Phylogenetic Insights into the Evolutionary History of the RSPO Gene Family in Metazoa
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Data Collection and Processing
2.2. Homologs Identification
2.3. Phylogenetic Analysis and Gene Family Clustering
2.4. Analysis of Structural Diversity of R-Spondins Protein
3. Results
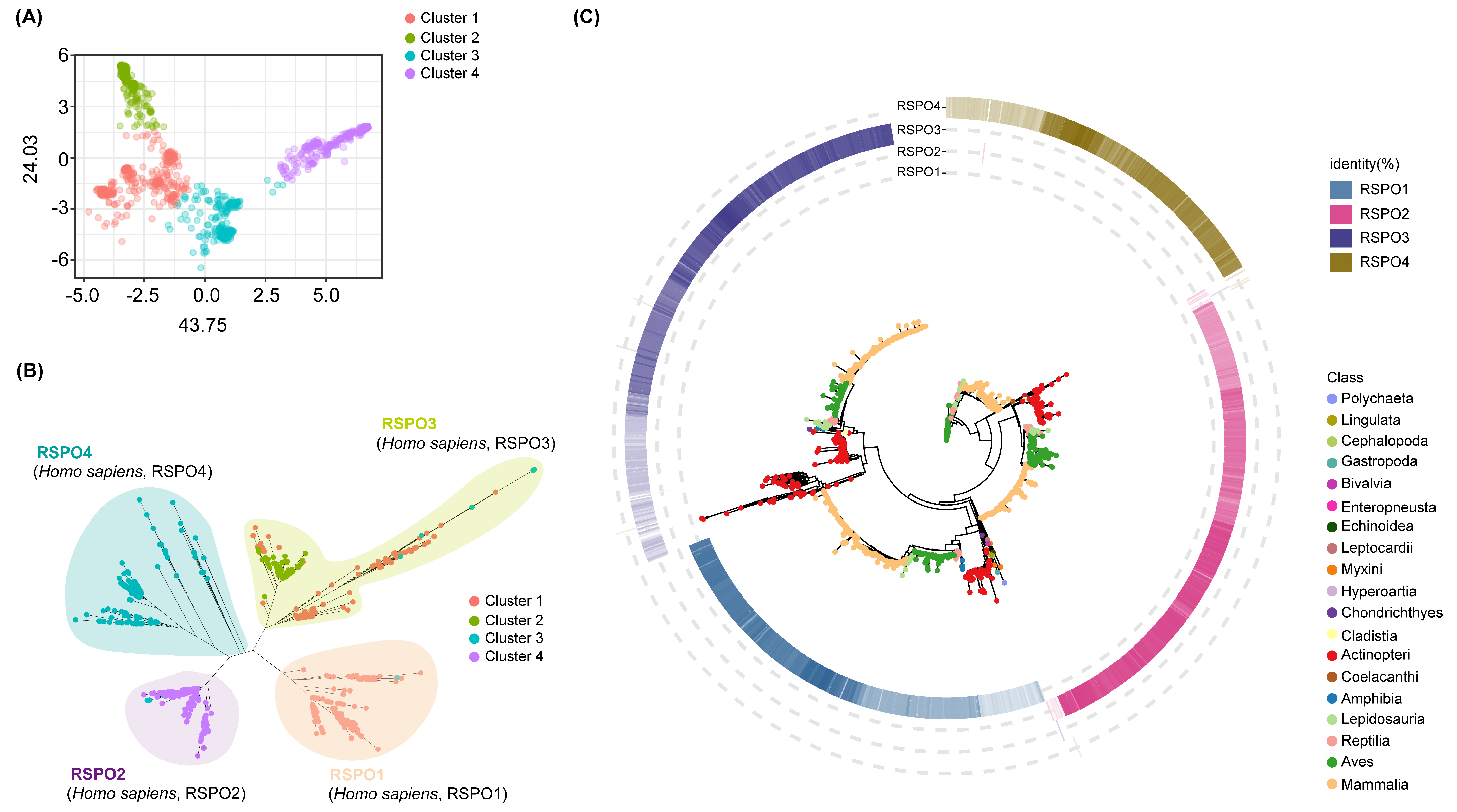
3.1. Phylogenetic Analysis and Gene Family Clustering Define the Four Subgroups of the RSPO Family
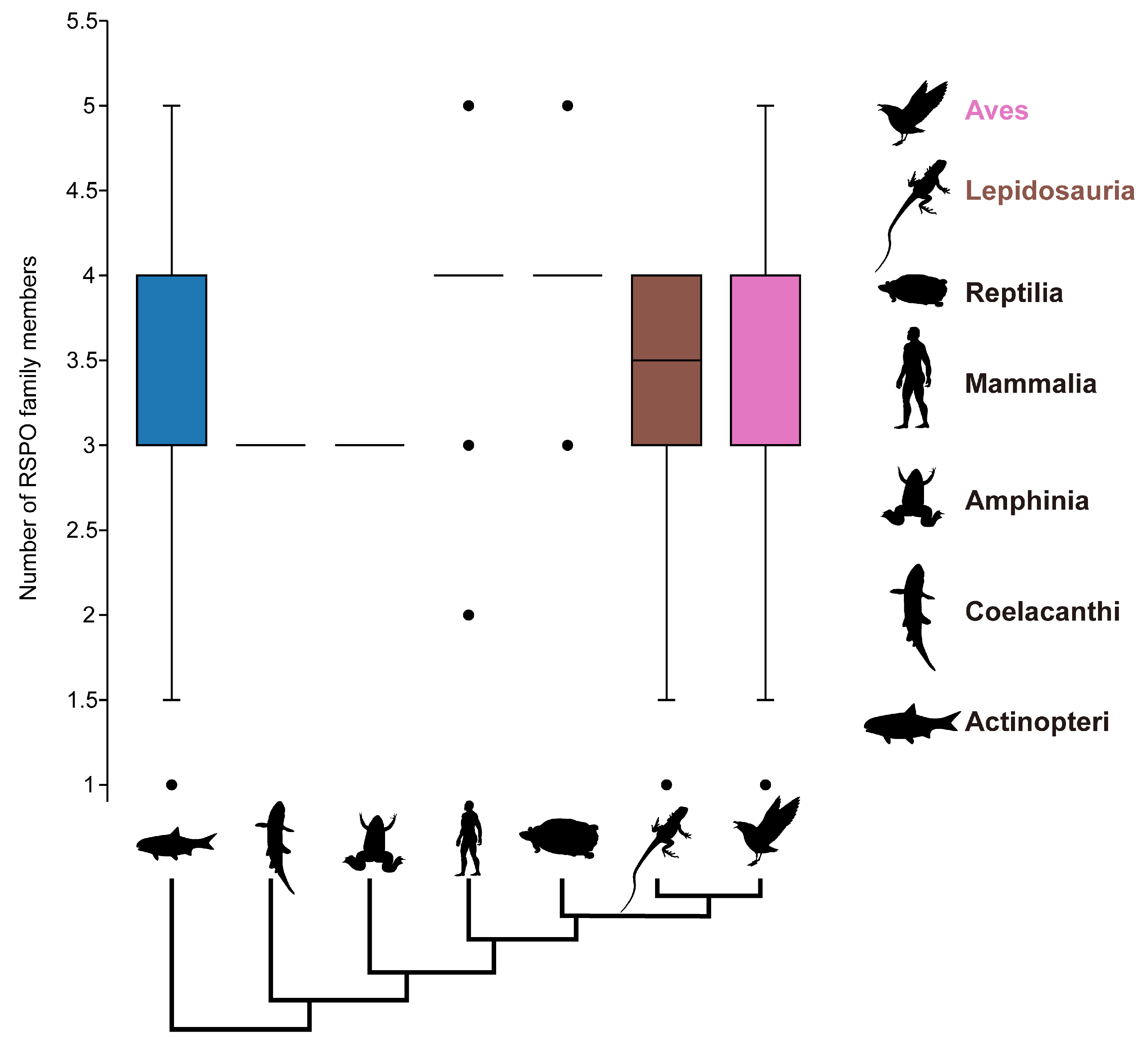
3.2. Four Subgroups Showed Different Evolutionary Patterns
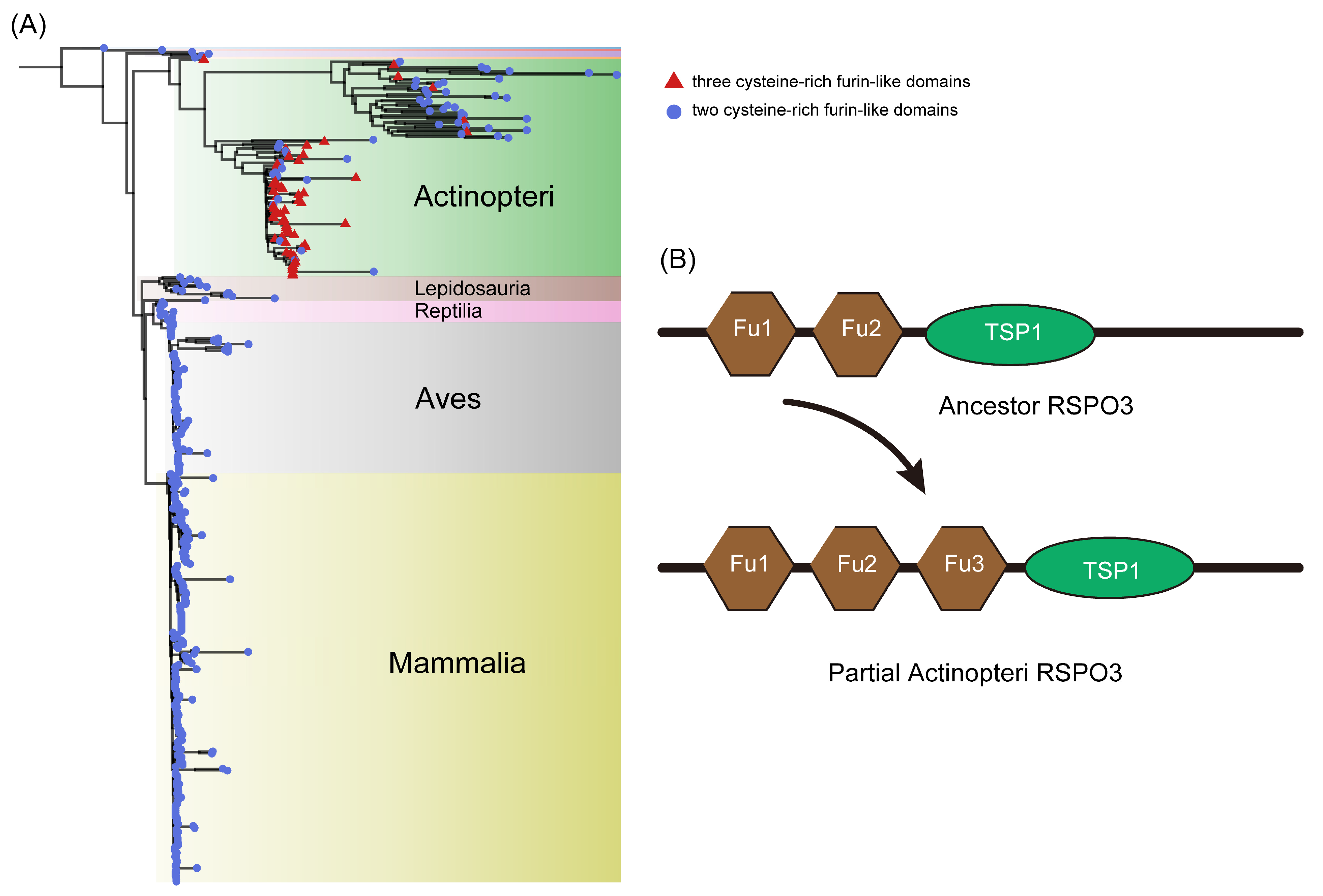
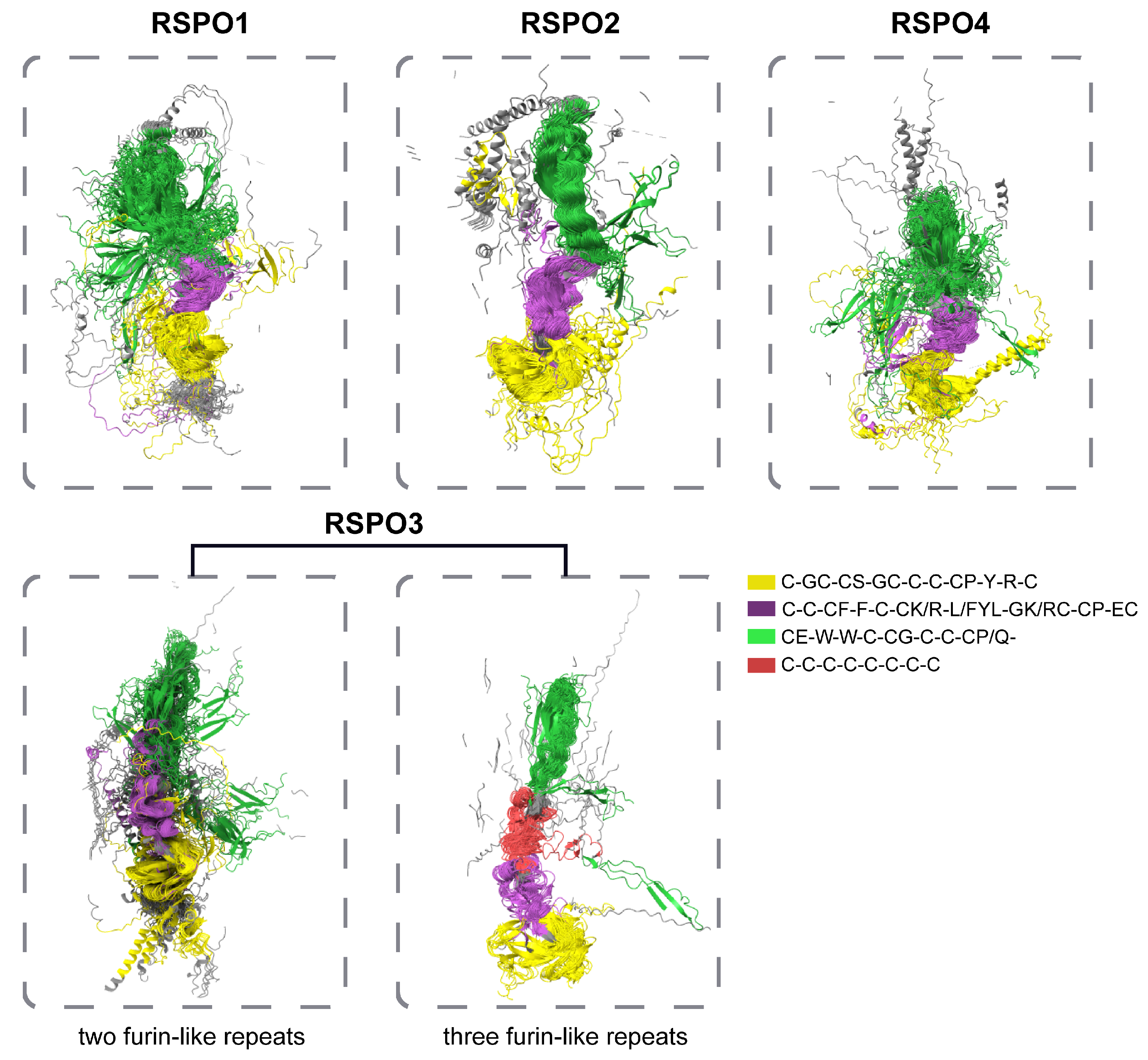
3.3. Some RSPO3 Members of Actinopteri Have Three Furin-like Repeats
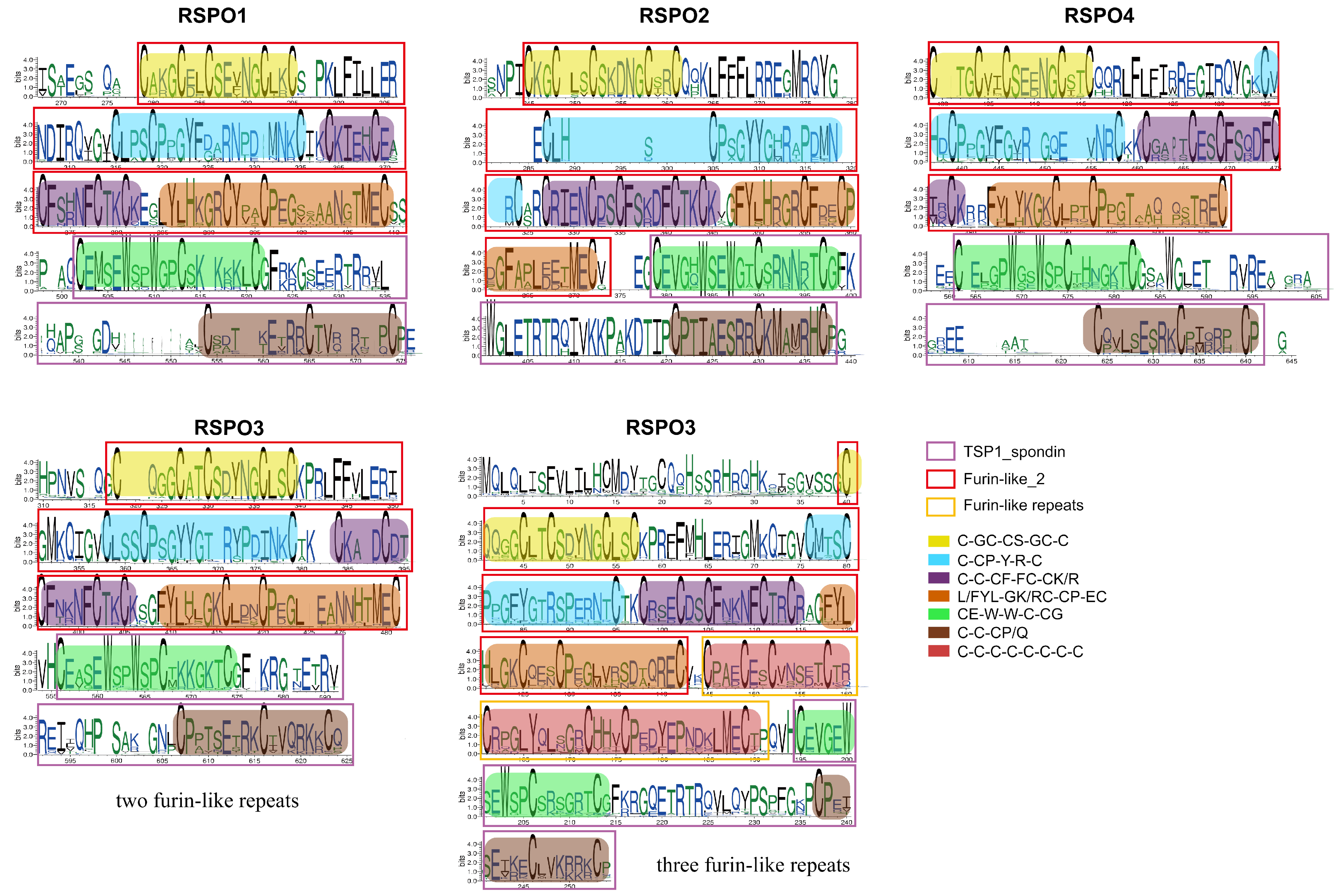
3.4. The Identification of Conservative Motif
3.5. The Analysis of Spatial Structure of RSPO Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RSPO | the R-spondin family |
| TSP domain | Thrombospondin Type 1 Repeat domain |
| FU domain | Repeated cysteine-rich furin-like domain |
References
- Pires-daSilva, A.; Sommer, R.J. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003, 4, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Gori, F.; Superti-Furga, A.; Baron, R. Bone Formation and the Wnt Signaling Pathway. N. Engl. J. Med. 2016, 375, 1902–1903. [Google Scholar] [PubMed]
- Pai, S.G.; Carneiro, B.A.; Mota, J.M.; Costa, R.; Leite, C.A.; Barroso-Sousa, R.; Kaplan, J.B.; Chae, Y.K.; Giles, F.J. Wnt/β-catenin pathway: Modulating anticancer immune response. J. Hematol. Oncol. 2017, 10, 101. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, L. The non-canonical Wnt pathway leads to aged dendritic cell differentiation. Cell. Mol. Immunol. 2018, 15, 871–872. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/β-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Moon, R.T.; Bowerman, B.; Boutros, M.; Perrimon, N. The promise and perils of Wnt signaling through β-catenin. Science 2002, 296, 1644–1646. [Google Scholar] [CrossRef]
- De, A. Wnt/Ca2+ signaling pathway: A brief overview. Acta Biochim. Biophys. Sin. 2011, 43, 745–756. [Google Scholar] [CrossRef]
- Shi, D.L. Wnt/planar cell polarity signaling controls morphogenetic movements of gastrulation and neural tube closure. Cell. Mol. Life Sci. 2022, 79, 586. [Google Scholar] [CrossRef]
- Hayat, R.; Manzoor, M.; Hussain, A. Wnt signaling pathway: A comprehensive review. Cell Biol. Int. 2022, 46, 863–877. [Google Scholar] [CrossRef]
- Ranes, M.; Zaleska, M.; Sakalas, S.; Knight, R.; Guettler, S. Reconstitution of the destruction complex defines roles of AXIN polymers and APC in β-catenin capture, phosphorylation, and ubiquitylation. Mol. Cell 2021, 81, 3246–3261.e3211. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.-X.; Xie, Y.; Zhang, Y.; Charlat, O.; Oster, E.; Avello, M.; Lei, H.; Mickanin, C.; Liu, D.; Ruffner, H. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012, 485, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.-K.; Spit, M.; Jordens, I.; Low, T.Y.; Stange, D.E.; Van De Wetering, M.; Van Es, J.H.; Mohammed, S.; Heck, A.J.; Maurice, M.M. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012, 488, 665–669. [Google Scholar] [CrossRef]
- Kazanskaya, O.; Glinka, A.; del Barco Barrantes, I.; Stannek, P.; Niehrs, C.; Wu, W. R-Spondin2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 2004, 7, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef]
- Chen, P.-H.; Chen, X.; Lin, Z.; Fang, D.; He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013, 27, 1345–1350. [Google Scholar] [CrossRef]
- Glinka, A.; Dolde, C.; Kirsch, N.; Huang, Y.L.; Kazanskaya, O.; Ingelfinger, D.; Boutros, M.; Cruciat, C.M.; Niehrs, C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011, 12, 1055–1061. [Google Scholar] [CrossRef]
- de Lau, W.; Barker, N.; Low, T.Y.; Koo, B.-K.; Li, V.S.W.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef]
- Zebisch, M.; Xu, Y.; Krastev, C.; MacDonald, B.T.; Chen, M.; Gilbert, R.J.C.; He, X.; Jones, E.Y. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun. 2013, 4, 2787. [Google Scholar] [CrossRef]
- Kim, K.A.; Zhao, J.; Andarmani, S.; Kakitani, M.; Oshima, T.; Binnerts, M.E.; Abo, A.; Tomizuka, K.; Funk, W.D. R-Spondin proteins: A novel link to β-catenin activation. Cell Cycle 2006, 5, 23–26. [Google Scholar] [CrossRef]
- Lebensohn, A.M.; Rohatgi, R. R-spondins can potentiate WNT signaling without LGRs. eLife 2018, 7, e33126. [Google Scholar] [CrossRef] [PubMed]
- Szenker-Ravi, E.; Altunoglu, U.; Leushacke, M.; Bosso-Lefèvre, C.; Khatoo, M.; Thi Tran, H.; Naert, T.; Noelanders, R.; Hajamohideen, A.; Beneteau, C. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature 2018, 557, 564–569. [Google Scholar] [CrossRef]
- Carmon, K.S.; Gong, X.; Yi, J.; Thomas, A.; Liu, Q. RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc. Natl. Acad. Sci. USA 2014, 111, E1221–E1229. [Google Scholar] [CrossRef]
- Ohkawara, B.; Glinka, A.; Niehrs, C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell 2011, 20, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Bezanilla, D.; Mardones, M.D.; Galassi, M.; Arredondo, S.B.; Santibanez, S.H.; Gutierrez-Jimenez, S.; Merino-Véliz, N.; Bustos, F.J.; Varela-Nallar, L. RSPO/LGR signaling regulates proliferation of adult hippocampal neural stem cells. Stem Cells 2024, 43, sxae065. [Google Scholar] [CrossRef]
- Dubey, R.; van Kerkhof, P.; Jordens, I.; Malinauskas, T.; Pusapati, G.V.; McKenna, J.K.; Li, D.; Carette, J.E.; Ho, M.; Siebold, C.; et al. R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling. eLife 2020, 9, e54469. [Google Scholar] [CrossRef]
- Lee, H.; Camuto, C.M.; Niehrs, C. R-Spondin 2 governs Xenopus left-right body axis formation by establishing an FGF signaling gradient. Nat. Commun. 2024, 15, 1003. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Z.; Wang, S.; Tang, R.; Yang, Q.-S.; Zhao, E.; Chao, Y.; Ying, K.; Xie, Y.; Mao, Y.-M. Cloning and identification of a cDNA that encodes a novel human protein with thrombospondin type I repeat domain, hPWTSR. Mol. Biol. Rep. 2002, 29, 287–292. [Google Scholar] [CrossRef]
- Kamata, T.; Katsube, K.-I.; Michikawa, M.; Yamada, M.; Takada, S.; Mizusawa, H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 2004, 1676, 51–62. [Google Scholar] [CrossRef]
- Nam, J.S.; Turcotte, T.J.; Smith, P.F.; Choi, S.; Yoon, J.K. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate β-catenin-dependent gene expression. J. Biol. Chem. 2006, 281, 13247–13257. [Google Scholar] [CrossRef]
- Wei, Q.; Yokota, C.; Semenov, M.V.; Doble, B.; Woodgett, J.; He, X. R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and β-catenin signaling. J. Biol. Chem. 2007, 282, 15903–15911. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Schreiner, C.M.; Wert, S.E.; Mucenski, M.L.; Scott, W.J.; Whitsett, J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 2008, 135, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Lv, C.; Sun, S.; Zhao, H.; Ling, H.; Li, M.; Qin, Y.; Zhang, J.; Wang, J.; Yang, X. TSP1 is the essential domain of SEMA5A involved in pannus formation in rheumatoid arthritis. Rheumatology 2021, 60, 5833–5842. [Google Scholar] [CrossRef]
- Zolfaghari, S.; Kaasbøll, O.J.; Monsen, V.T.; Sredic, B.; Hagelin, E.M.V.; Attramadal, H. The carboxyl-terminal TSP1-homology domain is the biologically active effector peptide of matricellular protein CCN5 that counteracts profibrotic CCN2. J. Biol. Chem. 2023, 299, 102803. [Google Scholar] [CrossRef]
- Rinta-Jaskari, M.M.; Naillat, F.; Ruotsalainen, H.J.; Koivunen, J.T.; Sasaki, T.; Pietilä, I.; Elamaa, H.P.; Kaur, I.; Manninen, A.; Vainio, S.J.; et al. Temporally and spatially regulated collagen XVIII isoforms are involved in ureteric tree development via the TSP1-like domain. Matrix Biol. 2023, 115, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, L. Molecular modelling of the TSR domain of R-spondin 4. Bioinformation 2008, 3, 119. [Google Scholar] [CrossRef][Green Version]
- Conboy, C.B.; Vélez-Reyes, G.L.; Rathe, S.K.; Abrahante, J.E.; Temiz, N.A.; Burns, M.B.; Harris, R.S.; Starr, T.K.; Largaespada, D.A. R-Spondins 2 and 3 Are Overexpressed in a Subset of Human Colon and Breast Cancers. DNA Cell Biol. 2021, 40, 70–79. [Google Scholar] [CrossRef]
- Conboy, C.B.; Vélez-Reyes, G.L.; Tschida, B.R.; Hu, H.; Kaufmann, G.; Koes, N.; Keller, B.; Alsinet, C.; Cornellà, H.; Pinyol, R.; et al. R-spondin 2 Drives Liver Tumor Development in a Yes-Associated Protein-Dependent Manner. Hepatol. Commun. 2019, 3, 1496–1509. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, L.; Chen, L.; Xiong, M.; Kazobinka, G.; Pang, Z.; Hou, T. RSPO3 promotes the aggressiveness of bladder cancer via Wnt/β-catenin and Hedgehog signaling pathways. Carcinogenesis 2019, 40, 360–369. [Google Scholar] [CrossRef]
- De Cian, M.C.; Pauper, E.; Bandiera, R.; Vidal, V.P.I.; Sacco, S.; Gregoire, E.P.; Chassot, A.A.; Panzolini, C.; Wilhelm, D.; Pailhoux, E.; et al. Amplification of R-spondin1 signaling induces granulosa cell fate defects and cancers in mouse adult ovary. Oncogene 2017, 36, 208–218. [Google Scholar] [CrossRef]
- da Silva, S.D.; Morand, G.B.; Diesel, L.; de Lima, J.M.; Bijian, K.; Kailasam, S.; Lefebvre, F.; Bourque, G.; Hier, M.; Alaoui-Jamali, M.A. Identification of R-Spondin Gene Signature Predictive of Metastatic Progression in BRAFV600E-Positive Papillary Thyroid Cancer. Cells 2023, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Kocer, A.; Pinheiro, I.; Pannetier, M.; Renault, L.; Parma, P.; Radi, O.; Kim, K.-A.; Camerino, G.; Pailhoux, E. R-spondin1 and FOXL2act into two distinct cellular types during goat ovarian differentiation. BMC Dev. Biol. 2008, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef]
- Tomizuka, K.; Horikoshi, K.; Kitada, R.; Sugawara, Y.; Iba, Y.; Kojima, A.; Yoshitome, A.; Yamawaki, K.; Amagai, M.; Inoue, A.; et al. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet. 2008, 17, 1278–1291. [Google Scholar] [CrossRef]
- Nam, J.-S.; Park, E.; Turcotte, T.J.; Palencia, S.; Zhan, X.; Lee, J.; Yun, K.; Funk, W.D.; Yoon, J.K. Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev. Biol. 2007, 311, 124–135. [Google Scholar] [CrossRef]
- Aoki, M.; Kiyonari, H.; Nakamura, H.; Okamoto, H. R-spondin2 expression in the apical ectodermal ridge is essential for outgrowth and patterning in mouse limb development. Dev. Growth Differ. 2008, 50, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Yamada, W.; Nagao, K.; Horikoshi, K.; Fujikura, A.; Ikeda, E.; Inagaki, Y.; Kakitani, M.; Tomizuka, K.; Miyazaki, H.; Suda, T.; et al. Craniofacial malformation in R-spondin2 knockout mice. Biochem. Biophys. Res. Commun. 2009, 381, 453–458. [Google Scholar] [CrossRef]
- Tachibana, N.; Chijimatsu, R.; Okada, H.; Oichi, T.; Taniguchi, Y.; Maenohara, Y.; Miyahara, J.; Ishikura, H.; Iwanaga, Y.; Arino, Y.; et al. RSPO2 defines a distinct undifferentiated progenitor in the tendon/ligament and suppresses ectopic ossification. Sci. Adv. 2022, 8, eabn2138. [Google Scholar] [CrossRef]
- Aoki, M.; Mieda, M.; Ikeda, T.; Hamada, Y.; Nakamura, H.; Okamoto, H. R-spondin3 is required for mouse placental development. Dev. Biol. 2007, 301, 218–226. [Google Scholar] [CrossRef]
- Kazanskaya, O.; Ohkawara, B.; Heroult, M.; Wu, W.; Maltry, N.; Augustin, H.G.; Niehrs, C. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development 2008, 135, 3655–3664. [Google Scholar] [CrossRef]
- Bergmann, C.; Senderek, J.; Anhuf, D.; Thiel, C.T.; Ekici, A.B.; Poblete-Gutiérrez, P.; van Steensel, M.; Seelow, D.; Nürnberg, G.; Schild, H.H.; et al. Mutations in the Gene Encoding the Wnt-Signaling Component R-Spondin 4 (RSPO4) Cause Autosomal Recessive Anonychia. Am. J. Hum. Genet. 2006, 79, 1105–1109. [Google Scholar] [CrossRef]
- Prud’Homme, B.; Lartillot, N.; Balavoine, G.; Adoutte, A.; Vervoort, M. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr. Biol. 2002, 12, 1395–1400. [Google Scholar] [CrossRef]
- Janssen, R.; Le Gouar, M.; Pechmann, M.; Poulin, F.; Bolognesi, R.; Schwager, E.E.; Hopfen, C.; Colbourne, J.K.; Budd, G.E.; Brown, S.J.; et al. Conservation, loss, and redeployment of Wnt ligands in protostomes: Implications for understanding the evolution of segment formation. BMC Evol. Biol. 2010, 10, 374. [Google Scholar] [CrossRef] [PubMed]
- Holzem, M.; Boutros, M.; Holstein, T.W. The origin and evolution of Wnt signalling. Nat. Rev. Genet. 2024, 25, 500–512. [Google Scholar] [CrossRef]
- Rim, E.Y.; Clevers, H.; Nusse, R. The Wnt Pathway: From Signaling Mechanisms to Synthetic Modulators. Annu. Rev. Biochem. 2022, 91, 571–598. [Google Scholar] [CrossRef]
- de Lau, W.B.M.; Snel, B.; Clevers, H.C. The R-spondin protein family. Genome Biol. 2012, 13, 242. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Sun, D.; Liu, J.; Liu, N.; Yu, Q. Molecular analysis shows differential expression of R-spondin1 in zebrafish (Danio rerio) gonads. Mol. Biol. Rep. 2011, 38, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Shoemaker, C.M.; Roeszler, K.N.; Queen, J.; Crews, D.; Sinclair, A.H. Cloning and expression of R-Spondin1in different vertebrates suggests a conserved role in ovarian development. BMC Dev. Biol. 2008, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Chen, C.; Zhou, P.; Zhou, Y.; Li, Y.; Lu, L.; Liu, Y.; Zhou, J.; Duan, C. R-Spondin 3 Regulates Dorsoventral and Anteroposterior Patterning by Antagonizing Wnt/β-Catenin Signaling in Zebrafish Embryos. PLoS ONE 2014, 9, e99514. [Google Scholar] [CrossRef]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 2025, 53, D30–D44. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Souza, J.S.M.; Lisboa, A.B.P.; Santos, T.M.; Andrade, M.V.S.; Neves, V.B.S.; Teles-Souza, J.; Jesus, H.N.R.; Bezerra, T.G.; Falcão, V.G.O.; Oliveira, R.C.; et al. The evolution of ADAM gene family in eukaryotes. Genomics 2020, 112, 3108–3116. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 2022, 1, e56. [Google Scholar] [CrossRef]
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.V.; Reis-Cunha, J.L.; Bartholomeu, D.C. dgfr: An R package to assess sequence diversity of gene families. BMC Bioinform. 2024, 25, 207. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shine, M.; Pyle, A.M.; Zhang, Y. US-align: Universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat. Methods 2022, 19, 1109–1115. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Meyer, A.; Schartl, M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999, 11, 699–704. [Google Scholar] [CrossRef]
- Dehal, P.; Boore, J.L. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic Timing of the Fish-Specific Genome Duplication Correlates with the Diversification of Teleost Fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Helfman, G.S.; Collette, B.B.; Facey, D.E.; Bowen, B.W. The Diversity of Fishes: Biology, Evolution, and Ecology; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- De Robertis, E.M.; Sasai, Y. A common plan for dorsoventral patterning in Bilateria. Nature 1996, 380, 37–40. [Google Scholar] [CrossRef]
- Ishii, Y.; Wajid, M.; Bazzi, H.; Fantauzzo, K.A.; Barber, A.G.; Blaydon, D.C.; Nam, J.-S.; Yoon, J.K.; Kelsell, D.P.; Christiano, A.M. Mutations in R-Spondin 4 (RSPO4) Underlie Inherited Anonychia. J. Investig. Dermatol. 2008, 128, 867–870. [Google Scholar] [CrossRef]
- Seitz, C.; Van Steensel, M.; Frank, J.; Senderek, J.; Zerres, K.; Hamm, H.; Bergmann, C. The Wnt signalling ligand RSPO4, causing inherited anonychia, is not mutated in a patient with congenital nail hypoplasia/aplasia with underlying skeletal defects. Br. J. Dermatol. 2007, 157, 801–802. [Google Scholar] [CrossRef]
- Park, S.; Cui, J.; Yu, W.; Wu, L.; Carmon, K.S.; Liu, Q.J. Differential activities and mechanisms of the four R-spondins in potentiating Wnt/β-catenin signaling. J. Biol. Chem. 2018, 293, 9759–9769. [Google Scholar] [CrossRef]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L. Zebrafish HOX clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Furmanek, A.; Hofsteenge, J. Protein C-mannosylation: Facts and questions. Acta Biochim. Pol. 2000, 47, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Blaydon, D.C.; Ishii, Y.; O’Toole, E.A.; Unsworth, H.C.; Teh, M.-T.; Rüschendorf, F.; Sinclair, C.; Hopsu-Havu, V.K.; Tidman, N.; Moss, C. The gene encoding R-spondin 4 (RSPO4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat. Genet. 2006, 38, 1245–1247. [Google Scholar] [CrossRef]
- Chishti, M.; Kausar, N.; Rafiq, M.; Amin, M.; Ahmad, W. A novel missense mutation in RSPO4 gene underlies autosomal recessive congenital anonychia in a consanguineous Pakistani family. Br. J. Dermatol. 2008, 158, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.N.; Klar, J.; Nawaz, S.; Jameel, M.; Tariq, M.; Malik, N.A.; Baig, S.M.; Dahl, N. Novel missense mutation in the RSPO4 gene in congenital hyponychia and evidence for a polymorphic initiation codon (p. M1I). BMC Med. Genet. 2012, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zamponi, R.; Charlat, O.; Ramones, M.; Swalley, S.; Jiang, X.; Rivera, D.; Tschantz, W.; Lu, B.; Quinn, L.; et al. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 2013, 14, 1120–1126. [Google Scholar] [CrossRef]
- Lu, W.; Kim, K.-A.; Liu, J.; Abo, A.; Feng, X.; Cao, X.; Li, Y. R-spondin1 synergizes with Wnt3A in inducing osteoblast differentiation and osteoprotegerin expression. FEBS Lett. 2008, 582, 643–650. [Google Scholar] [CrossRef]
- Cambier, L.; Plate, M.; Sucov, H.M.; Pashmforoush, M. Nkx2-5 regulates cardiac growth through modulation of Wnt signaling by R-spondin3. Development 2014, 141, 2959–2971. [Google Scholar] [CrossRef]
- Sharma, A.R.; Chakraborty, C.; Lee, S.-S.; Sharma, G.; Yoon, J.K.; George Priya Doss, C.; Song, D.-K.; Nam, J.-S. Computational Biophysical, Biochemical, and Evolutionary Signature of Human R-Spondin Family Proteins, the Member of Canonical Wnt/β-Catenin Signaling Pathway. BioMed Res. Int. 2014, 2014, 974316. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Peña, M.I.; Capella-Gutiérrez, S.; Antó, M.; Gabaldón, T.; Ruiz-Trillo, I.; Sabidó, E. High-Throughput Proteomics Reveals the Unicellular Roots of Animal Phosphosignaling and Cell Differentiation. Dev. Cell 2016, 39, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Sebé-Pedrós, A.; Ballaré, C.; Parra-Acero, H.; Chiva, C.; Tena, J.J.; Sabidó, E.; Gómez-Skarmeta, J.L.; Di Croce, L.; Ruiz-Trillo, I. The Dynamic Regulatory Genome of Capsaspora and the Origin of Animal Multicellularity. Cell 2016, 165, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Richter, D.J.; Fozouni, P.; Eisen, M.B.; King, N. Gene family innovation, conservation and loss on the animal stem lineage. eLife 2018, 7, e34226. [Google Scholar] [CrossRef]
- Kasperski, A.; Heng, H.H. The Spiral Model of Evolution: Stable Life Forms of Organisms and Unstable Life Forms of Cancers. Int. J. Mol. Sci. 2024, 25, 9163. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A. Life Entrapped in a Network of Atavistic Attractors: How to Find a Rescue. Int. J. Mol. Sci. 2022, 23, 4017. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Lee, V.H.; Yang, L.; Jiang, Y.; Kong, F.S. Radiation Therapy for Thoracic Malignancies. Hematol. Oncol. Clin. 2020, 34, 109–125. [Google Scholar] [CrossRef]
- Davies, P.C.; Lineweaver, C.H. Cancer tumors as Metazoa 1.0: Tapping genes of ancient ancestors. Phys. Biol. 2011, 8, 015001. [Google Scholar] [CrossRef]
- Lineweaver, C.H.; Davies, P.C.; Vincent, M.D. Targeting cancer’s weaknesses (not its strengths): Therapeutic strategies suggested by the atavistic model. Bioessays 2014, 36, 827–835. [Google Scholar] [CrossRef]
- Cui, J.; Toh, Y.; Park, S.; Yu, W.; Tu, J.; Wu, L.; Li, L.; Jacob, J.; Pan, S.; Carmon, K.S.; et al. Drug Conjugates of Antagonistic R-Spondin 4 Mutant for Simultaneous Targeting of Leucine-Rich Repeat-Containing G Protein-Coupled Receptors 4/5/6 for Cancer Treatment. J. Med. Chem. 2021, 64, 12572–12581. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Yang, L.; Wang, S.; Luo, K.; Luo, S.; Dong, Y.; Ning, Y.; Wang, W. Phylogenetic Insights into the Evolutionary History of the RSPO Gene Family in Metazoa. Genes 2025, 16, 477. https://doi.org/10.3390/genes16050477
Cheng J, Yang L, Wang S, Luo K, Luo S, Dong Y, Ning Y, Wang W. Phylogenetic Insights into the Evolutionary History of the RSPO Gene Family in Metazoa. Genes. 2025; 16(5):477. https://doi.org/10.3390/genes16050477
Chicago/Turabian StyleCheng, Jia, Ling Yang, Shiping Wang, Kaiyong Luo, Senlin Luo, Yang Dong, Ya Ning, and Weibin Wang. 2025. "Phylogenetic Insights into the Evolutionary History of the RSPO Gene Family in Metazoa" Genes 16, no. 5: 477. https://doi.org/10.3390/genes16050477
APA StyleCheng, J., Yang, L., Wang, S., Luo, K., Luo, S., Dong, Y., Ning, Y., & Wang, W. (2025). Phylogenetic Insights into the Evolutionary History of the RSPO Gene Family in Metazoa. Genes, 16(5), 477. https://doi.org/10.3390/genes16050477





