The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Strains
2.2. Flow Cytometry
2.3. Synthetic Lethality Assays
3. Results
3.1. Transcription Is a Major Cause of Chromosome Degradation in recA Cells
3.2. Rapid Chromosome Duplication Has a Greater Requirement for Rep than for RecA
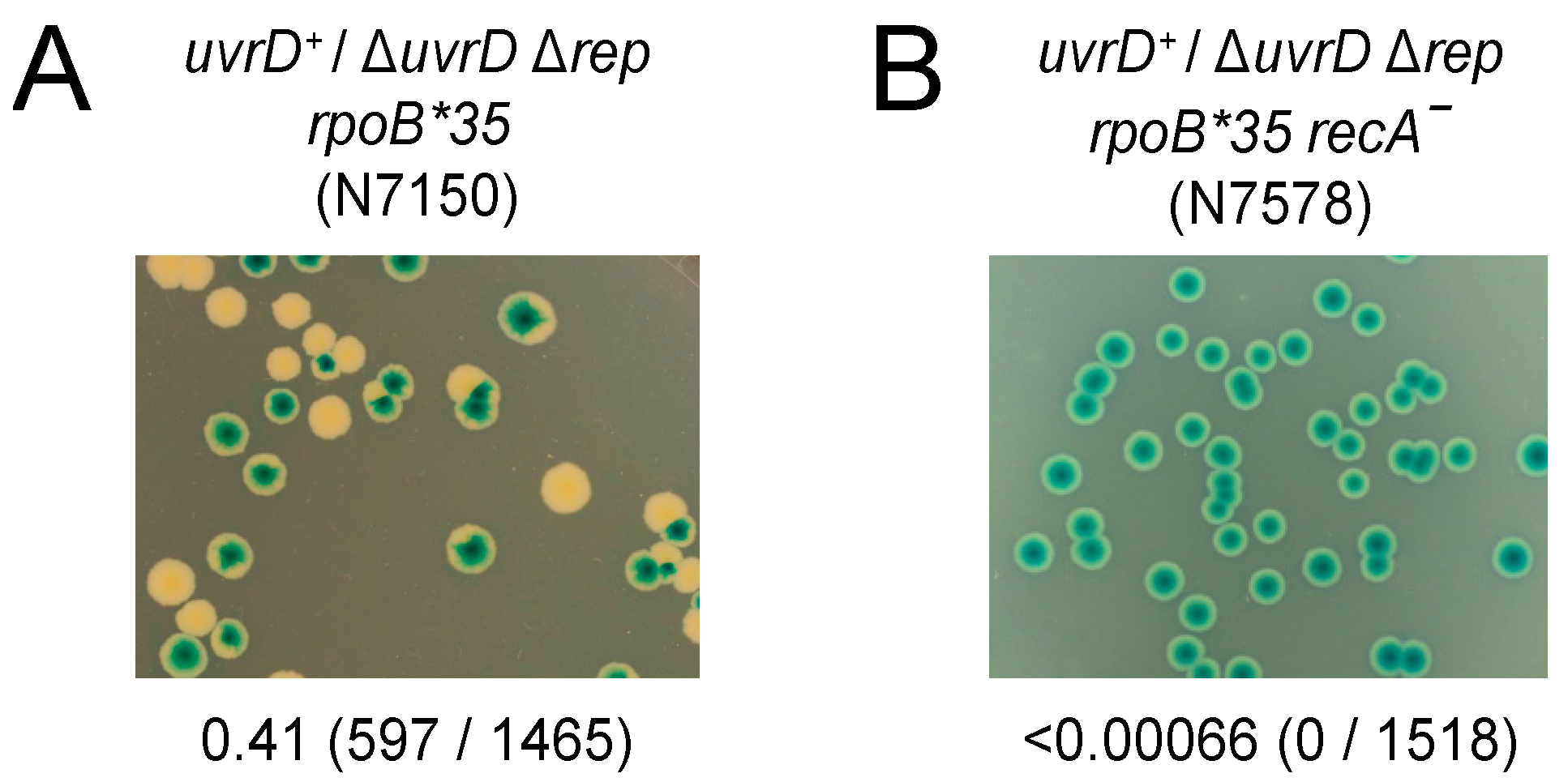
3.3. Both RecA and RecBCD Are Needed in the Absence of Accessory Helicase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yeeles, J.T.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Garcia-Muse, T. Causes of genome instability. Ann. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Shyy, S.H.; Wang, J.C.; Liu, L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell 1988, 53, 433–440. [Google Scholar] [CrossRef]
- Olavarrieta, L.; Hernandez, P.; Krimer, D.B.; Schvartzman, J.B. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 2002, 322, 1–6. [Google Scholar] [CrossRef]
- McGlynn, P.; Savery, N.J.; Dillingham, M.S. The conflict between DNA replication and transcription. Mol. Microbiol. 2012, 85, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Guy, C.P.; Yeeles, J.T.; Atkinson, J.; Bell, H.; Lloyd, R.G.; Marians, K.J.; McGlynn, P. Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 7252–7257. [Google Scholar] [CrossRef] [PubMed]
- Syeda, A.H.; Hawkins, M.; McGlynn, P. Recombination and replication. Cold Spring Harb. Perspect. Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Marians, K.J.; Hiasa, H.; Kim, D.R.; McHenry, C.S. Role of the core DNA polymerase III subunits at the replication fork. α Is the only subunit required for processive replication. J. Biol. Chem. 1998, 273, 2452–2457. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Guy, C.P. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J. Mol. Biol. 2008, 381, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Mettrick, K.A.; Grainge, I. Stability of blocked replication forks in vivo. Nucleic Acids Res. 2015, 44, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Rupp, W.D.; Wilde, C.E., III; Reno, D.L.; Howard-Flanders, P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 1971, 61, 25–44. [Google Scholar] [CrossRef]
- Lehmann, A.R. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol. 1972, 66, 319–337. [Google Scholar] [CrossRef]
- Smith, K.C. Recombinational DNA repair: The ignored repair systems. Bioessays 2004, 26, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Marians, K.J. The Escherichia coli replisome is inherently DNA damage tolerant. Science 2011, 334, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Marians, K.J. Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell 2013, 52, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Gabbai, C.B.; Yeeles, J.T.; Marians, K.J. Replisome-mediated translesion synthesis and leading strand template lesion skipping are competing bypass mechanisms. J. Biol. Chem. 2014, 289, 32811–32823. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, R.P.; Fujii, S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.F.; Woodgate, R. Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Payne, B.T.; van Knippenberg, I.C.; Bell, H.; Filipe, S.R.; Sherratt, D.J.; McGlynn, P. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res. 2006, 34, 5194–5202. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, R.T.; O’Donnell, M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 2008, 456, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.P.; Atkinson, J.; Gupta, M.K.; Mahdi, A.A.; Gwynn, E.J.; Rudolph, C.J.; Moon, P.B.; van Knippenberg, I.C.; Cadman, C.J.; Dillingham, M.S.; et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 2009, 36, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Yancey-Wrona, J.E.; Matson, S.W. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992, 20, 6713–6721. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.K.; Raney, K.D. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004, 11, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, A.S.; Zhou, J.Q.; Zakian, V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 2000, 100, 479–489. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Zhou, J.Q.; Schulz, V.P.; Monson, E.K.; Zakian, V.A. Saccharomyces Rrm3p, a 5’ to 3’ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002, 16, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Azvolinsky, A.; Dunaway, S.; Torres, J.Z.; Bessler, J.B.; Zakian, V.A. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006, 20, 3104–3116. [Google Scholar] [CrossRef] [PubMed]
- Lane, H.E.; Denhardt, D.T. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J. Mol. Biol. 1975, 97, 99–112. [Google Scholar] [PubMed]
- Boubakri, H.; de Septenville, A.L.; Viguera, E.; Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010, 29, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.; Gupta, M.K.; Rudolph, C.J.; Bell, H.; Lloyd, R.G.; McGlynn, P. Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic Acids Res. 2011, 39, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, C.N.; Brewer, B.J.; Merrikh, H. The B. subtilis accessory helicase PcrA facilitates DNA replication through transcription units. PLoS Genet. 2015, 11, e1005289. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.H.; Derry, K.L.; Kolodner, R.D. Saccharomyces cerevisiae RRM3, a 5’ to 3’ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 2002, 277, 45331–45337. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.; Gupta, M.K.; McGlynn, P. Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 2011, 39, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.G.; Howard, J.A.; McGlynn, P. Use of streptavidin bound to biotinylated DNA structures as model substrates for analysis of nucleoprotein complex disruption by helicases. Methods 2016. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.A.; Dervyn, E.; Rose, M.; Entian, K.D.; McGovern, S.; Ehrlich, S.D.; Bruand, C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 1998, 29, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Denhardt, D.T.; Dressler, D.H.; Hathaway, A. The abortive replication of fX174 DNA in a recombination-deficient mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 1967, 57, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Keil, R.L.; McWilliams, A.D. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 1993, 135, 711–718. [Google Scholar] [PubMed]
- Bochman, M.L.; Sabouri, N.; Zakian, V.A. Unwinding the functions of the Pif1 family helicases. DNA Repair 2010, 9, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.G.; Howard, J.L.; McGlynn, P. Accessory replicative helicases and the replication of protein-bound DNA. J. Mol. Biol. 2014, 426, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell 1988, 53, 679–686. [Google Scholar] [CrossRef]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, V.A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Machon, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petryk, N.; Kahli, M.; d’Aubenton-Carafa, Y.; Jaszczyszyn, Y.; Shen, Y.; Silvain, M.; Thermes, C.; Chen, C.L.; Hyrien, O. Replication landscape of the human genome. Nat. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Nudler, E.; Tora, L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013, 20, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Seigneur, M.; Ehrlich, S.D.; Michel, B. RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol. Microbiol. 2000, 38, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Helleday, T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010, 11, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Hanawalt, P.C. RecA-dependent recovery of arrested DNA replication forks. Ann. Rev. Genet. 2003, 37, 611–646. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Ray Chaudhuri, A.; Lopes, M.; Costanzo, V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010, 17, 1305–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Donaldson, J.R.; Chow, K.H.; Courcelle, C.T. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 2003, 299, 1064–1067. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, C.T.; Chow, K.H.; Casey, A.; Courcelle, J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 9154–9159. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [PubMed]
- Skarstad, K.; Boye, E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 1993, 175, 5505–5509. [Google Scholar] [PubMed]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A.; Stahl, F.W. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol. 1997, 179, 880–888. [Google Scholar] [PubMed]
- Seigneur, M.; Bidnenko, V.; Ehrlich, S.D.; Michel, B. RuvAB acts at arrested replication forks. Cell 1998, 95, 419–430. [Google Scholar] [CrossRef]
- Lambert, S.; Watson, A.; Sheedy, D.M.; Martin, B.; Carr, A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 2005, 121, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Saveson, C.J.; Lovett, S.T. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 1997, 146, 457–470. [Google Scholar] [PubMed]
- Vilette, D.; Uzest, M.; Ehrlich, S.D.; Michel, B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 1992, 11, 3629–3634. [Google Scholar] [PubMed]
- Prado, F.; Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Duch, A.; Felipe-Abrio, I.; Barroso, S.; Yaakov, G.; Garcia-Rubio, M.; Aguilera, A.; de Nadal, E.; Posas, F. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 2013, 493, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, A.A.; Buckman, C.; Harris, L.; Lloyd, R.G. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 2006, 20, 2135–2147. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.H.; Kolodner, R.D. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 18196–18201. [Google Scholar] [CrossRef] [PubMed]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef] [PubMed]
- Veaute, X.; Delmas, S.; Selva, M.; Jeusset, J.; le Cam, E.; Matic, I.; Fabre, F.; Petit, M.A. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005, 24, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.; Lesage, G.; Bader, G.D.; Ding, H.; Xu, H.; Xin, X.; Young, J.; Berriz, G.F.; Brost, R.L.; Chang, M.; et al. Global mapping of the yeast genetic interaction network. Science 2004, 303, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.H.; Kolodner, R.D. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004, 24, 3213–3226. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.Z.; Schnakenberg, S.L.; Zakian, V.A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004, 24, 3198–3212. [Google Scholar] [CrossRef] [PubMed]
- Uzest, M.; Ehrlich, S.D.; Michel, B. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 1995, 17, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Michel, B.; Ehrlich, S.D.; Uzest, M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997, 16, 430–438. [Google Scholar] [CrossRef] [PubMed]
- De Septenville, A.L.; Duigou, S.; Boubakri, H.; Michel, B. Replication fork reversal after replication-transcription collision. PLoS Genet. 2012, 8, e1002622. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A.; Stahl, F.W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999, 13, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Taucher-Scholtz, G.; Abdel-Monem, M.; Hoffmann-Berling, H. Functions of helicases in E. coli. In Mechanisms of DNA Replication and Recombination; Cozzarelli, N.R., Ed.; Alan R. Liss Inc.: New York, NY, USA, 1983; pp. 65–76. [Google Scholar]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Ann. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Denapoli, J.; Tehranchi, A.K.; Wang, J.D. Dose-dependent reduction of replication elongation rate by (p)ppGpp in Escherichia coli and Bacillus subtilis. Mol. Microbiol. 2013, 88, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Lestini, R.; Duigou, S.; Michel, B. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol. Microbiol. 2010, 77, 324–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGlynn, P.; Lloyd, R.G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 2000, 101, 35–45. [Google Scholar] [CrossRef]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.A.; Ehrlich, D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002, 21, 3137–3147. [Google Scholar] [CrossRef] [PubMed]
- Lestini, R.; Michel, B. UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J. Bacteriol. 2008, 190, 5995–6001. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.; McGlynn, P.; Ngo, H.P.; Sharples, G.J.; Lloyd, R.G. The RdgC protein of Escherichia coli binds DNA and counters a toxic effect of RecFOR in strains lacking the replication restart protein PriA. EMBO J. 2003, 22, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Magner, D.B.; Blankschien, M.D.; Lee, J.A.; Pennington, J.M.; Lupski, J.R.; Rosenberg, S.M. RecQ promotes toxic recombination in cells lacking recombination intermediate-removal proteins. Mol. Cell 2007, 26, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Capaldo-Kimball, F.; Barbour, S.D. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J. Bacteriol. 1971, 106, 204–212. [Google Scholar] [PubMed]
- Lloyd, R.G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 1991, 173, 5414–5418. [Google Scholar] [PubMed]
- Bernhardt, T.G.; de Boer, P.A. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 2004, 52, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Trautinger, B.W.; Lloyd, R.G. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002, 21, 6944–6953. [Google Scholar] [CrossRef] [PubMed]
- Lane, H.E.; Denhardt, D.T. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J. Bacteriol. 1974, 120, 805–814. [Google Scholar] [PubMed]
- Howard-Flanders, P.; Bardwell, E. Effects of recB21, recF143, and uvrD152 on recombination in lambda bacteriophage-prophage and Hfr by F-crosses. J. Bacteriol. 1981, 148, 739–743. [Google Scholar] [PubMed]
- Bidnenko, V.; Lestini, R.; Michel, B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol. Microbiol. 2006, 62, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Centore, R.C.; Sandler, S.J. UvrD limits the number and intensities of RecA-green fluorescent protein structures in Escherichia coli K-12. J. Bacteriol. 2007, 189, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Marians, K.J. PriA-directed assembly of a primosome on D loop DNA. J. Biol. Chem. 1999, 274, 25033–25041. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999, 63, 751–813. [Google Scholar] [PubMed]







© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syeda, A.H.; Atkinson, J.; Lloyd, R.G.; McGlynn, P. The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication. Genes 2016, 7, 42. https://doi.org/10.3390/genes7080042
Syeda AH, Atkinson J, Lloyd RG, McGlynn P. The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication. Genes. 2016; 7(8):42. https://doi.org/10.3390/genes7080042
Chicago/Turabian StyleSyeda, Aisha H., John Atkinson, Robert G. Lloyd, and Peter McGlynn. 2016. "The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication" Genes 7, no. 8: 42. https://doi.org/10.3390/genes7080042
APA StyleSyeda, A. H., Atkinson, J., Lloyd, R. G., & McGlynn, P. (2016). The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication. Genes, 7(8), 42. https://doi.org/10.3390/genes7080042





