Epitranscriptomic Role of m6A in Obesity-Associated Disorders and Cancer Metabolic Reprogramming
Abstract
1. Introduction
2. Overview of m6A Methylation Modification
3. The Role of m6A in Regulating Obesity
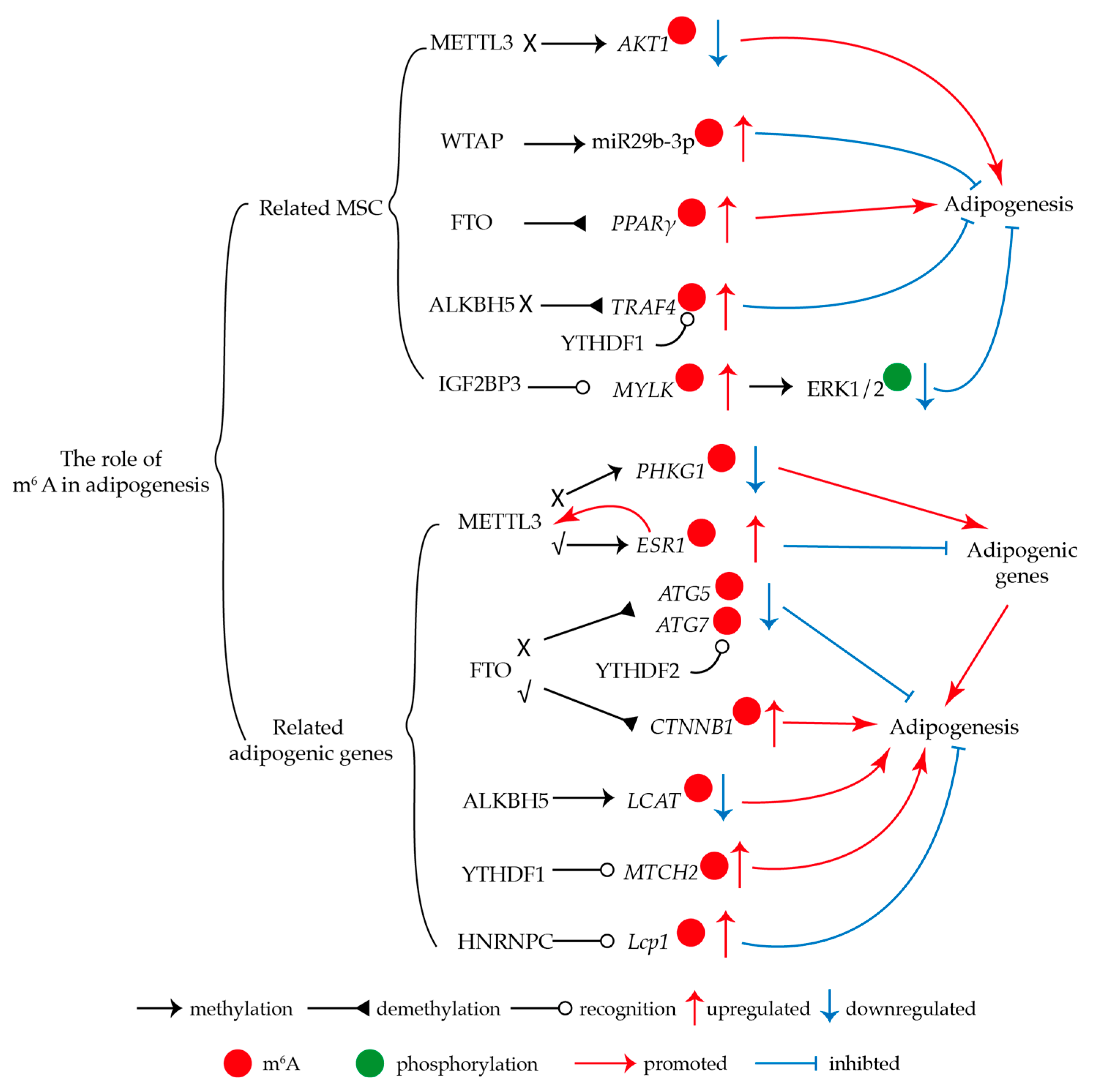
3.1. M6A Regulates Adipogenesis
3.2. M6A Regulates Lipid Metabolism
3.3. M6A Regulates White Adipose Tissue Beiging
4. The Role of m6A in Glucose Metabolism-Related Diseases
5. The Role of m6A in Glucose and Lipid Metabolism in Cancer Cells
5.1. M6A Regulates Glycolysis in Cancer Cells
5.2. M6A Affects Cancer Cell Lipogenesis
6. Clinical Research on m6A Targeted Therapy Needs Further Investigation
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALKBH5 | AlkB homolog H5 |
| ATG | Autophagy protein |
| BAT | Brown adipose tissue |
| C/EBP | CCAAT/enhancer-binding protein |
| EIF3 | Eukaryotic initiation factor 3 |
| FTO | Fat mass and obesity associated |
| HIF1A | Hypoxia-inducible factor 1-α |
| HNRNP | Heterogeneous nuclear ribonucleoproteins |
| IGF2BP | Insulin-like growth factor 2 mRNA-binding protein |
| iWAT | Inguinal white adipose tissue |
| METTL | Methyltransferase like |
| MP3C | Methyl piperidine-3-carboxylate |
| mRNA | Messenger RNA |
| m6A | N6-methyladenosine |
| m6A-IP | m6A-specific immunoprecipitation |
| MSCs | Mesenchymal stem cells |
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| NAFLD | Non-alcoholic fatty liver disease |
| RBM15 | RNA binding motif protein 15 |
| UCP1 | Uncoupling protein 1 |
| UTR | Untranslated region |
| VIRMA | Vir like M6A methyltransferase associated |
| WAT | White adipose tissue |
| WTAP | WT1-associated protein |
| YTHDC | YT521-B homology domain containing |
| YTHDF | YT521-B homology domain family |
| ZC3H13 | Zinc finger CCCH-type containing 13 |
References
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, Regional, and National Prevalence of Overweight and Obesity in Children and Adults during 1980–2013: A Systematic Analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Scully, T. Public Health: Society at Large. Nature 2014, 508, S50–S51. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Flavell, R.A.; Li, H.-B. RNA m6A Modification and Its Function in Diseases. Front. Med. 2018, 12, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 12 March 2025).
- Saxton, S.N.; Clark, B.J.; Withers, S.B.; Eringa, E.C.; Heagerty, A.M. Mechanistic Links Between Obesity, Diabetes, and Blood Pressure: Role of Perivascular Adipose Tissue. Physiol. Rev. 2019, 99, 1701–1763. [Google Scholar] [CrossRef]
- Müller, T.D.; Blüher, M.; Tschöp, M.H.; DiMarchi, R.D. Anti-Obesity Drug Discovery: Advances and Challenges. Nat. Rev. Drug Discov. 2022, 21, 201–223. [Google Scholar] [CrossRef]
- McAllister, E.J.; Dhurandhar, N.V.; Keith, S.W.; Aronne, L.J.; Barger, J.; Baskin, M.; Benca, R.M.; Biggio, J.; Boggiano, M.M.; Eisenmann, J.C.; et al. Ten Putative Contributors to the Obesity Epidemic. Crit. Rev. Food Sci. Nutr. 2009, 49, 868–913. [Google Scholar] [CrossRef]
- Heng, J.; Tian, M.; Zhang, W.; Chen, F.; Guan, W.; Zhang, S. Maternal Heat Stress Regulates the Early Fat Deposition Partly through Modification of m6A RNA Methylation in Neonatal Piglets. Cell Stress Chaperones 2019, 24, 635–645. [Google Scholar] [CrossRef]
- Noh, J. The Effect of Circadian and Sleep Disruptions on Obesity Risk. J. Obes. Metab. Syndr. 2018, 27, 78–83. [Google Scholar] [CrossRef]
- Cappuccio, F.P.; Taggart, F.M.; Kandala, N.-B.; Currie, A.; Peile, E.; Stranges, S.; Miller, M.A. Meta-Analysis of Short Sleep Duration and Obesity in Children and Adults. Sleep 2008, 31, 619–626. [Google Scholar] [CrossRef]
- Quarta, C.; Schneider, R.; Tschöp, M.H. Epigenetic ON/OFF Switches for Obesity. Cell 2016, 164, 341–342. [Google Scholar] [CrossRef][Green Version]
- Lefterova, M.I.; Lazar, M.A. New Developments in Adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Rohde, K.; Keller, M.; La Cour Poulsen, L.; Blüher, M.; Kovacs, P.; Böttcher, Y. Genetics and Epigenetics in Obesity. Metabolism 2019, 92, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, X. Epigenetic Regulation of Adipose Tissue Expansion and Adipogenesis by N6-methyladenosine. Obes. Rev. 2021, 22, e13124. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, X. Epigenetic Regulation of N6-methyladenosine Modifications in Obesity. J. Diabetes Investig. 2021, 12, 1306–1315. [Google Scholar] [CrossRef]
- He, C. Grand Challenge Commentary: RNA Epigenetics? Nat. Chem. Biol. 2010, 6, 863–865. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Liu, J.; Harada, B.T.; He, C. Regulation of Gene Expression by N-Methyladenosine in Cancer. Trends Cell Biol. 2019, 29, 487–499. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 Mediates Nuclear Export of N6-Methyladenosine Methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Bodi, Z.; Button, J.D.; Grierson, D.; Fray, R.G. Yeast Targets for mRNA Methylation. Nucleic Acids Res. 2010, 38, 5327–5335. [Google Scholar] [CrossRef]
- Haugland, R.A.; Cline, M.G. Post-transcriptional Modifications of Oat Coleoptile Ribonucleic Acids: 5′-Terminal Capping and Methylation of Internal Nucleosides in Poly(A)-Rich RNA. Eur. J. Biochem. 1980, 104, 271–277. [Google Scholar] [CrossRef]
- Hongay, C.F.; Orr-Weaver, T.L. Drosophila Inducer of MEiosis 4 (IME4) Is Required for Notch Signaling during Oogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 14855–14860. [Google Scholar] [CrossRef]
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 2017, 171, 877–889.e17. [Google Scholar] [CrossRef]
- Dubin, D.T.; Taylor, R.H. The Methylation State of Poly A-Containing-Messenger RNA from Cultured Hamster Cells. Nucl. Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Lim, Y.C.; Chia, S.Y.; Jin, S.; Han, W.; Ding, C.; Sun, L. Dynamic DNA Methylation Landscape Defines Brown and White Cell Specificity during Adipogenesis. Mol. Metab. 2016, 5, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6 A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Selberg, S.; Blokhina, D.; Aatonen, M.; Koivisto, P.; Siltanen, A.; Mervaala, E.; Kankuri, E.; Karelson, M. Discovery of Small Molecules That Activate RNA Methylation through Cooperative Binding to the METTL3-14-WTAP Complex Active Site. Cell Rep. 2019, 26, 3762–3771.e5. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Granadino, B.; Campuzano, S.; Sánchez, L. The Drosophila Melanogaster Fl(2)d Gene Is Needed for the Female-Specific Splicing of Sex-Lethal RNA. EMBO J. 1990, 9, 2597–2602. [Google Scholar] [CrossRef]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ Tumor 1-Associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of Factors Required for m6 A mRNA Methylation in Arabidopsis Reveals a Role for the Conserved E3 Ubiquitin Ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible Methylation of m6Am in the 5′ Cap Controls mRNA Stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Liu, N.; Pan, T. N6-Methyladenosine–Encoded Epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef]
- Nachtergaele, S.; He, C. The Emerging Biology of RNA Post-Transcriptional Modifications. RNA Biol. 2017, 14, 156–163. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Lasman, L.; Krupalnik, V.; Viukov, S.; Mor, N.; Aguilera-Castrejon, A.; Schneir, D.; Bayerl, J.; Mizrahi, O.; Peles, S.; Tawil, S.; et al. Context-Dependent Functional Compensation between Ythdf m6 A Reader Proteins. Genes Dev. 2020, 34, 1373–1391. [Google Scholar] [CrossRef]
- Luo, S.; Tong, L. Molecular Basis for the Recognition of Methylated Adenines in RNA by the Eukaryotic YTH Domain. Proc. Natl. Acad. Sci. USA 2014, 111, 13834–13839. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 Destabilizes m6A-Containing RNA through Direct Recruitment of the CCR4–NOT Deadenylase Complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A Reader YTHDF3 Promotes mRNA Translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Zhang, Z.; Theler, D.; Kaminska, K.H.; Hiller, M.; De La Grange, P.; Pudimat, R.; Rafalska, I.; Heinrich, B.; Bujnicki, J.M.; Allain, F.H.-T.; et al. The YTH Domain Is a Novel RNA Binding Domain. J. Biol. Chem. 2010, 285, 14701–14710. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 Is an N6-Methyladenosine Binding Protein That Regulates Mammalian Spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA Helicase YTHDC2 Promotes Cancer Metastasis via the Enhancement of the Efficiency by Which HIF-1α mRNA Is Translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef]
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular Basis for the Specific and Multivariant Recognitions of RNA Substrates by Human hnRNP A2/B1. Nat. Commun. 2018, 9, 420. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Van Eenige, R.; Van Der Stelt, M.; Rensen, P.C.N.; Kooijman, S. Regulation of Adipose Tissue Metabolism by the Endocannabinoid System. Trends Endocrinol. Metab. 2018, 29, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Scuteri, A.; Sanna, S.; Chen, W.-M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Fernyhough, M.E.; Okine, E.; Hausman, G.; Vierck, J.L.; Dodson, M.V. PPARγ and GLUT-4 Expression as Developmental Regulators/Markers for Preadipocyte Differentiation into an Adipocyte. Domest. Anim. Endocrinol. 2007, 33, 367–378. [Google Scholar] [CrossRef]
- Otto, T.C.; Lane, M.D. Adipose Development: From Stem Cell to Adipocyte. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 229–242. [Google Scholar] [CrossRef]
- Cristancho, A.G.; Lazar, M.A. Forming Functional Fat: A Growing Understanding of Adipocyte Differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef]
- Yao, Y.; Bi, Z.; Wu, R.; Zhao, Y.; Liu, Y.; Liu, Q.; Wang, Y.; Wang, X. METTL3 Inhibits BMSC Adipogenic Differentiation by Targeting the JAK1/STAT5/C/EBPβ Pathway via an m6 A-YTHDF2–Dependent Manner. FASEB J. 2019, 33, 7529–7544. [Google Scholar] [CrossRef]
- Liao, X.; Cai, D.; Liu, J.; Hu, H.; You, R.; Pan, Z.; Chen, S.; Xu, K.; Dai, W.; Zhang, S.; et al. Deletion of Mettl3 in Mesenchymal Stem Cells Promotes Acute Myeloid Leukemia Resistance to Chemotherapy. Cell Death Dis. 2023, 14, 796. [Google Scholar] [CrossRef]
- Liu, J.; You, Y.; Sun, Z.; Zhang, L.; Li, X.; Dai, Z.; Ma, J.; Chen, Y.; Jiao, G. WTAP-Mediated m6A RNA Methylation Regulates the Differentiation of Bone Marrow Mesenchymal Stem Cells via the miR-29b-3p/HDAC4 Axis. Stem Cells Transl. Med. 2023, 12, 307–321. [Google Scholar] [CrossRef]
- Shen, G.-S.; Zhou, H.-B.; Zhang, H.; Chen, B.; Liu, Z.-P.; Yuan, Y.; Zhou, X.-Z.; Xu, Y.-J. The GDF11-FTO-PPARγ Axis Controls the Shift of Osteoporotic MSC Fate to Adipocyte and Inhibits Bone Formation during Osteoporosis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 3644–3654. [Google Scholar] [CrossRef] [PubMed]
- Cen, S.; Li, J.; Cai, Z.; Pan, Y.; Sun, Z.; Li, Z.; Ye, G.; Zheng, G.; Li, M.; Liu, W.; et al. TRAF4 Acts as a Fate Checkpoint to Regulate the Adipogenic Differentiation of MSCs by Activating PKM2. EBioMedicine 2020, 54, 102722. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; He, W.; Fan, S.; Li, H.; Ye, G. IGF2BP3-Mediated Enhanced Stability of MYLK Represses MSC Adipogenesis and Alleviates Obesity and Insulin Resistance in HFD Mice. Cell. Mol. Life Sci. 2024, 81, 17. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, L.; Chen, J.; Wang, Y. mRNA m6A Methylation Downregulates Adipogenesis in Porcine Adipocytes. Biochem. Biophys. Res. Commun. 2015, 459, 201–207. [Google Scholar] [CrossRef]
- Chao, M.; Wang, M.; Han, H.; Liu, Y.; Sun, X.; Tian, T.; Pang, W.; Cai, R. Profiling of m6A Methylation in Porcine Intramuscular Adipocytes and Unravelling PHKG1 Represses Porcine Intramuscular Lipid Deposition in an m6A-Dependent Manner. Int. J. Biol. Macromol. 2024, 272, 132728. [Google Scholar] [CrossRef]
- Zhou, H.; Feng, S.; Cai, J.; Shao, X.; Zhu, S.; Zhou, H.; Cao, Y.; Wang, R.; Lin, X.; Wang, J. Oestrogen Suppresses the Adipogenesis of Fibro/Adipogenic Progenitors through Reactivating the METTL3–ESR1-mediated Loop in Post-menopausal Females. Clin. Transl. Med. 2025, 15, e70206. [Google Scholar] [CrossRef]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. M6 A mRNA Methylation Controls Autophagy and Adipogenesis by Targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235. [Google Scholar] [CrossRef]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m6A Demethylase FTO Regulate CTNNB1 to Promote Adipogenesis of Chicken Preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.-C.; Yang, Q. Author Correction: NADP Modulates RNA m6A Methylation and Adipogenesis via Enhancing FTO Activity. Nat. Chem. Biol. 2023, 19, 1286. [Google Scholar] [CrossRef]
- Chao, X.; Guo, L.; Ye, C.; Liu, A.; Wang, X.; Ye, M.; Fan, Z.; Luan, K.; Chen, J.; Zhang, C.; et al. ALKBH5 Regulates Chicken Adipogenesis by Mediating LCAT mRNA Stability Depending on m6A Modification. BMC Genom. 2024, 25, 634. [Google Scholar] [CrossRef]
- Jiang, Q.; Sun, B.; Liu, Q.; Cai, M.; Wu, R.; Wang, F.; Yao, Y.; Wang, Y.; Wang, X. MTCH2 Promotes Adipogenesis in Intramuscular Preadipocytes via an m6 A-YTHDF1-dependent Mechanism. FASEB J. 2019, 33, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Cui, Y.; Yue, L.; Zhang, T.; Huang, C.; Yu, X.; Ma, D.; Liu, D.; Cheng, R.; Zhao, X.; et al. m6A Reader HNRNPC Facilitates Adipogenesis by Regulating Cytoskeletal Remodeling through Enhanced Lcp1 mRNA Stability. Aging Dis. 2024, 16, 1080. [Google Scholar] [CrossRef]
- Gong, H.; Gong, T.; Liu, Y.; Wang, Y.; Wang, X. Profiling of N6-Methyladenosine Methylation in Porcine Longissimus Dorsi Muscle and Unravelling the Hub Gene ADIPOQ Promotes Adipogenesis in an m6A-YTHDF1-Dependent Manner. J. Anim. Sci. Biotechnol. 2023, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.K.; Rajasekharan, R. The m6A Methyltransferase Ime4 Epitranscriptionally Regulates Triacylglycerol Metabolism and Vacuolar Morphology in Haploid Yeast Cells. J. Biol. Chem. 2017, 292, 13727–13744. [Google Scholar] [CrossRef]
- Wang, X.; Sun, B.; Jiang, Q.; Wu, R.; Cai, M.; Yao, Y.; Liu, Q.; Shi, H.; Feng, J.; Wang, Y. mRNA m6A Plays Opposite Role in Regulating UCP2 and PNPLA2 Protein Expression in Adipocytes. Int. J. Obes. 2018, 42, 1912–1924. [Google Scholar] [CrossRef]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO Reduces Mitochondria and Promotes Hepatic Fat Accumulation through RNA Demethylation. J. Cell. Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef]
- Xie, W.; Ma, L.L.; Xu, Y.Q.; Wang, B.H.; Li, S.M. METTL3 Inhibits Hepatic Insulin Sensitivity via N6-Methyladenosine Modification of Fasn mRNA and Promoting Fatty Acid Metabolism. Biochem. Biophys. Res. Commun. 2019, 518, 120–126. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y.; et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cell Rep. 2018, 25, 1816–1828.e4. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, R.; Chen, W.; Liu, Y.; Liao, X.; Zeng, B.; Guo, G.; Lou, F.; Xiang, Y.; Wang, Y.; et al. Curcumin Prevents Obesity by Targeting TRAF4-induced Ubiquitylation in m6 A-dependent Manner. EMBO Rep. 2021, 22, e52146. [Google Scholar] [CrossRef]
- Cui, L.; Wu, Y.; Chen, Z.; Li, B.; Cai, J.; Chang, Z.; Xiao, W.; Wang, Y.; Yang, N.; Wang, Y.; et al. N6-Methyladenosine Modification-Tuned Lipid Metabolism Controls Skin Immune Homeostasis via Regulating Neutrophil Chemotaxis. Sci. Adv. 2024, 10, eadp5332. [Google Scholar] [CrossRef]
- Wang, W.; Seale, P. Control of Brown and Beige Fat Development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Boström, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.-H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Lizcano, F. The Beige Adipocyte as a Therapy for Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 5058. [Google Scholar] [CrossRef]
- Xie, R.; Yan, S.; Zhou, X.; Gao, Y.; Qian, Y.; Hou, J.; Chen, Z.; Lai, K.; Gao, X.; Wei, S. Activation of METTL3 Promotes White Adipose Tissue Beiging and Combats Obesity. Diabetes 2023, 72, 1083–1094. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Zhang, T.; Ping, X.; Wang, D.; Chen, Y.; Yu, J.; Liu, C.; Liu, Z.; Zheng, Y.; et al. RNA M6 a Methylation Regulates Glycolysis of Beige Fat and Contributes to Systemic Metabolic Homeostasis. Adv. Sci. 2023, 10, e2300436. [Google Scholar] [CrossRef]
- Church, C.; Lee, S.; Bagg, E.A.L.; McTaggart, J.S.; Deacon, R.; Gerken, T.; Lee, A.; Moir, L.; Mecinović, J.; Quwailid, M.M.; et al. A Mouse Model for the Metabolic Effects of the Human Fat Mass and Obesity Associated FTO Gene. PLoS Genet. 2009, 5, e1000599. [Google Scholar] [CrossRef]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto Gene Protects from Obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of Entacapone as a Chemical Inhibitor of FTO Mediating Metabolic Regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef]
- Wu, R.; Chen, Y.; Liu, Y.; Zhuang, L.; Chen, W.; Zeng, B.; Liao, X.; Guo, G.; Wang, Y.; Wang, X. m6A Methylation Promotes White-to-Beige Fat Transition by Facilitating Hif1a Translation. EMBO Rep. 2021, 22, e52348. [Google Scholar] [CrossRef]
- Yan, S.; Zhou, X.; Wu, C.; Gao, Y.; Qian, Y.; Hou, J.; Xie, R.; Han, B.; Chen, Z.; Wei, S.; et al. Adipocyte YTH N(6)-Methyladenosine RNA-Binding Protein 1 Protects against Obesity by Promoting White Adipose Tissue Beiging in Male Mice. Nat. Commun. 2023, 14, 1379. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Y.; Liu, J.; Chen, Y.; Wang, X. Acute Exercise Promotes WAT Browning by Remodeling mRNA m6A Methylation. Life Sci. 2025, 361, 123269. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, Z.; Jiang, S.; Hu, W.; Li, T.; Di, S.; Wang, D.; Yang, Y. A Global Perspective on FOXO1 in Lipid Metabolism and Lipid-Related Diseases. Prog. Lipid Res. 2017, 66, 42–49. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, F.; Huang, W.; Qin, S.; Huang, J.-T.; Sergi, C.; Yuan, B.-F.; Liu, S.-M. Glucose Is Involved in the Dynamic Regulation of m6A in Patients With Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2019, 104, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Li, L.; Liu, Y.-H.; Hu, L.-K.; Yan, Y.-X. Identification of Metabolism-Related Proteins as Biomarkers of Insulin Resistance and Potential Mechanisms of m6A Modification. Nutrients 2023, 15, 1839. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.; Sun, X.; Wu, Y.; Chen, Z. METTL3 Is Required for Maintaining β-Cell Function. Metabolism 2021, 116, 154702. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Cui, G.; Zhao, F.; Tian, X.; Sun, B.-F.; Yang, Y.; Li, W. m6A Regulates Liver Metabolic Disorders and Hepatogenous Diabetes. Genom. Proteom. Bioinform. 2020, 18, 371–383. [Google Scholar] [CrossRef]
- Liu, J.; Luo, G.; Sun, J.; Men, L.; Ye, H.; He, C.; Ren, D. METTL14 Is Essential for β-Cell Survival and Insulin Secretion. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 2138–2148. [Google Scholar] [CrossRef]
- Xiao, L.; De Jesus, D.F.; Ju, C.-W.; Wei, J.-B.; Hu, J.; DiStefano-Forti, A.; Gonzales, V.S.; Tsuji, T.; Wei, S.; Blüher, M.; et al. Divergent Roles of m6A in Orchestrating Brown and White Adipocyte Transcriptomes and Systemic Metabolism. Nat. Commun. 2025, 16, 533. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, J.; Yang, X.; Wang, K.; Sun, K.; Yang, Z.; Zhang, L.; Yang, L.; Gu, C.; Huang, X.; et al. Dysregulated m6A Modification Promotes Lipogenesis and Development of Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. Mol. Ther. 2022, 30, 2342–2353. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Li, Z.; Wang, Y.; Qiao, J.; Chen, Z. Deficiency of WTAP in Islet β Cells Results in β Cell Failure and Diabetes in Mice. Diabetologia 2023, 66, 1084–1096. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Chen, Z. Downregulation of the m6A Reader Protein YTHDC1 Leads to Islet β-Cell Failure and Diabetes. Metabolism 2023, 138, 155339. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, C.; Xu, L.; Yuan, Y.; Zhao, J.; Zhao, W.; Chen, Y.; Qiu, J.; Meng, M.; Zheng, Y.; et al. N6 -Methyladenosine Reader Protein YT521-B Homology Domain-Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 2021, 73, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Gong, Y.; Wang, X.; He, W.; Wu, L.; Zhang, L.; Xiong, L.; Huang, Y.; Su, L.; Shi, P.; et al. METTL3-m6A-Rubicon Axis Inhibits Autophagy in Nonalcoholic Fatty Liver Disease. Mol. Ther. 2022, 30, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, W.; Wang, Z.; Liu, Z.; Yi, X.; Wu, J. Mettl3-m6A-YTHDF1 Axis Promotion of Mitochondrial Dysfunction in Metabolic Dysfunction-Associated Steatotic Liver Disease. Cell Signal 2024, 121, 111303. [Google Scholar] [CrossRef]
- Kang, Q.; Zhu, X.; Ren, D.; Ky, A.; MacDougald, O.A.; O’Rourke, R.W.; Rui, L. Adipose METTL14-Elicited N6 -Methyladenosine Promotes Obesity, Insulin Resistance, and NAFLD Through Suppressing β Adrenergic Signaling and Lipolysis. Adv. Sci. 2023, 10, 2301645. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Li, Z.; Liu, Y.; Tan, J.; Yin, H.; Zhang, Z.; Piao, X.; Ruan, M.; Dai, Z.; et al. METTL14 Downregulation Drives S100A4+ Monocyte-Derived Macrophages via MyD88/NF-κB Pathway to Promote MAFLD Progression. Sig Transduct. Target. Ther. 2024, 9, 91. [Google Scholar] [CrossRef]
- Wang, W.; Yan, J.; Han, L.; Zou, Z.-L.; Xu, A.-L. Silencing METTL14 Alleviates Liver Injury in Non-Alcoholic Fatty Liver Disease by Regulating Mitochondrial Homeostasis. Biomol. Biomed. 2024, 24, 505–519. [Google Scholar] [CrossRef]
- Hu, Y.; Feng, Y.; Zhang, L.; Jia, Y.; Cai, D.; Qian, S.-B.; Du, M.; Zhao, R. GR-Mediated FTO Transactivation Induces Lipid Accumulation in Hepatocytes via Demethylation of m6A on Lipogenic mRNAs. RNA Biol. 2020, 17, 930–942. [Google Scholar] [CrossRef]
- Ding, K.; Zhang, Z.; Han, Z.; Shi, L.; Li, X.; Liu, Y.; Li, Z.; Zhao, C.; Cui, Y.; Zhou, L.; et al. Liver ALKBH5 Regulates Glucose and Lipid Homeostasis Independently through GCGR and mTORC1 Signaling. Science 2025, 387, eadp4120. [Google Scholar] [CrossRef]
- Li, L.; Sun, Y.; Li, L.; Zheng, W.; Zha, W.; Zhao, T.; Zhu, G.; Li, H. The Deficiency of ALKBH5 Contributes to Hepatic Lipid Deposition by Impairing VPS11-Dependent Autophagic Flux. FEBS J. 2024, 291, 5256–5275. [Google Scholar] [CrossRef]
- Huang, C.; Luo, Y.; Liu, Y.; Liu, J.; Chen, Y.; Zeng, B.; Liao, X.; Liu, Y.; Wang, X. DNA Hypermethylation-Induced Suppression of ALKBH5 Is Required for Folic Acid to Alleviate Hepatic Lipid Deposition by Enhancing Autophagy in an ATG12-Dependent Manner. J. Nutr. Biochem. 2025, 140, 109870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, X.; Wang, Z.; Liu, D.; Zhao, X.; Sun, X.; Tu, Z.; Li, Z.; Zhao, Y.; Zheng, S.; et al. A Novel Peptide Encoded by circ-SLC9A6 Promotes Lipid Dyshomeostasis through the Regulation of H4K16ac-mediated CD36 Transcription in NAFLD. Clin. Transl. Med. 2024, 14, e1801. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, S.; Nagarajan, S. GRP78 and next Generation Cancer Hallmarks: An Underexplored Molecular Target in Cancer Chemoprevention Research. Biochimie 2020, 175, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Ajith, A.; Singh, S.; Panday, R.K.; Samaiya, A.; Shukla, S. PAK2–c-Myc–PKM2 Axis Plays an Essential Role in Head and Neck Oncogenesis via Regulating Warburg Effect. Cell Death Dis. 2018, 9, 825. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, Q.; Halim, A.; Song, G. Targeting Lipid Metabolism of Cancer Cells: A Promising Therapeutic Strategy for Cancer. Cancer Lett. 2017, 401, 39–45. [Google Scholar] [CrossRef]
- Hsu, P.P.; Sabatini, D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef]
- Shen, C.; Xuan, B.; Yan, T.; Ma, Y.; Xu, P.; Tian, X.; Zhang, X.; Cao, Y.; Ma, D.; Zhu, X.; et al. m6A-Dependent Glycolysis Enhances Colorectal Cancer Progression. Mol. Cancer 2020, 19, 72. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, X.; Li, L.; Gao, Z.; Su, X.; Ji, M.; Liu, J. N6-Methyladenosine METTL3 Promotes Cervical Cancer Tumorigenesis and Warburg Effect through YTHDF1/HK2 Modification. Cell Death Dis. 2020, 11, 911. [Google Scholar] [CrossRef]
- Yang, F.; Liu, Y.; Xiao, J.; Li, B.; Chen, Y.; Hu, A.; Zeng, J.; Liu, Z.; Liu, H. Circ-CTNNB1 Drives Aerobic Glycolysis and Osteosarcoma Progression via m6A Modification through Interacting with RBM15. Cell Prolif. 2023, 56, e13344. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, K.; Zeng, H.; Li, Z.; Chen, K.; Zhang, Z.; Li, E.; Wu, Z. N6-Methyladenosine (m6A) Methyltransferase WTAP Accelerates the Warburg Effect of Gastric Cancer through Regulating HK2 Stability. Biomed. Pharmacother. 2021, 133, 111075. [Google Scholar] [CrossRef]
- Ye, M.; Chen, J.; Lu, F.; Zhao, M.; Wu, S.; Hu, C.; Yu, P.; Kan, J.; Bai, J.; Tian, Y.; et al. Down-Regulated FTO and ALKBH5 Co-Operatively Activates FOXO Signaling through m6A Methylation Modification in HK2 mRNA Mediated by IGF2BP2 to Enhance Glycolysis in Colorectal Cancer. Cell Biosci. 2023, 13, 148. [Google Scholar] [CrossRef]
- Miao, D.; Wang, Q.; Shi, J.; Lv, Q.; Tan, D.; Zhao, C.; Xiong, Z.; Zhang, X. N6-Methyladenosine-Modified DBT Alleviates Lipid Accumulation and Inhibits Tumor Progression in Clear Cell Renal Cell Carcinoma through the ANXA2/YAP Axis-Regulated Hippo Pathway. Cancer Commun. 2023, 43, 480–502. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Fan, B.; Othmane, B.; Hu, J.; Li, H.; Cui, Y.; Ou, Z.; Chen, J.; Zu, X. m6A-Induced lncDBET Promotes the Malignant Progression of Bladder Cancer through FABP5-Mediated Lipid Metabolism. Theranostics 2022, 12, 6291–6307. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Sheng, L.; Gao, Q.; Xiong, Q.; Zhang, H.; Wu, M.; Liang, Y.; Zhu, F.; Zhang, Y.; Zhang, X.; et al. The m6A Methyltransferase METTL3 Promotes Bladder Cancer Progression via AFF4/NF-κB/MYC Signaling Network. Oncogene 2019, 38, 3667–3680. [Google Scholar] [CrossRef]
- Yu, H.; Yang, X.; Tang, J.; Si, S.; Zhou, Z.; Lu, J.; Han, J.; Yuan, B.; Wu, Q.; Lu, Q.; et al. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol. Ther. Nucleic Acids 2021, 23, 27–41. [Google Scholar] [CrossRef]
- Li, Z.; Peng, Y.; Li, J.; Chen, Z.; Chen, F.; Tu, J.; Lin, S.; Wang, H. N6-Methyladenosine Regulates Glycolysis of Cancer Cells through PDK4. Nat. Commun. 2020, 11, 2578. [Google Scholar] [CrossRef]
- Ray, U.; Roy, S.S. Aberrant Lipid Metabolism in Cancer Cells—The Role of Oncolipid-activated Signaling. FEBS J. 2018, 285, 432–443. [Google Scholar] [CrossRef]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-Mediated Upregulation of LINC00958 Increases Lipogenesis and Acts as a Nanotherapeutic Target in Hepatocellular Carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef]
- Cai, X.; Li, X.; Zhang, M.; Dong, Z.; Weng, Y.; Yu, W. RBM15 Promotes Lipogenesis and Malignancy in Gastric Cancer by Regulating N6-Methyladenosine Modification of ACLY mRNA in an IGF2BP2-Dependent Manner. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2025, 1870, 159580. [Google Scholar] [CrossRef]
- Tang, Z.; Sun, C.; Yan, Y.; Niu, Z.; Li, Y.; Xu, X.; Zhang, J.; Wu, Y.; Li, Y.; Wang, L.; et al. Aberrant Elevation of FTO Levels Promotes Liver Steatosis by Decreasing the m6A Methylation and Increasing the Stability of SREBF1 and ChREBP mRNAs. J. Mol. Cell Biol. 2023, 14, mjac061. [Google Scholar] [CrossRef]
- Duan, X.; Yang, L.; Wang, L.; Liu, Q.; Zhang, K.; Liu, S.; Liu, C.; Gao, Q.; Li, L.; Qin, G.; et al. m6A Demethylase FTO Promotes Tumor Progression via Regulation of Lipid Metabolism in Esophageal Cancer. Cell Biosci. 2022, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ye, M.; Bai, J.; Gong, Z.; Yan, L.; Gu, D.; Hu, C.; Lu, F.; Yu, P.; Xu, L.; et al. ALKBH5 Enhances Lipid Metabolism Reprogramming by Increasing Stability of FABP5 to Promote Pancreatic Neuroendocrine Neoplasms Progression in an m6A-IGF2BP2-Dependent Manner. J. Transl. Med. 2023, 21, 741. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Hu, C.; Liu, T.; Sun, Y.; Hu, F.; He, Y.; Zhang, J.; Chen, J.; Ding, J.; Fan, J.; et al. IGF2BP3 Enhances Lipid Metabolism in Cervical Cancer by Upregulating the Expression of SCD. Cell Death Dis. 2024, 15, 138. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; Meng, X.; Wang, W.; Duan, F.; Chen, S.; Zhang, Y.; Sheng, Z.; Gao, Y.; Zhou, L. METTL16 Inhibits Papillary Thyroid Cancer Tumorigenicity through m6A/YTHDC2/SCD1-Regulated Lipid Metabolism. Cell. Mol. Life Sci. 2024, 81, 81. [Google Scholar] [CrossRef]
- Zhong, S.; Guo, Q.; Chen, X.; Luo, X.; Long, Y.; Chong, T.; Ye, M.; He, H.; Lu, A.; Ao, K.; et al. The Inhibition of YTHDF3/m6A/LRP6 Reprograms Fatty Acid Metabolism and Suppresses Lymph Node Metastasis in Cervical Cancer. Int. J. Biol. Sci. 2024, 20, 916–936. [Google Scholar] [CrossRef]
- Du, W.; Tan, S.; Peng, Y.; Lin, S.; Wu, Y.; Ding, K.; Chen, C.; Liu, R.; Cao, Y.; Li, Z.; et al. Histone Lactylation-Driven YTHDC1 Promotes Hepatocellular Carcinoma Progression via Lipid Metabolism Remodeling. Cancer Lett. 2024, 611, 217426. [Google Scholar] [CrossRef]
- Chen, B.; Ye, F.; Yu, L.; Jia, G.; Huang, X.; Zhang, X.; Peng, S.; Chen, K.; Wang, M.; Gong, S.; et al. Development of Cell-Active N6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 2012, 134, 17963–17971. [Google Scholar] [CrossRef]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.-F.; Luo, C.; et al. Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef]
- Toh, J.D.W.; Sun, L.; Lau, L.Z.M.; Tan, J.; Low, J.J.A.; Tang, C.W.Q.; Cheong, E.J.Y.; Tan, M.J.H.; Chen, Y.; Hong, W.; et al. A Strategy Based on Nucleotide Specificity Leads to a Subfamily-Selective and Cell-Active Inhibitor of N6 -Methyladenosine Demethylase FTO. Chem. Sci. 2015, 6, 112–122. [Google Scholar] [CrossRef]
- Zheng, G.; Cox, T.; Tribbey, L.; Wang, G.Z.; Iacoban, P.; Booher, M.E.; Gabriel, G.J.; Zhou, L.; Bae, N.; Rowles, J.; et al. Synthesis of a FTO Inhibitor with Anticonvulsant Activity. ACS Chem. Neurosci. 2014, 5, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Qing, Y.; Dong, L.; Gao, L.; Li, C.; Li, Y.; Han, L.; Prince, E.; Tan, B.; Deng, X.; Wetzel, C.; et al. R-2-Hydroxyglutarate Attenuates Aerobic Glycolysis in Leukemia by Targeting the FTO/m6A/PFKP/LDHB Axis. Mol. Cell 2021, 81, 922–939.e9. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Hong, F.; Liu, H.; Fang, Y.; Wang, H.; Song, N.; Ning, Y.; Lu, Z.; Jin, S.; Dai, Y.; et al. FTO Aggravates Podocyte Injury and Diabetic Nephropathy Progression via m6A-Dependent Stabilization of ACC1 mRNA and Promoting Fatty Acid Metabolism. Biochem. Pharmacol. 2025, 235, 116819. [Google Scholar] [CrossRef]
- Huang, Y.; Xia, W.; Dong, Z.; Yang, C.-G. Chemical Inhibitors Targeting the Oncogenic m6A Modifying Proteins. Acc. Chem. Res. 2023, 56, 3010–3022. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-Molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.-W.; Zhu, H.-L.; Zhang, J.; Geng, H.; Tan, L.-L.; Zheng, X.-M.; Li, H.; Fan, L.-L.; Wang, X.-R.; Zhang, X.-D.; et al. Multigenerational Paternal Obesity Enhances the Susceptibility to Male Subfertility in Offspring via Wt1 N6-Methyladenosine Modification. Nat. Commun. 2024, 15, 1353. [Google Scholar] [CrossRef]
- Wu, Z.; Smith, A.R.; Qian, Z.; Zheng, G. Patent Landscape of Small Molecule Inhibitors of METTL3 (2020–Present). Expert. Opin. Ther. Pat. 2024, 35, 305–320. [Google Scholar] [CrossRef]
- Hong, Y.-G.; Yang, Z.; Chen, Y.; Liu, T.; Zheng, Y.; Zhou, C.; Wu, G.-C.; Chen, Y.; Xia, J.; Wen, R.; et al. The RNA m6A Reader YTHDF1 Is Required for Acute Myeloid Leukemia Progression. Cancer Res. 2023, 83, 845–860. [Google Scholar] [CrossRef]
- Deng, L.-J.; Deng, W.-Q.; Fan, S.-R.; Chen, M.-F.; Qi, M.; Lyu, W.-Y.; Qi, Q.; Tiwari, A.K.; Chen, J.-X.; Zhang, D.-M.; et al. m6A Modification: Recent Advances, Anticancer Targeted Drug Discovery and Beyond. Mol. Cancer 2022, 21, 52. [Google Scholar] [CrossRef]
- Sakers, A.; De Siqueira, M.K.; Seale, P.; Villanueva, C.J. Adipose-Tissue Plasticity in Health and Disease. Cell 2022, 185, 419–446. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ohsugi, M.; Sasako, T.; Awazawa, M.; Umehara, T.; Iwane, A.; Kobayashi, N.; Okazaki, Y.; Kubota, N.; Suzuki, R.; et al. The RNA Methyltransferase Complex of WTAP, METTL3, and METTL14 Regulates Mitotic Clonal Expansion in Adipogenesis. Mol. Cell. Biol. 2018, 38, e00116-18. [Google Scholar] [CrossRef] [PubMed]
- Rønningen, T.; Dahl, M.B.; Valderhaug, T.G.; Cayir, A.; Keller, M.; Tönjes, A.; Blüher, M.; Böttcher, Y. m6A Regulators in Human Adipose Tissue—Depot-Specificity and Correlation With Obesity. Front. Endocrinol. 2021, 12, 778875. [Google Scholar] [CrossRef] [PubMed]
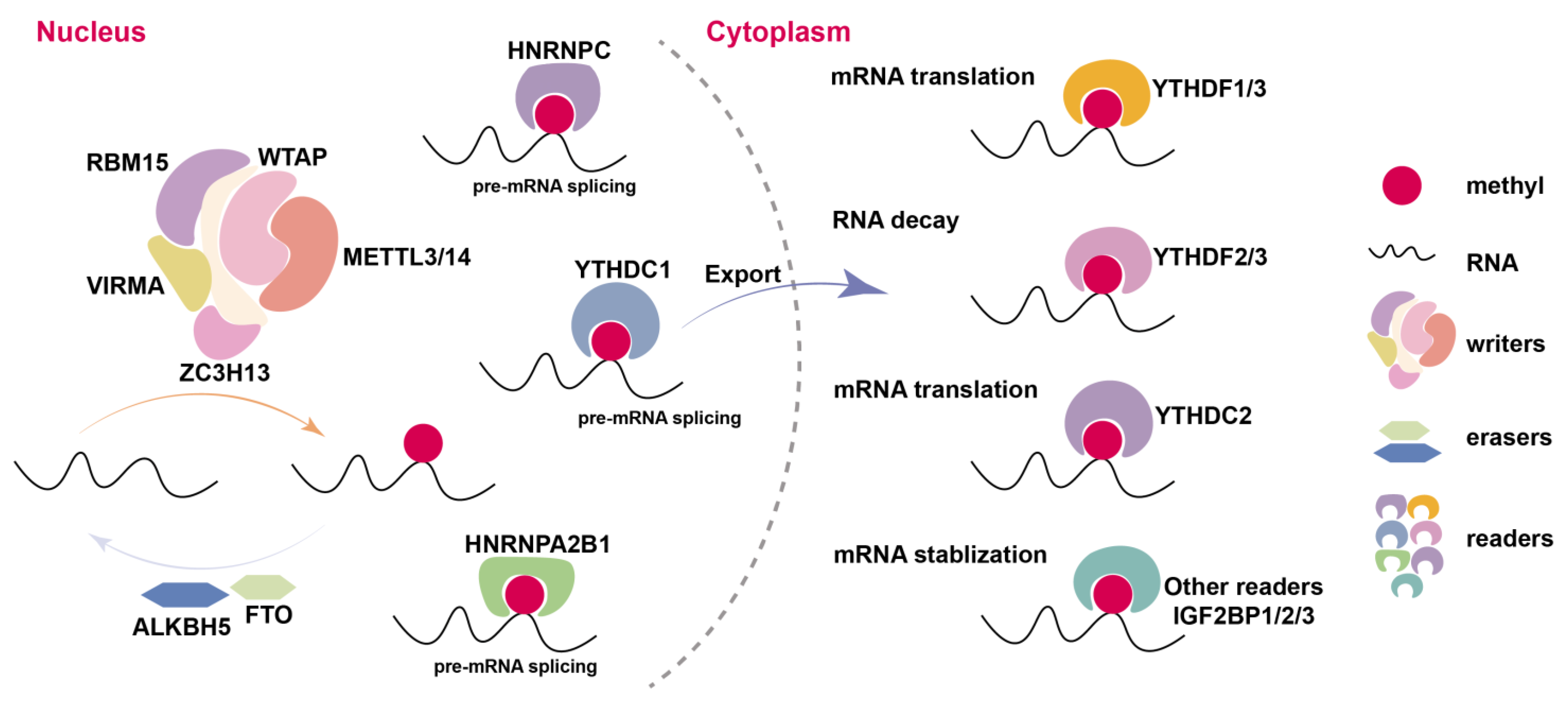
| m6A Regulators | m6A “Readers” | Key Mechanism | References |
|---|---|---|---|
| METTL3 | - | ↓ the thermogenic mRNAs (including Klf9) degradation | [95] |
| IGF2BP2 | ↑ mRNA stability of key glycolytic genes in beige adipocytes | [96] | |
| FTO | - | Inhibitor of FTO → ↑ the m6A of FOXO1 → ↑ white adipose tissue beiging | [99] |
| YTHDC2 | FTO deficiency → ↑ the m6A of Hif1a → YTHDC2 recognized → ↑ HIF1A protein → ↑ white adipose tissue beiging | [100] | |
| - | YTHDF1 | Recognized the m6A of Bmp8b → ↑ white adipose tissue beiging | [101] |
| m6A Regulators | m6A “Readers” | Mechanism | Cancer | Reference |
|---|---|---|---|---|
| METTL3 | - | ↑ mRNA level of FASN → ↑ fatty acid metabolism | Insulin sensitivity | [88] |
| METTL3/14 | - | ↑ protein level of ACLY and SCD1 → ↑ lipid droplets | NAFLD | [110] |
| METTL3 | YTHDF1 | ↑ RUBICON → ↓ lipid metabolism | NAFLD | [114] |
| METTL14 | - | ↓ ADRB signaling and lipolysis → ↑ obesity and MASLD | MASLD | [116,117] |
| FTO | - | ↑ lipogenic gene → ↑ lipid accumulation | NAFLD | [119] |
| ALKBH5 | - | ↓ cAMP and EGFR-PI3K-AKT-mTORC1 signaling → ↑ glucose tolerance | MAFLD | [120] |
| ↑ the translation of VPS11 → ↓ lipid deposition | NAFLD | [121] | ||
| - | YTHDC2 | ↓ mRNA stability of lipogenic genes → ↑ liver steatosis | Insulin resistance and NAFLD | [113] |
| m6A Regulators | m6A “Readers” | Mechanism | Cancer | Reference |
|---|---|---|---|---|
| METTL3 | IGF2BP2/3 | ↑ mRNA stability of GLUT1 → ↑ glycolysis pathway | Colorectal cancer | [128] |
| IGF2BP2 | ↑ mRNA stability of HK2 → ↑ glycolysis pathway | |||
| YTHDF1 | ↑ mRNA stability of HK2 → ↑ Warburg effect | Cervical cancer | [129] | |
| RBM15 | - | RBM15 interacted with Circ-CTNNB1 → ↑ HK2 expression→ ↑glycolysis | Osteosarcoma | [130] |
| WTAP | YTHDF1 | ↑ mRNA stability of HK2 → Recognized by YTHDF1 → ↑ HK2 protein → ↑ Warburg effect | Gastric cancer | [131] |
| FTO and ALKBH5 | IGF2BP2 | ↑ FOXO signaling → ↑ HK2 recognized by IGF2BP2 → ↑ glycolysis | Colorectal cancer | [132] |
| m6A Regulators | m6A “Readers” | Mechanism | Cancer | Reference |
|---|---|---|---|---|
| METTL3 | - | ↑ mRNA stability of LINC00958 → ↑ lipogenesis | Hepatocellular carcinoma | [139] |
| METTL3 | YTHDF1 | Regulating lipid metabolism via the autophagy pathway | Nonalcoholic fatty liver disease | [114] |
| METTL14 | IGF2BP3 | ↑ SCD → lipid metabolism | Cervical cancer | [144] |
| RBM15 | IGF2BP2 | ↑ ACLY → recognized by IGF2BP2 → ↑ lipogenesis | Gastric cancer | [140] |
| FTO | - | ↑ mRNA stability of lipogenic genes | [141] | |
| - | ↑ HSD17B11→ ↑ lipogenesis | Esophageal cancer | [142] | |
| ALKBH5 | IGF2BP2 | ↑ FABP5 expression → disorder the lipid metabolism | Pancreatic neuroendocrine neoplasms | [143] |
| - | YTHDF3 | ↑ LRP6 expression → ↑ FASN and ACC1 expression → lipid metabolism | Cervical cancer | [146] |
| - | YTHDC1 | ↑ mRNA stability of NEAT1 → lipid metabolism | Hepatocellular carcinoma | [147] |
| - | YTHDC2 | SCD1-activated lipid metabolism | Thyroid cancer | [145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, S.; Wen, W.; Mo, Z.; Gu, S.; Chen, Z. Epitranscriptomic Role of m6A in Obesity-Associated Disorders and Cancer Metabolic Reprogramming. Genes 2025, 16, 498. https://doi.org/10.3390/genes16050498
Yan S, Wen W, Mo Z, Gu S, Chen Z. Epitranscriptomic Role of m6A in Obesity-Associated Disorders and Cancer Metabolic Reprogramming. Genes. 2025; 16(5):498. https://doi.org/10.3390/genes16050498
Chicago/Turabian StyleYan, Sujun, Weijing Wen, Zhe Mo, Simeng Gu, and Zhijian Chen. 2025. "Epitranscriptomic Role of m6A in Obesity-Associated Disorders and Cancer Metabolic Reprogramming" Genes 16, no. 5: 498. https://doi.org/10.3390/genes16050498
APA StyleYan, S., Wen, W., Mo, Z., Gu, S., & Chen, Z. (2025). Epitranscriptomic Role of m6A in Obesity-Associated Disorders and Cancer Metabolic Reprogramming. Genes, 16(5), 498. https://doi.org/10.3390/genes16050498






