherited and Multiple de Novo Mutations in Autism/Developmental Delay Risk Genes Suggest a Multifactorial Model. Mol. Autism 2018, 9, 64. [Google Scholar] [CrossRef]
- Bourgeron, T. From the Genetic Architecture to Synaptic Plasticity in Autism Spectrum Disorder. Nat. Rev. Neurosci. 2015, 16, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef] [PubMed]
- Zaslavsky, K.; Zhang, W.-B.; McCready, F.P.; Rodrigues, D.C.; Deneault, E.; Loo, C.; Zhao, M.; Ross, P.J.; El Hajjar, J.; Romm, A.; et al. SHANK2 Mutations Associated with Autism Spectrum Disorder Cause Hyperconnectivity of Human Neurons. Nat. Neurosci. 2019, 22, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Chen, S.; Chen, X.; Zheng, J.; Xu, Z.; Doostparast Torshizi, A.; Gong, S.; Chen, Q.; Ma, X.; Yu, J.; et al. Uncovering the Functional Link Between SHANK3 Deletions and Deficiency in Neurodevelopment Using IPSC-Derived Human Neurons. Front. Neuroanat. 2019, 13, 23. [Google Scholar] [CrossRef]
- Qin, L.; Ma, K.; Wang, Z.-J.; Hu, Z.; Matas, E.; Wei, J.; Yan, Z. Social Deficits in Shank3-Deficient Mouse Models of Autism Are Rescued by Histone Deacetylase (HDAC) Inhibition. Nat. Neurosci. 2018, 21, 564–575. [Google Scholar] [CrossRef]
- Ronan, J.L.; Wu, W.; Crabtree, G.R. From Neural Development to Cognition: Unexpected Roles for Chromatin. Nat. Rev. Genet. 2013, 14, 347–359. [Google Scholar] [CrossRef]
- Suetterlin, P.; Hurley, S.; Mohan, C.; Riegman, K.L.H.; Pagani, M.; Caruso, A.; Ellegood, J.; Galbusera, A.; Crespo-Enriquez, I.; Michetti, C.; et al. Altered Neocortical Gene Expression, Brain Overgrowth and Functional Over-Connectivity in Chd8 Haploinsufficient Mice. Cereb. Cortex 2018, 28, 2192–2206. [Google Scholar] [CrossRef]
- Fernandes, I.R.; Cruz, A.C.P.; Ferrasa, A.; Phan, D.; Herai, R.H.; Muotri, A.R. Genetic Variations on SETD5 Underlying Autistic Conditions. Dev. Neurobiol. 2018, 78, 500–518. [Google Scholar] [CrossRef]
- de Masfrand, S.; Cogné, B.; Nizon, M.; Deb, W.; Goldenberg, A.; Lecoquierre, F.; Nicolas, G.; Bournez, M.; Vitobello, A.; Mau-Them, F.T.; et al. Penetrance, Variable Expressivity and Monogenic Neurodevelopmental Disorders. Eur. J. Med. Genet. 2024, 69, 104932. [Google Scholar] [CrossRef]
- Mollon, J.; Almasy, L.; Jacquemont, S.; Glahn, D.C. The Contribution of Copy Number Variants to Psychiatric Symptoms and Cognitive Ability. Mol. Psychiatry 2023, 28, 1480–1493. [Google Scholar] [CrossRef]
- Čiuladaitė, Ž.; Kasnauskienė, J.; Cimbalistienė, L.; Preikšaitienė, E.; Patsalis, P.C.; Kučinskas, V. Mental Retardation and Autism Associated with Recurrent 16p11.2 Microdeletion: Incomplete Penetrance and Variable Expressivity. J. Appl. Genet. 2011, 52, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, E.Z.; Brown, K.; Byler, M.C.; D’haenens, E.; Dheedene, A.; Henderson, L.B.; Humberson, J.B.; van Jaarsveld, R.H.; Kanani, F.; Lebel, R.R.; et al. Expanding the Molecular Spectrum and the Neurological Phenotype Related to CAMTA1 Variants. Clin. Genet. 2021, 99, 259–268. [Google Scholar] [CrossRef] [PubMed]
- van Rahden, V.A.; Fernandez-Vizarra, E.; Alawi, M.; Brand, K.; Fellmann, F.; Horn, D.; Zeviani, M.; Kutsche, K. Mutations in NDUFB11, Encoding a Complex I Component of the Mitochondrial Respiratory Chain, Cause Microphthalmia with Linear Skin Defects Syndrome. Am. J. Hum. Genet. 2015, 96, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Latchman, K.; Calder, M.; Morel, D.; Rhodes, L.; Juusola, J.; Tekin, M. Autosomal Dominant Inheritance in a Recently Described ZMIZ1-Related Neurodevelopmental Disorder: Case Report of Siblings and an Affected Parent. Am. J. Med. Genet. A 2020, 182, 548–552. [Google Scholar] [CrossRef]
- Kendall, K.M.; Bracher-Smith, M.; Fitzpatrick, H.; Lynham, A.; Rees, E.; Escott-Price, V.; Owen, M.J.; O’Donovan, M.C.; Walters, J.T.R.; Kirov, G. Cognitive Performance and Functional Outcomes of Carriers of Pathogenic Copy Number Variants: Analysis of the UK Biobank. Br. J. Psychiatry 2019, 214, 297–304. [Google Scholar] [CrossRef]
- Furukawa, S.; Kushima, I.; Aleksic, B.; Ozaki, N. Case Reports of Two Siblings with Autism Spectrum Disorder and 15q13.3 Deletions. Neuropsychopharmacol. Rep. 2023, 43, 462–466. [Google Scholar] [CrossRef]
- Goh, S.; Thiyagarajan, L.; Dudding-Byth, T.; Pinese, M.; Kirk, E.P. A Systematic Review and Pooled Analysis of Penetrance Estimates of Copy-Number Variants Associated with Neurodevelopment. Genet. Med. 2025, 27, 101227. [Google Scholar] [CrossRef]
- Lombardo, B.; Ceglia, C.; Verdesca, F.; Vitale, A.; Perrotta, C.; Leggiero, E.; Pastore, L. CGH Array for the Identification of a Compound Heterozygous Mutation in the CYP1B1 Gene in a Patient with Bilateral Anterior Segment Dysgenesis. Clin. Chem. Lab. Med. 2019, 57, e63–e66. [Google Scholar] [CrossRef]
- Toro, R.; Konyukh, M.; Delorme, R.; Leblond, C.; Chaste, P.; Fauchereau, F.; Coleman, M.; Leboyer, M.; Gillberg, C.; Bourgeron, T. Key Role for Gene Dosage and Synaptic Homeostasis in Autism Spectrum Disorders. Trends Genet. 2010, 26, 363–372. [Google Scholar] [CrossRef]
- Lim, E.T.; Raychaudhuri, S.; Sanders, S.J.; Stevens, C.; Sabo, A.; MacArthur, D.G.; Neale, B.M.; Kirby, A.; Ruderfer, D.M.; Fromer, M.; et al. Rare Complete Knockouts in Humans: Population Distribution and Significant Role in Autism Spectrum Disorders. Neuron 2013, 77, 235–242. [Google Scholar] [CrossRef]
- Lin, B.D.; Colas, F.; Nijman, I.J.; Medic, J.; Brands, W.; Parr, J.R.; van Eijk, K.R.; Klauck, S.M.; Chiocchetti, A.G.; Freitag, C.M.; et al. The Role of Rare Compound Heterozygous Events in Autism Spectrum Disorder. Transl. Psychiatry 2020, 10, 204. [Google Scholar] [CrossRef] [PubMed]
- Hochstenbach, R.; Poot, M.; Nijman, I.J.; Renkens, I.; Duran, K.J.; van’T Slot, R.; van Binsbergen, E.; van der Zwaag, B.; Vogel, M.J.; Terhal, P.A.; et al. Discovery of Variants Unmasked by Hemizygous Deletions. Eur. J. Hum. Genet. 2012, 20, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Bacchelli, E.; Ceroni, F.; Pinto, D.; Lomartire, S.; Giannandrea, M.; D’Adamo, P.; Bonora, E.; Parchi, P.; Tancredi, R.; Battaglia, A.; et al. A CTNNA3 Compound Heterozygous Deletion Implicates a Role for AT-Catenin in Susceptibility to Autism Spectrum Disorder. J. Neurodev. Disord. 2014, 6, 17. [Google Scholar] [CrossRef]
- Malerba, N.; Towner, S.; Keating, K.; Squeo, G.M.; Wilson, W.; Merla, G. A NGS-Targeted Autism/ID Panel Reveals Compound Heterozygous GNB5 Variants in a Novel Patient. Front. Genet. 2018, 9, 626. [Google Scholar] [CrossRef]
- Casey, J.P.; Magalhaes, T.; Conroy, J.M.; Regan, R.; Shah, N.; Anney, R.; Shields, D.C.; Abrahams, B.S.; Almeida, J.; Bacchelli, E.; et al. A Novel Approach of Homozygous Haplotype Sharing Identifies Candidate Genes in Autism Spectrum Disorder. Hum. Genet. 2012, 131, 565–579. [Google Scholar] [CrossRef]

| CNV Locus | Clinical Features Associated with CNV |
|---|---|
| 1q21.1 deletion | Mild-moderate ID, SCZ, mild facial dysmorphic features, microcephaly, cataracts |
| 1q21.1 duplication | Mild-moderate ID, ADHD, mild dysmorphic features, macrocephaly, hypotonia |
| 3q29 deletion | Mild-moderate ID, SCZ, mild facial dysmorphic features |
| 5q35 deletion (Sotos Syndrome) | Developmental delay, microcephaly |
| 7q11.23 duplication (William Syndrome) | ID, SCZ, brain MRI abnormalities, variable dysmorphic features |
| 15q11q13 duplication (Prader-Willi Syndrome) | Mild-moderate ID, epilepsy, ataxia, behavioral problems, hypotonia |
| 15q13.3 deletion | Mild-moderate ID, epilepsy, learning disabilities, ADHD, variable dysmorphic features |
| 16p11.2 deletion | Mild-moderate ID, epilepsy, MCA, variable dysmorphic features, macrocephaly, obesity |
| 16p11.2 duplication | Mild-moderate ID, ADHD, microcephaly, dysmorphic features |
| 16p12.1 deletion | Mild-moderate ID, ADHD, and dysmorphic features |
| 16p13.1 deletion | ID, SCZ, epilepsy, MCA, dysmorphic features |
| 17p11.2 deletion | ID, speech delay, deafness, sleep disturbance, hypotonia |
| 17q12 deletion | Mild-moderate ID, SCZ, epilepsy, facial dysmorphic features |
| 22q11.2 deletion (DiGeorge Syndrome) | ID, SCZ, learning difficulties, MCA, dysmorphic features |
| 22q11.2 duplication | ID, SCZ, speech difficulties, learning difficulties, dysmorphic features, microcephaly |
| Genetic Alteration | Clinical Effect | References |
|---|---|---|
| 16p11.2 deletion | Identification of secondary CNVs affecting genes associated with the risk of ASD and intellectual disability, such as CACNA2D3, TRIO, and KATNAL2. Identification of additional rare SNVs that correlate with increased disease severity, especially in multiplex families. | Girirajan et al. [81] Duyzend et al. [82,83] Pizzo et al. [83] |
| 16p12.1 deletion | Secondary genetic events transform the deletion phenotype from normal to severe:
| Veltman et al. [84] Servetti et al. [89] |
| Category | Gene | Biological Function | Associated Syndrome/ OMIM Number |
|---|---|---|---|
| Synaptic receptors and cell adhesion molecules | NRXN1 | Cell-surface receptors that bind neuroligins to form Ca(2+)-dependent neurexin/neuroligin complexes at synapses in the central nervous system. | Pitt-hopkins-like syndrome 2 (#614325) |
| NLGN1 | Neuronal cell surface proteins that may act as splice site-specific ligands for β-neurexins and may be involved in the formation and remodeling of central nervous system synapses. | Susceptibility to autism (#618830) | |
| Scaffolding proteins and the actin cytoskeleton | SHANK2 | Scaffold protein that localizes to postsynaptic sites of excitatory synapses in the brain. | Susceptibility to autism (#613436) |
| SHANK3 | Scaffolding protein that is enriched in postsynaptic densities of excitatory synapses. | Schizophrenia (#613950) and Phelan-McDermid syndrome (#606232) | |
| Chromatin remodeling and transcription | MECP2 | Chromatin-associated protein that can both activate and repress transcription. It is required for the maturation of neurons and is developmentally regulated. | Intellectual developmental disorder X-linked syndromic (#300055), Rett syndrome (#312750) and Susceptibility to autism (#300496) |
| SETD5 | Methyltransferase that targets histone H3 lys36 for trimethylation (H3K36me3) and thereby controls the transcriptional landscape in neural progenitors and their derivatives. | Intellectual developmental disorder (#615761) | |
| CHD8 | ATP-dependent chromatin-remodeling factor that regulates transcription of β-catenin (CTNNB1) target genes. | Intellectual developmental disorder with autism and macrocephaly (#615032) | |
| ASH1L | Histone methyltransferase is involved in epigenetic modification of chromatin and plays important roles in development. | Intellectual developmental disorder (#617796) |
| Reference | Evidence |
|---|---|
| de Masfrand et al. [102] | 12 patients with NDD presented loss-of-function variants in 11 genes, including CAMTA1, NDUFB11, and ZMIZ1; all variants were inherited from healthy parents, showing phenotypic variability in the same family. |
| Kendall et al. [108] | Carriers may show intermediate cognitive profiles compared to non-carriers and affected patients. CNV carriers often have lower educational levels and less skilled jobs, despite appearing clinically unaffected. |
| Furukawa et al. [109] | 15q13.3 deletion in affected siblings and a healthy mother who shared the same CHRNA7 gene deletion, and represents a clear example of incomplete penetrance |
| Mollon et al. [103] | Additional rare variants influence patient phenotype, and incomplete penetrance interferes with phenotype variability in individuals with CNVs such as 1q21.1, 7q11.23, 16p12.1, 16p11.2, and 22q11.2. |
| Goh et al. [110] | Penetrance estimates for 92 CNVs using gnomAD v4.0:
|
| Study Details | Reference |
|---|---|
| 550 genes sequenced in 149 ASD probands and their deletion-transmitting parents: 13.4% of the probands and 8.1% of the parents had co-occurring variations. The pathogenic effect of a deletion may be triggered by compound heterozygosity. | Lin et al. [114] |
| The proband with ASD has a compound heterozygous deletion in the CTNNA3 gene, which is implicated in cell adhesion and is essential in ASD, and his sister acquired only one deletion and is unaffected. | Bacchelli et al. [116] |
| A 2.5-year-old girl with ASD had compound heterozygous mutations in the GNB5 gene: a frameshift (paternal) and a missense (maternal) variant. In contrast to homozygous mutations, which result in more severe consequences, compound heterozygosity of GNB5 might induce milder symptoms of ASD. The severity of mutations in the GNB5 gene varies. | Malerba et al. [117] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Monica, I.; Di Iorio, M.R.; Sica, A.; Rufino, F.; Sotira, C.; Pastore, L.; Lombardo, B. Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns. Genes 2025, 16, 478. https://doi.org/10.3390/genes16050478
La Monica I, Di Iorio MR, Sica A, Rufino F, Sotira C, Pastore L, Lombardo B. Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns. Genes. 2025; 16(5):478. https://doi.org/10.3390/genes16050478
Chicago/Turabian StyleLa Monica, Ilaria, Maria Rosaria Di Iorio, Antonia Sica, Francesca Rufino, Chiara Sotira, Lucio Pastore, and Barbara Lombardo. 2025. "Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns" Genes 16, no. 5: 478. https://doi.org/10.3390/genes16050478
APA StyleLa Monica, I., Di Iorio, M. R., Sica, A., Rufino, F., Sotira, C., Pastore, L., & Lombardo, B. (2025). Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns. Genes, 16(5), 478. https://doi.org/10.3390/genes16050478





