Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli
Abstract
:1. Introduction
2. DNA Replication
2.1. Initiation of DNA Replication
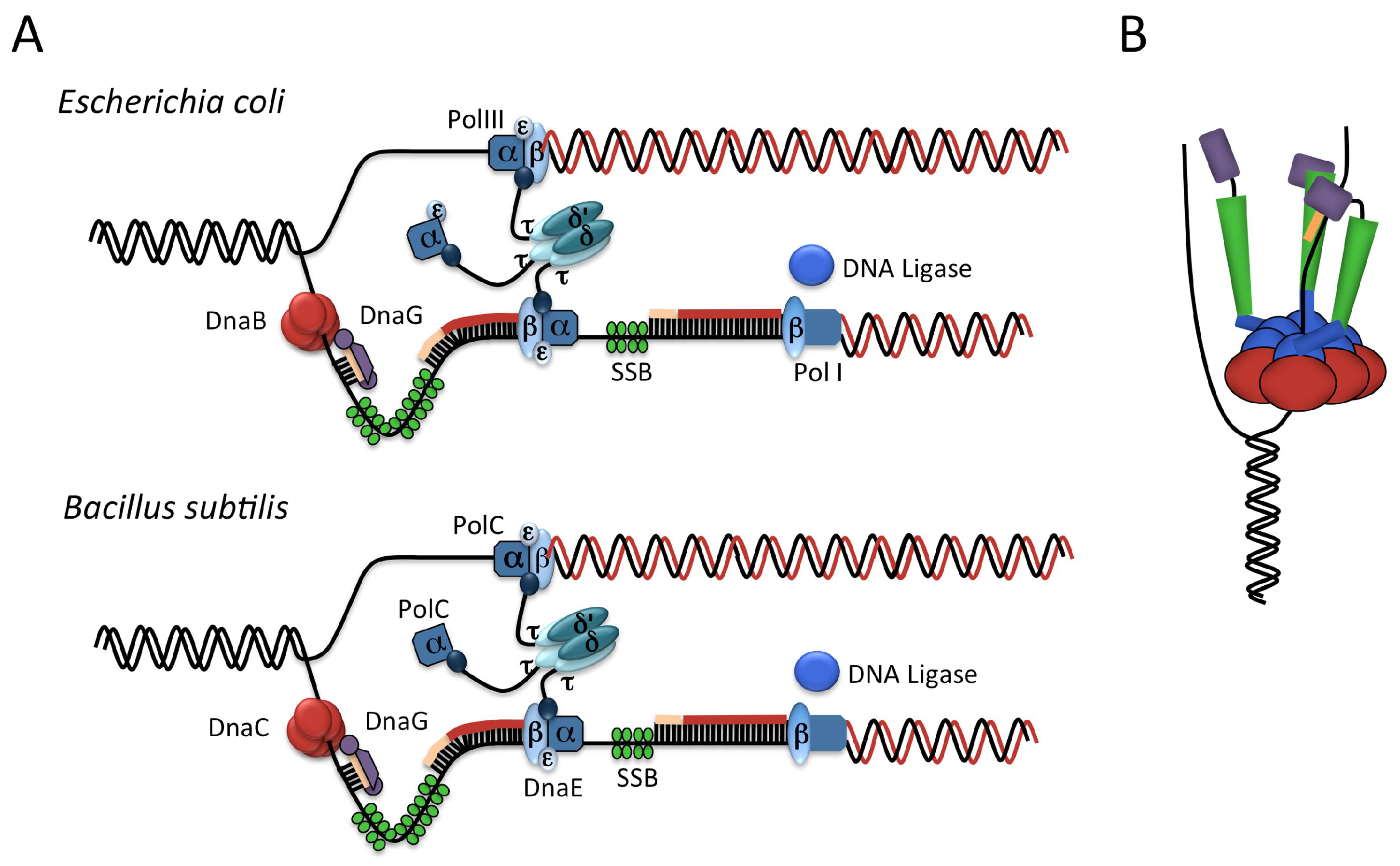
2.2. The Elongation Phase
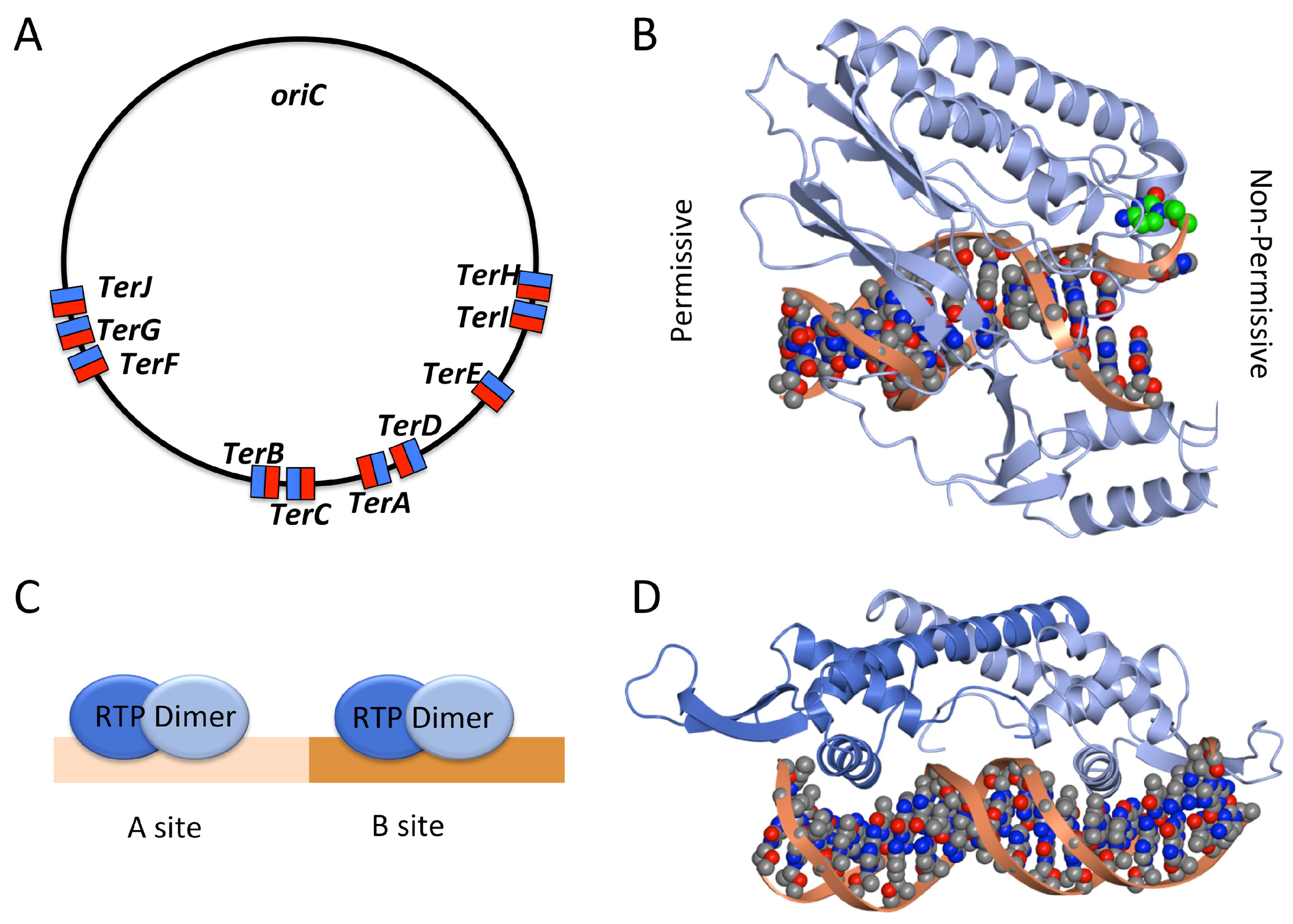
2.3. Termination of DNA Replication
3. Initiation of DNA Replication
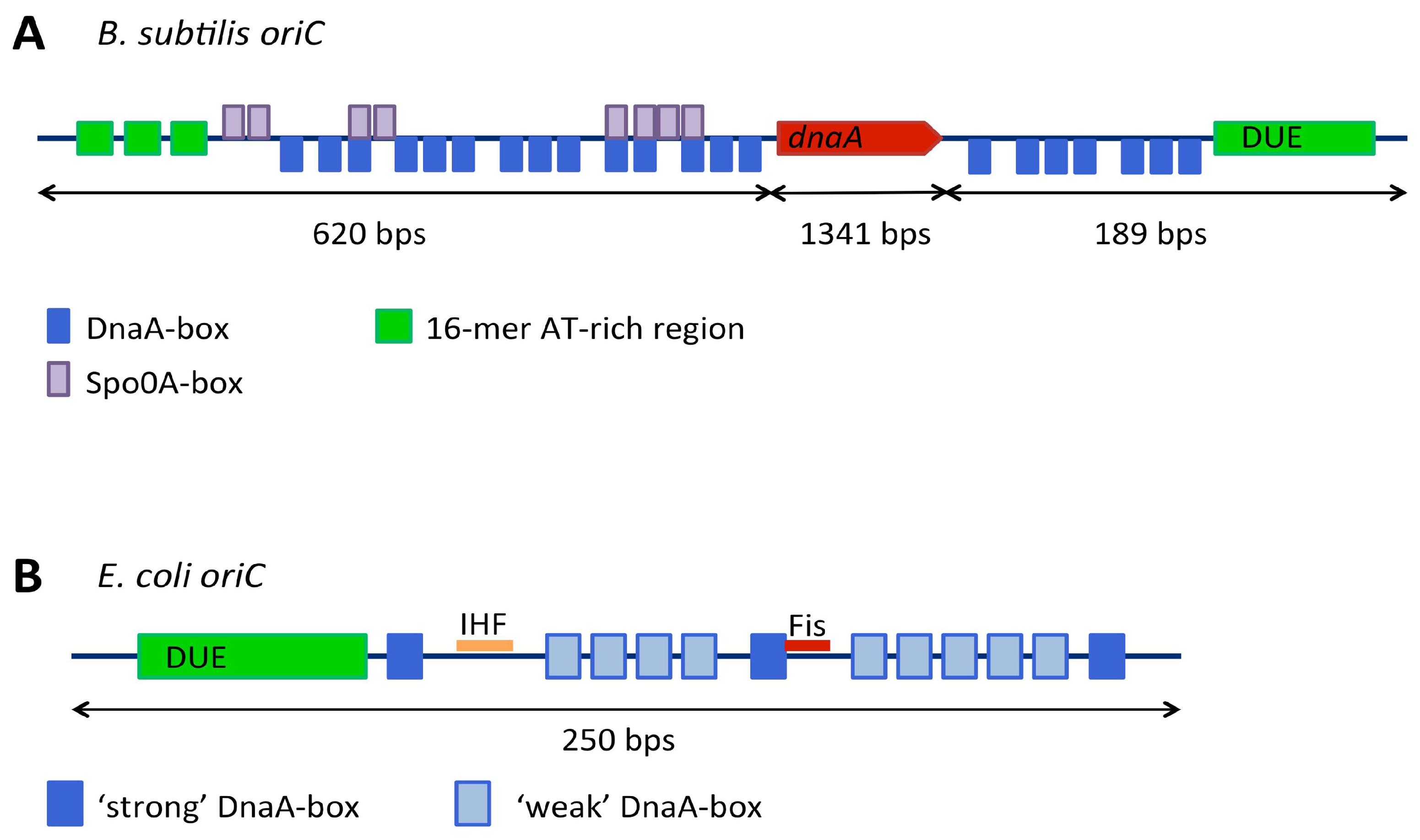
3.1. Replication Origins
3.1.1. Genetic Context of Replication Origins
3.1.2. Continuous and Bipartite Origins
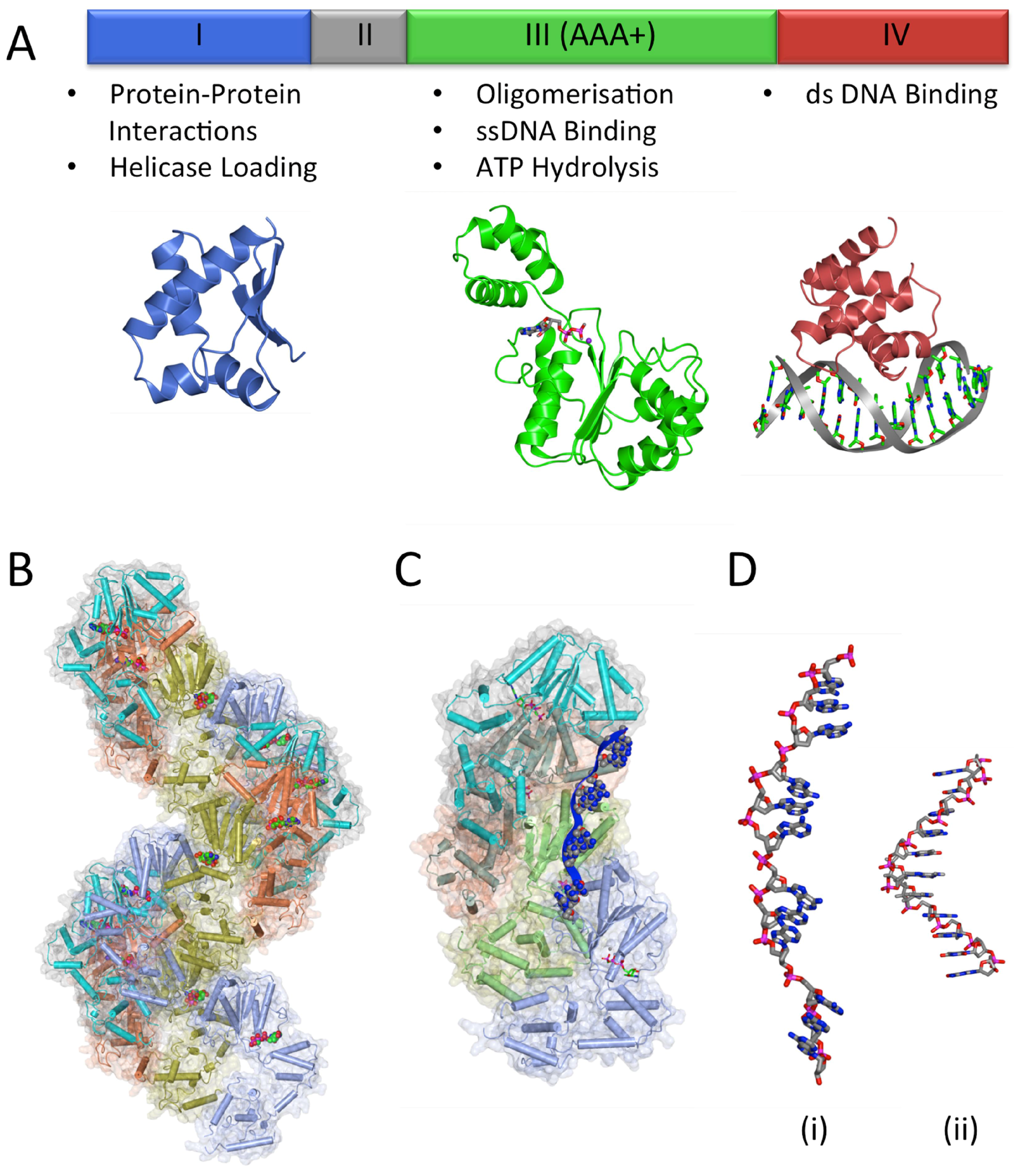
3.2. The DNA Replication Initiator, DnaA
3.2.1. DnaA-Box Recognition by DnaA
3.2.2. Variable Affinity of DnaA-Boxes
3.2.3. DnaA Oligomerisation
3.2.4. DNA Unwinding and ssDNA Binding
3.2.5. Bacillus DnaA
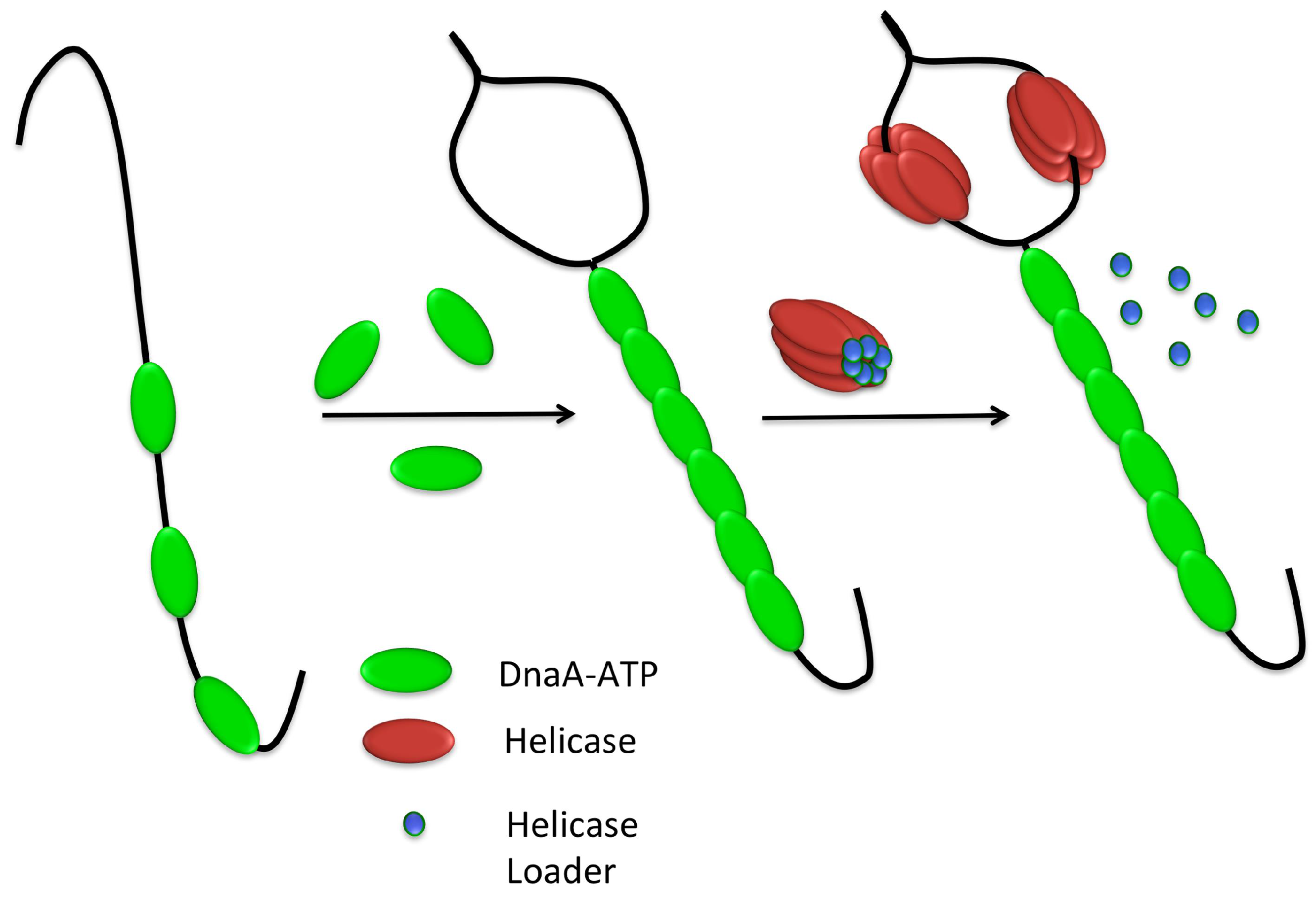
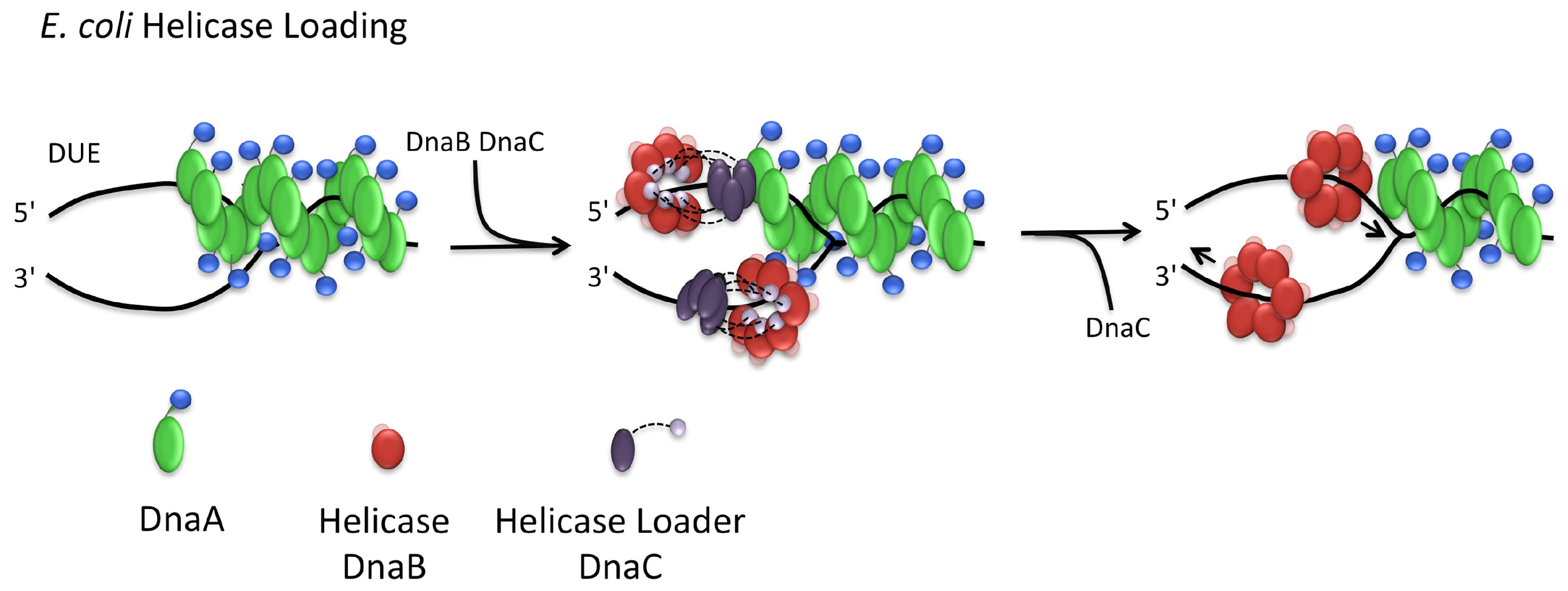
3.3. Helicase Loading
3.4. Bacillus Initiation Proteins DnaD and DnaB
3.5. Regulation of DNA Replication
3.5.1. During Vegetative Growth in B. subtilis
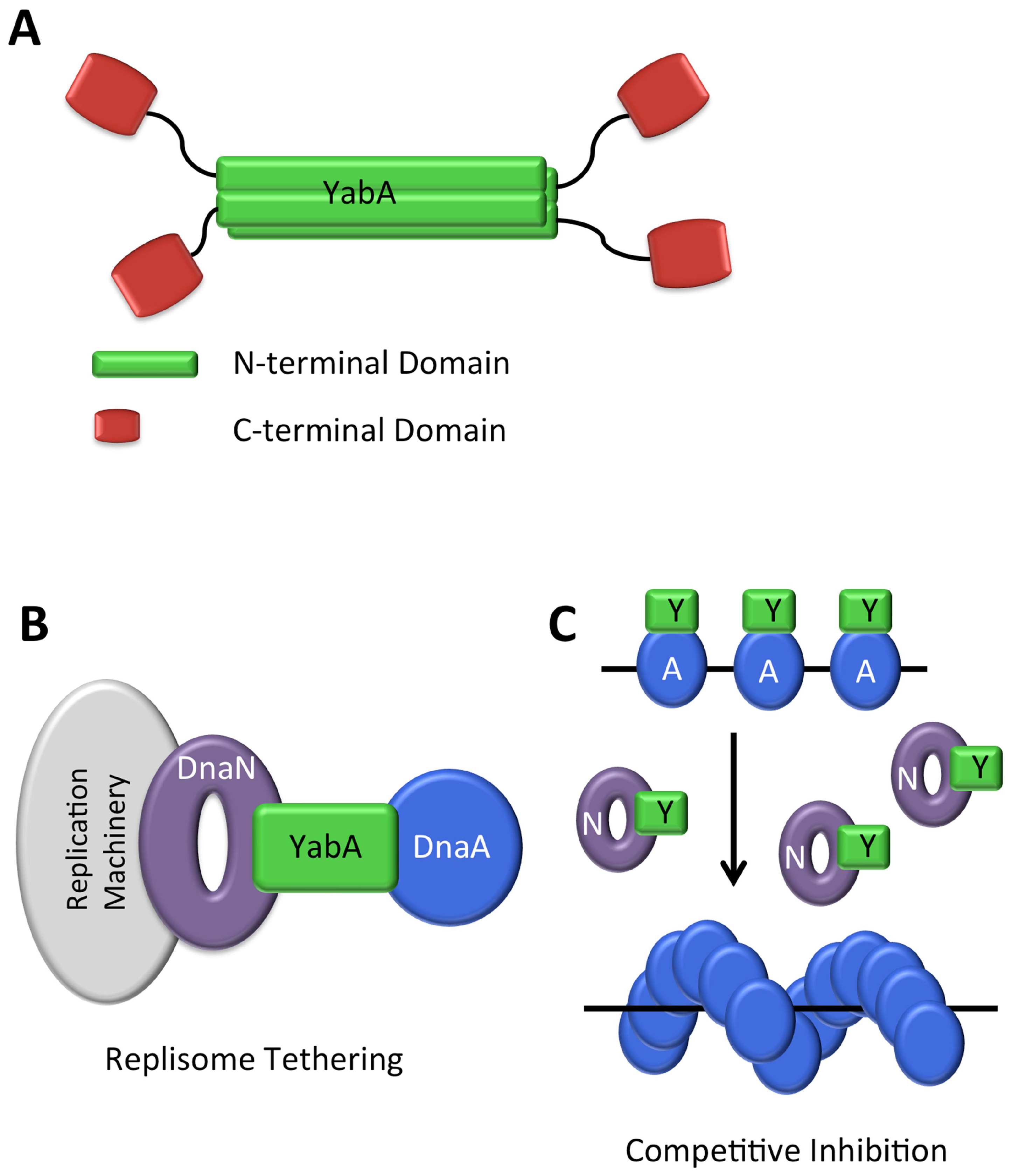
YabA
Soj/Spo0J
DnaD
DnaA-Box Clusters
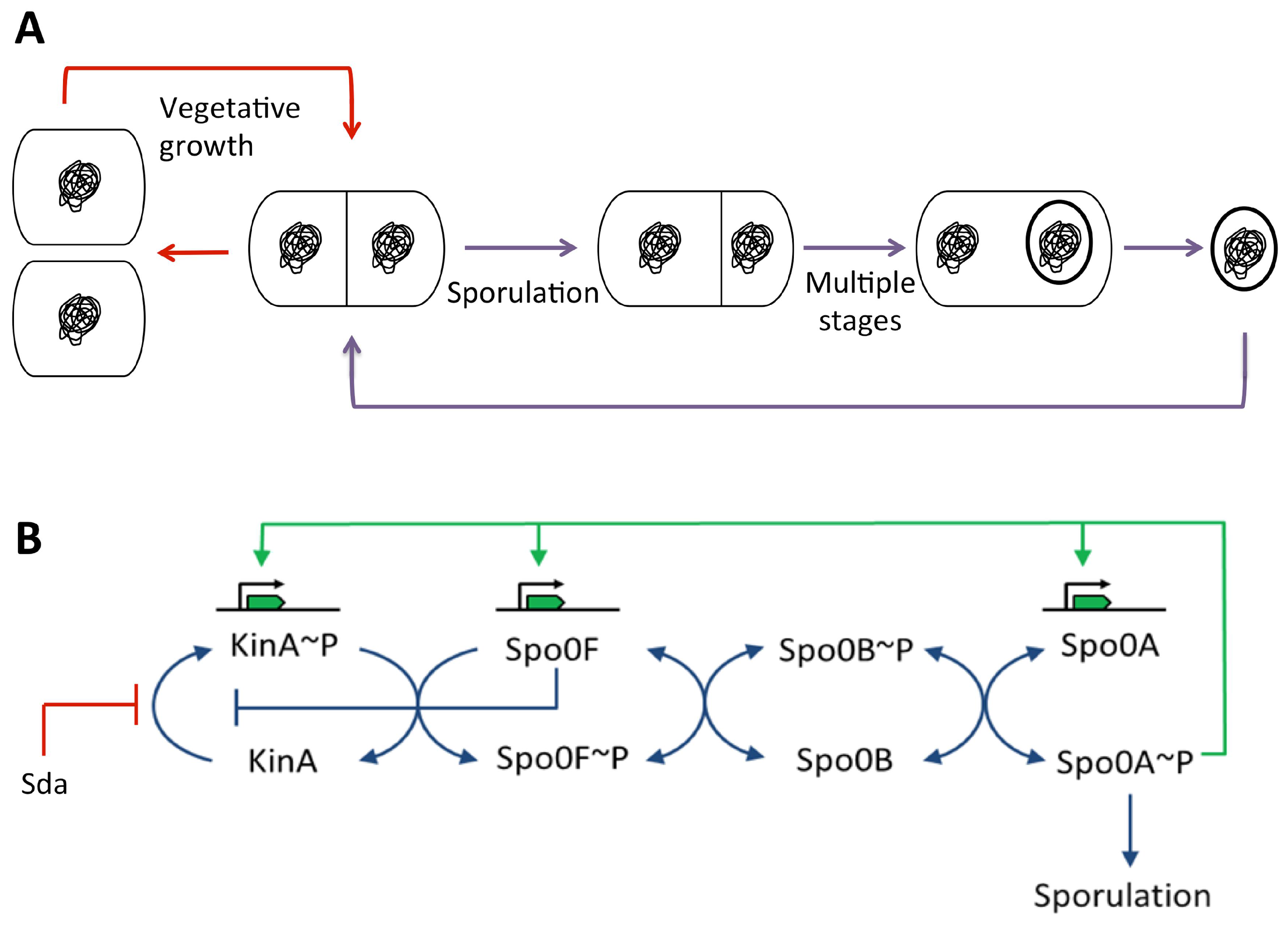
3.5.2. During Sporulation in Bacillus subtilis
Spo0A~P Pulsing
SirA
A Direct Role for Spo0A~P
4. In E. coli
4.1. Regulatory Inactivation of DnaA (RIDA)
4.2. IHF and Fis
4.3. DnaA-Box Sequences: datA and DARS
4.4. SeqA
4.5. DiaA (and HobA)
4.6. Lysine Acetylation of DnaA
5. Regulatory Mechanisms for DNA Replication Regulation: B. subtilis vs. E. coli
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kaguni, J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011, 15, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Ozaki, S.; Keyamura, K.; Fujimitsu, K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010, 8, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, G.; Veening, J.-W.; Murray, H. DnaA and ORC: More than DNA replication initiators. Trends Cell Biol. 2011, 21, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Mott, M.L.; Berger, J.M. DNA replication initiation: Mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007, 5, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.C.; Grimwade, J.E. Regulation of DnaA assembly and activity: Taking directions from the genome. Annu. Rev. Microbiol. 2011, 65, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Briggs, G.S.; Smits, W.K.; Soultanas, P. Chromosomal replication initiation machinery of low-G+C-content Firmicutes. J. Bacteriol. 2012, 194, 5162–5170. [Google Scholar] [CrossRef] [PubMed]
- Zorman, S.; Seitz, H.; Sclavi, B.; Strick, T.R. Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 2012, 40, 7375–7383. [Google Scholar] [CrossRef] [PubMed]
- Erzberger, J.P.; Mott, M.L.; Berger, J.M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006, 13, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R.S.; Funnell, B.E.; Kornberg, A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 1984, 38, 889–900. [Google Scholar] [CrossRef]
- Kowalski, D.; Eddy, M.J. The DNA unwinding element: A novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989, 8, 4335–4344. [Google Scholar] [PubMed]
- Bramhill, D.; Kornberg, A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 1988, 52, 743–755. [Google Scholar] [CrossRef]
- Marszalek, J.; Kaguni, J.M. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 1994, 269, 4883–4890. [Google Scholar] [PubMed]
- Koboris, J.A.; Kornberg, A. Escherichia coli dnaC Gene Product. Biol. Chem. 1982, 257, 13770–13775. [Google Scholar]
- Fang, L.; Davey, M.J.; O’Donnell, M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 1999, 4, 541–553. [Google Scholar] [CrossRef]
- Bruand, C.; Ehrlich, S.D.; Jannière, L. Primosome assembly site in Bacillus subtilis. EMBO J. 1995, 14, 2642–2650. [Google Scholar] [PubMed]
- Zhang, W.; Carneiro, M.J.V.M.; Turner, I.J.; Allen, S.; Roberts, C.J.; Soultanas, P. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J. Mol. Biol. 2005, 351, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K.; Goranov, A.I.; Grossman, A.D. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 2010, 75, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.; Eliason, W.K.; Steitz, T.A. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 2007, 318, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Sherratt, D.J.; Leake, M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 2010, 328, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.; Causer, R.J.; Dixon, N.E. Architecture and Conservation of the Bacterial DNA Replication Machinery, an Underexploited Drug Target. Curr. Drug Targets 2012, 13, 352–372. [Google Scholar] [CrossRef] [PubMed]
- Beattie, T.R.; Reyes-Lamothe, R. A Replisome’s journey through the bacterial chromosome. Front. Microbiol. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G. DNA Replication, Repair, and Recombination. In Biochemistry; Wiley: Hoboken, NJ, USA, 2011; pp. 1171–1259. [Google Scholar]
- Corn, J.E.; Pelton, J.G.; Berger, J.M. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat. Struct. Mol. Biol. 2008, 15, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Corn, J.E.; Berger, J.M. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 2006, 34, 4082–4088. [Google Scholar] [CrossRef] [PubMed]
- Corn, J.E.; Pease, P.J.; Hura, G.L.; Berger, J.M. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 2005, 20, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Dervyn, E.; Suski, C.; Daniel, R.; Bruand, C.; Chapuis, J.; Errington, J.; Jannière, L.; Ehrlich, S.D. Two essential DNA polymerases at the bacterial replication fork. Science 2001, 294, 1716–1719. [Google Scholar] [CrossRef] [PubMed]
- Sanders, G.M.; Dallmann, H.G.; McHenry, C.S. Reconstitution of the B. subtilis Replisome with 13 Proteins Including Two Distinct Replicases. Mol. Cell 2010, 37, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.M.; Henson, J.M.; Kuempel, P.L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc. Natl. Acad. Sci. USA 1987, 84, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, M.; Kobayashi, T.; Takenaka, S.; Takeya, H.; Horiuchi, T. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J. Biol. Chem. 1989, 264, 21031–21037. [Google Scholar] [PubMed]
- Neylon, C.; Kralicek, A.V.; Hill, T.M.; Dixon, N.E. Replication Termination in Escherichia coli: Structure and Antihelicase Activity of the Tus-Ter Complex. Microbiol. Mol. Biol. Rev. 2005, 69, 501–526. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Horiuchi, T.; Ohsumi, K.; Shimamoto, N.; Morikawa, K. Structure of a replication-terminator protein complexed with DNA. Nature 1996, 383, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Mulcair, M.D.; Schaeffer, P.M.; Oakley, A.J.; Cross, H.F.; Neylon, C.; Hill, T.M.; Dixon, N.E. A Molecular Mousetrap Determines Polarity of Termination of DNA Replication in E. coli. Cell 2006, 125, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, B.A.; Dulin, D.; Xu, Z.-Q.; van Laar, T.; Cross, B.; Janissen, R.; Jergic, S.; Dixon, N.E.; Depken, M.; Dekker, N.H. Strand separation establishes a sustained lock at the Tus–Ter replication fork barrier. Nat. Chem. Biol. 2015, 11, 579–585. [Google Scholar] [CrossRef] [PubMed]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: the CCP4mg molecular-graphics software. Acta. Cryst. D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar]
- Hill, T.M. Arrest of bacterial DNA replication. Annu. Rev. Microbiol. 1992, 46, 603–633. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.J.; Ralston, G.B.; Christopherson, R.I.; Wake, R.G. Identification of the replication terminator protein binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding. J. Mol. Biol. 1990, 214, 73–84. [Google Scholar] [CrossRef]
- Vivian, J.P.; Porter, C.J.; Wilce, J.A.; Wilce, M.C.J. An Asymmetric Structure of the Bacillus subtilis Replication Terminator Protein in Complex with DNA. J. Mol. Biol. 2007, 370, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Langley, D.B.; Smith, M.T.; Lewis, P.J.; Wake, R.G. Protein-nucleoside contacts in the interaction between the replication terminator protein of Bacillus subtilis and the DNA terminator. Mol. Microbiol. 1993, 10, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Wolanski, M.; Donczew, R.; Zawilak-Pawlik, A.; Zakrzewska-Czerwinska, J. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2015, 5, 1–14. [Google Scholar]
- Leonard, A.C.; Méchali, M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013, 5, a010116. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, N.; Yoshikawa, H. Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 1992, 6, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Moriya, S.; Atlung, T.; Hansen, F.G.; Yoshikawa, H.; Ogasawara, N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 1992, 6, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Donczew, R.; Weigel, C.; Lurz, R.; Zakrzewska-Czerwińska, J.; Zawilak-Pawlik, A. Helicobacter pylori oriC-the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 2012, 40, 9647–9660. [Google Scholar] [CrossRef] [PubMed]
- Krause, M.; Rückert, B.; Lurz, R.; Messer, W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 1997, 274, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Messer, W.; Blaesing, F.; Majka, J.; Nardmann, J.; Schaper, S.; Schmidt, A.; Seitz, H.; Speck, C.; Tüngler, D.; Wegrzyn, G.; et al. Functional domains of DnaA proteins. Biochimie 1999, 81, 819–825. [Google Scholar] [CrossRef]
- Sutton, M.D.; Kaguni, J.M. The Escherichia coli dnaA gene: Four functional domains. J. Mol. Biol. 1997, 274, 546–561. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, T.; Moriya, S.; Yoshikawa, H.; Ogasawara, N. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 1990, 107, 732–739. [Google Scholar] [PubMed]
- Nishida, S. A Nucleotide Switch in the Escherichia coli DnaA Protein Initiates Chromosomal Replication. J. Biol. Chem. 2002, 277, 14986–14995. [Google Scholar] [CrossRef] [PubMed]
- Sekimizu, K.; Bramhill, D.; Kornberg, A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 1987, 50, 259–265. [Google Scholar] [CrossRef]
- Roth, A.; Messer, W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995, 14, 2106–2111. [Google Scholar] [PubMed]
- Fujikawa, N.; Kurumizaka, H.; Nureki, O.; Terada, T. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003, 31, 2077–2086. [Google Scholar] [CrossRef] [PubMed]
- Kaguni, J.M. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006, 60, 351–375. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, S.; Ogawa, T. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 2008, 154, 3379–3384. [Google Scholar] [CrossRef] [PubMed]
- Rahn-Lee, L.; Merrikh, H.; Grossman, A.D.; Losick, R. The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J. Bacteriol. 2011, 193, 1302–1307. [Google Scholar] [CrossRef] [PubMed]
- Zawilak-Pawlik, A.; Kois, A.; Stingl, K.; Boneca, I.G.; Skrobuk, P.; Piotr, J.; Lurz, R.; Zakrzewska-Czerwińska, J.; Labigne, A. HobA—A novel protein involved in initiation of chromosomal replication in Helicobacter pylori. Mol. Microbiol. 2007, 65, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Akimitsu, N.; Kashioka, T.; Hatano, M.; Kubota, T.; Ogata, Y.; Sekimizu, K.; Katayama, T. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 2004, 279, 45546–45555. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.; Weigel, C.; Messer, W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 2000, 37, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Margulies, C.; Kaguni, J.M. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 1996, 271, 17035–17040. [Google Scholar] [CrossRef] [PubMed]
- Langer, U.; Richter, S.; Roth, A.; Weigel, C.; Messer, W. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol. Microbiol. 1996, 21, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Schaper, S.; Messer, W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995, 270, 17622–17626. [Google Scholar] [CrossRef] [PubMed]
- Blaesing, F.; Weigel, C.; Welzeck, M.; Messer, W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 2000, 36, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.T.; Grimwade, J.E.; Betteridge, T.; Rozgaja, T.; Torgue, J.J.-C.; Leonard, A.C. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. USA 2009, 106, 18479–18484. [Google Scholar] [CrossRef] [PubMed]
- Rozgaja, T.A.; Grimwade, J.E.; Iqbal, M.; Czerwonka, C.; Vora, M.; Leonard, A.C. Two oppositely oriented arrays of low affinity recognition sites in oriC guide progressive binding of DnaA during E. coli pre-RC assembly. Mol. Microbiol. 2012, 82, 475–488. [Google Scholar] [CrossRef] [PubMed]
- McGarry, K.C.; Ryan, V.T.; Grimwade, J.E.; Leonard, A.C. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. USA 2004, 101, 2811–2816. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Vora, M.P.; Czerwonka, C.A.; Rozgaja, T.A.; Grimwade, J.E.; Leonard, A.C. Building the bacterial orisome: High affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014, 91, 1148–1163. [Google Scholar] [CrossRef] [PubMed]
- Erzberger, J.P.; Pirruccello, M.M.; Berger, J.M. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002, 21, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Whiteheart, S.W.; Wilkinson, A.J. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004, 146, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Mott, M.L.; Crisona, N.J.; Chuang, K.; Yang, H.; Berger, J.M. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010, 285, 28229–28239. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Chuang, K.; Berger, J.M. DNA stretching by bacterial initiators promotes replication origin opening. Nature 2011, 478, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Speck, C.; Messer, W. Mechanism of origin unwinding: Sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001, 20, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.T.; Harran, O.; Murray, H. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 2016, 534, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, G.; Errington, J.; Murray, H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012, 31, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Grossman, A.D. Control of the replication initiator DnaA by an anti-cooperativity factor. Mol. Microbiol. 2011, 82, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Rotoli, S.M.; Biswas-Fiss, E.; Biswas, S.B. Quantitative analysis of the mechanism of DNA binding by Bacillus DnaA protein. Biochimie 2012, 94, 2764–2775. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.D.; Carr, K.M.; Vicente, M.; Kaguni, J.M. Escherichia coli DnaA Protein. J. Biol. Chem. 1998, 273, 34255–34262. [Google Scholar] [CrossRef] [PubMed]
- Keyamura, K.; Abe, Y.; Higashi, M.; Ueda, T.; Katayama, T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009, 284, 25038–25050. [Google Scholar] [CrossRef] [PubMed]
- Su’etsugu, M.; Harada, Y.; Keyamura, K.; Matsunaga, C.; Kasho, K.; Abe, Y.; Ueda, T.; Katayama, T. The DnaA N-terminal domain interacts with Hda to facilitate replicase clamp-mediated inactivation of DnaA. Environ. Microbiol. 2013, 15, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Carr, K.M.; Kaguni, J.M. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 2001, 276, 44919–44925. [Google Scholar] [CrossRef] [PubMed]
- Weigel, C.; Seitz, H. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol. Microbiol. 2002, 46, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Felczak, M.M.; Simmons, L.A.; Kaguni, J.M. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005, 280, 24627–24633. [Google Scholar] [CrossRef] [PubMed]
- Simmons, L.A.; Felczak, M.; Kaguni, J.M. DnaA Protein of Escherichia coli: Oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 2003, 49, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Natrajan, G.; Noirot-Gros, M.-F.; Zawilak-Pawlik, A.; Kapp, U.; Terradot, L. The structure of a DnaA/HobA complex from Helicobacter pylori provides insight into regulation of DNA replication in bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 21115–21120. [Google Scholar] [CrossRef] [PubMed]
- Lowery, T.J.; Pelton, J.G.; Chandonia, J.-M.; Kim, R.; Yokota, H.; Wemmer, D.E. NMR structure of the N-terminal domain of the replication initiator protein DnaA. J. Struct. Funct. Genom. 2007, 8, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Jo, T.; Matsuda, Y.; Matsunaga, C.; Katayama, T.; Ueda, T. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007, 282, 17816–17827. [Google Scholar] [CrossRef] [PubMed]
- Molt, K.L.; Sutera, V.A.; Moore, K.K.; Lovett, S.T. A role for nonessential domain II of initiator protein, DnaA, in replication control. Genetics 2009, 183, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Arias-Palomo, E.; O’Shea, V.L.; Hood, I.V.; Berger, J.M. The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 2013, 153, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Marszalek, J.; Zhang, W.; Hupp, T.R.; Margulies, C.; Carr, K.M.; Cherry, S.; Kaguni, J.M. Domains of DnaA Protein Involved in Interaction with DnaB Protein, and in Unwinding the Escherichia coli Chromosomal Origin. J. Biol. Chem. 1996, 271, 18535–18542. [Google Scholar]
- Mott, M.L.; Erzberger, J.P.; Coons, M.M.; Berger, J.M. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 2008, 135, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Makowska-Grzyska, M.; Kaguni, J.M. Primase Directs the Release of DnaC from DnaB. Mol. Cell 2010, 37, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Kaguni, J.M. Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Soultanas, P. Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 2012, 84, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Velten, M.; McGovern, S.; Marsin, S.; Ehrlich, S.D.; Noirot, P.; Polard, P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 2003, 11, 1009–1020. [Google Scholar] [CrossRef]
- Ioannou, C.; Schaeffer, P.M.; Dixon, N.E.; Soultanas, P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006, 34, 5247–5258. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Eliason, W.K.; Steitz, T.A. Structure of a helicase-helicase loader complex reveals insights into the mechanism of bacterial primosome assembly. Nat. Commun. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T.; McKenzie, T.; Schmidt, S.; Tanaka, T.; Sueoka, N. Nucleotide sequence of Bacillus subtilis dnaB: A gene essential for DNA replication initiation and membrane attachment. Proc. Natl. Acad. Sci. USA 1987, 84, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Marston, F.Y.; Grainger, W.H.; Smits, W.K.; Hopcroft, N.H.; Green, M.; Hounslow, A.M.; Grossman, A.D.; Craven, C.J.; Soultanas, P. When simple sequence comparison fails: The cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD. Nucleic Acids Res. 2010, 38, 6930–6942. [Google Scholar] [CrossRef] [PubMed]
- Bruand, C.; Farache, M.; McGovern, S.; Ehrlich, S.D.; Polard, P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 2001, 42, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Ishigo-oka, D.; Ogasawara, N.; Moriya, S. DnaD Protein of Bacillus subtilis Interacts with DnaA, the Initiator Protein of Replication. J. Bacteriol. 2001, 183, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Allen, S.; Roberts, C.J.; Soultanas, P. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. J. Bacteriol. 2006, 188, 5487–5493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Machón, C.; Orta, A.; Phillips, N.; Roberts, C.J.; Allen, S.; Soultanas, P. Single-molecule atomic force spectroscopy reveals that DnaD forms scaffolds and enhances duplex melting. J. Mol. Biol. 2008, 377, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.; Zhang, W.; Soultanas, P.; Paoli, M. Structure of the N-Terminal Oligomerization Domain of DnaD Reveals a Unique Tetramerization Motif and Provides Insights into Scaffold Formation. J. Mol. Biol. 2008, 376, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.J.V.M.; Zhang, W.; Ioannou, C.; Scott, D.J.; Allen, S.; Roberts, C.J.; Soultanas, P. The DNA-remodelling activity of DnaD is the sum of oligomerization and DNA-binding activities on separate domains. Mol. Microbiol. 2006, 60, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Bruand, C.; Velten, M.; McGovern, S.; Marsin, S.; Sérèna, C.; Dusko Ehrlich, S.; Polard, P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 2005, 55, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Collier, C.; Machón, C.; Briggs, G.S.; Smits, W.K.; Soultanas, P. Untwisting of the DNA helix stimulates the endonuclease activity of Bacillus subtilis Nth at AP sites. Nucleic Acids Res. 2012, 40, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Rannou, O.; Le Chatelier, E.; Larson, M.A.; Nouri, H.; Dalmais, B.; Laughton, C.; Jannière, L.; Soultanas, P. Functional interplay of DnaE polymerase, DnaG primase and DnaC helicase within a ternary complex, and primase to polymerase hand-off during lagging strand DNA replication in Bacillus subtilis. Nucleic Acids Res. 2013, 41, 5303–5320. [Google Scholar] [CrossRef] [PubMed]
- Rokop, M.E.; Auchtung, J.M.; Grossman, A.D. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 2004, 52, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Grainger, W.H.; Machón, C.; Scott, D.J.; Soultanas, P. DnaB proteolysis in vivo regulates oligomerization and its localization at oriC in Bacillus subtilis. Nucleic Acids Res. 2010, 38, 2851–2864. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Ogura, Y.; Harry, E.J.; Ogasawara, N.; Moriya, S. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 2005, 247, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Noirot-Gros, M.-F.; Dervyn, E.; Wu, L.J.; Mervelet, P.; Errington, J.; Ehrlich, S.D.; Noirot, P. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 2002, 99, 8342–8347. [Google Scholar] [CrossRef] [PubMed]
- Noirot-Gros, M.-F.; Velten, M.; Yoshimura, M.; McGovern, S.; Morimoto, T.; Ehrlich, S.D.; Ogasawara, N.; Polard, P.; Noirot, P. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2006, 103, 2368–2373. [Google Scholar] [CrossRef] [PubMed]
- Felicori, L.; Jameson, K.H.; Roblin, P.; Fogg, M.J.; Cherrier, V.; Bazin, A.; Garcia-garcia, T.; Ventroux, M.; Noirot, P.; Wilkinson, A.J.; et al. Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners. Nucleic Acids Res. 2016, 44, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, G.; Murray, H. YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA. Mol. Microbiol. 2013, 90, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Ogasawara, N.; Ishikawa, S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet. Syst. 2008, 83, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Murray, H.; Errington, J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 2008, 135, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, G.; Whiting, R.; Errington, J.; Murray, H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 2011, 79, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Leonard, T.A.; Butler, P.J.; Löwe, J. Bacterial chromosome segregation: Structure and DNA binding of the Soj dimer—A conserved biological switch. EMBO J. 2005, 24, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Ringgaard, S.; van Zon, J.; Howard, M.; Gerdes, K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl. Acad. Sci. USA 2009, 106, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Howard, M.; Szardenings, F. Pushing and pulling in prokaryotic DNA segregation. Cell 2010, 141, 927–942. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Grossman, A.D. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 2006, 60, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, N.L.; Marquis, K.A.; Rudner, D.Z. Recruitment of SMC by ParB-parS Organizes the Origin Region and Promotes Efficient Chromosome Segregation. Cell 2009, 137, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Gruber, S.; Errington, J. Recruitment of Condensin to Replication Origin Regions by ParB/SpoOJ Promotes Chromosome Segregation in B. subtilis. Cell 2009, 137, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, C.Y.; Grossman, A.D. The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis. J. Bacteriol. 2012, 194, 5110–5117. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Yoshimura, M.; Ueki, M.; Oshima, T.; Ogasawara, N.; Ishikawa, S. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 2012, 40, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Errington, J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003, 1, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.; Dworkin, J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012, 36, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Kay, D.; Warren, S.C. Sporulation in Bacillus subtilis. Morphological changes. Biochem. J. 1968, 109, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Shao, W.; Perego, M.; Hoch, J.A. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000, 38, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Molle, V.; Fujita, M.; Jensen, S.T.; Eichenberger, P.; González-Pastor, J.E.; Liu, J.S.; Losick, R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003, 50, 1683–1701. [Google Scholar] [CrossRef] [PubMed]
- Veening, J.-W.; Murray, H.; Errington, J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009, 23, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Mandelstam, J.; Higgs, S.A. Induction of sporulation during synchronized chromosome replication in Bacillus subtilis. J. Bacteriol. 1974, 120, 38–42. [Google Scholar] [PubMed]
- Dunn, B.G.; Jeffs, P.; Ma, N.H. The Relationship Between DNA Replication and the Induction of Sporulation in Bacillus subtilis. Microbiology 1978, 108, 189–195. [Google Scholar] [CrossRef]
- Narula, J.; Kuchina, A.; Lee, D.D.; Fujita, M.; Süel, G.M.; Igoshin, O.A. Chromosomal Arrangement of Phosphorelay Genes Couples Sporulation and DNA Replication. Cell 2015, 162, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; González-Pastor, J.E.; Gonza, E.; Losick, R. High- and Low-Threshold Genes in the Spo0A Regulon of Bacillus subtilis. J. Bacteriol. 2005, 187, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Losick, R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005, 19, 2236–2244. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, W.F.; Kurtser, I.; Grossman, A.D. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 2001, 104, 269–279. [Google Scholar] [CrossRef]
- Bick, M.J.; Lamour, V.; Rajashankar, K.R.; Gordiyenko, Y.; Robinson, C.V.; Darst, S.A. How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 2009, 386, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Whitten, A.E.; Jacques, D.A.; Hammouda, B.; Hanley, T.; King, G.F.; Guss, J.M.; Trewhella, J.; Langley, D.B. The Structure of the KinA-Sda Complex Suggests an Allosteric Mechanism of Histidine Kinase Inhibition. J. Mol. Biol. 2007, 368, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, M.V.; Mach, K.E.; Burkholder, W.F. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol. Microbiol. 2006, 60, 1490–1508. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.H.; Fontes, M.E.; Dworkin, J.; Elowitz, M.B. Pulsed feedback defers cellular differentiation. PLoS Biol. 2012, 10, e1001252. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.W.; Piggot, P.J. Analysis of the inhibition of sporulation of Bacillus subtilis caused by increasing the number of copies of the spo0F gene. J. Gen. Microbiol. 1987, 133, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Grimshaw, C.E.; Huang, S.; Hanstein, C.G.; Strauch, M.A.; Burbulys, D.; Wang, L.; Hoch, J.A.; Whiteley, J.M. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 1998, 37, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Rahn-Lee, L.; Gorbatyuk, B.; Skovgaard, O.; Losick, R. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 2009, 191, 3736–3739. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.K.; Marquis, K.A.; Rudner, D.Z. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 2009, 73, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Jameson, K.H.; Rostami, N.; Fogg, M.J.; Turkenburg, J.P.; Grahl, A.; Murray, H.; Wilkinson, A.J. Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA. Mol. Microbiol. 2014, 93, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Huey, J.D.; Herman, J.K. The DnaA inhibitor SirA acts in the same pathway as Soj (ParA) to facilitate oriC segregation during Bacillus subtilis sporulation. Mol. Microbiol. 2016, 102, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, M.; de Jong, I.G.; Scholefield, G.; Murray, H.; Kuipers, O.P.; Veening, J.-W.W. Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol. Microbiol. 2013, 87, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Castilla-Llorente, V.; Muñoz-Espín, D.; Villar, L.; Salas, M.; Meijer, W.J.J. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J. 2006, 25, 3890–3899. [Google Scholar] [CrossRef] [PubMed]
- Camara, J.E.; Breier, A.M.; Brendler, T.; Austin, S.; Cozzarelli, N.R.; Crooke, E. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 2005, 6, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kubota, T.; Kurokawa, K.; Crooke, E.; Sekimizu, K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 1998, 94, 61–71. [Google Scholar] [CrossRef]
- Kato, J.; Katayama, T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001, 20, 4253–4262. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.; Kubota, T.; Matsuda, Y.; Katayama, T. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: Interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells 2004, 9, 509–522. [Google Scholar]
- Kurz, M.; Dalrymple, B.; Wijffels, G.; Kongsuwan, K. Interaction of the Sliding Clamp Beta-Subunit and Hda, a DnaA-Related Protein. J. Bacteriol. 2004, 186, 3508–3515. [Google Scholar] [CrossRef] [PubMed]
- Su’etsugu, M.; Shimuta, T.-R.R.; Ishida, T.; Kawakami, H.; Katayama, T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 2005, 280, 6528–6536. [Google Scholar] [CrossRef] [PubMed]
- Fujimitsu, K.; Su’etsugu, M.; Yamaguchi, Y.; Mazda, K.; Fu, N.; Kawakami, H.; Katayama, T. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 2008, 190, 5368–5381. [Google Scholar] [CrossRef] [PubMed]
- Su’etsugu, M.; Nakamura, K.; Keyamura, K.; Kudo, Y.; Katayama, T. Hda Monomerization by ADP Binding Promotes Replicase Clamp-mediated DnaA-ATP Hydrolysis. J. Biol. Chem. 2008, 283, 36118–36131. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; McMullan, D.; Abdubek, P.; Astakhova, T.; Carlton, D.; Chen, C.; Chiu, H.-J.; Clayton, T.; Das, D.; Deller, M.C.; et al. A structural basis for the regulatory inactivation of DnaA. J. Mol. Biol. 2009, 385, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Katayama, T. Novel essential residues of Hda for interaction with DnaA in the regulatory inactivation of DnaA: Unique roles for Hda AAA + Box VI and VII motifs. Mol. Microbiol. 2010, 76, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Keyamura, K.; Katayama, T. DnaA Protein DNA-binding Domain Binds to Hda Protein to Promote Inter-AAA+ Domain Interaction Involved in Regulatory Inactivation of DnaA. J. Biol. Chem. 2011, 286, 29336–29346. [Google Scholar] [CrossRef] [PubMed]
- Ryan, V.T.; Grimwade, J.E.; Camara, J.E.; Crooke, E.; Leonard, A.C. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 2004, 51, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Kasho, K.; Fujimitsu, K.; Matoba, T.; Oshima, T.; Katayama, T. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 2014, 42, 13134–13149. [Google Scholar] [CrossRef] [PubMed]
- Kasho, K.; Katayama, T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. USA 2012, 110, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Filutowicz, M.; Ross, W.; Wild, J.; Gourse, R.L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J. Bacteriol. 1992, 174, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Pratt, T.S.; Steiner, T.; Feldman, L.S.; Walker, K.A.; Osuna, R. Deletion analysis of the fis promoter region in Escherichia coli: Antagonistic effects of integration host factor and Fis. J. Bacteriol. 1997, 179, 6367–6377. [Google Scholar] [CrossRef] [PubMed]
- Flåtten, I.; Skarstad, K. The Fis protein has a stimulating role in initiation of replication in Escherichia coli in vivo. PLoS ONE 2013, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cassler, M.R.; Grimwade, J.E.; Leonard, A.C. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995, 14, 5833–5841. [Google Scholar] [PubMed]
- Grimwade, J.E.; Ryan, V.T.; Leonard, A.C. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 2000, 35, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, R.; Mitsuki, H.; Okazaki, T.; Ogawa, T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 1996, 19, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, R.; Ozaki, T.; Moriya, S.; Ogawa, T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998, 12, 3032–3043. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Yamada, Y.; Kuroda, T.; Kishi, T.; Moriya, S. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 2002, 44, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, S.; Yamada, Y.; Ogawa, T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells 2009, 14, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Fujimitsu, K.; Senriuchi, T.; Katayama, T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009, 23, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Frimodt-Møller, J.; Charbon, G.; Krogfelt, K.A.; Løbner-Olesen, A. DNA Replication Control Is Linked to Genomic Positioning of Control Regions in Escherichia coli. PLoS Genet. 2016, 12, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Tanaka, H.; Kasho, K.; Fujimitsu, K.; Oshima, T.; Katayama, T. Chromosomal location of the DnaA-reactivating sequence DARS2 is important to regulate timely initiation of DNA replication in Escherichia coli. Genes Cells 2016, 21, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Slater, S.; Wold, S.; Lu, M.; Boye, E.; Skarstad, K.; Kleckner, N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 1995, 82, 927–936. [Google Scholar] [CrossRef]
- Lu, M.; Campbell, J.L.; Boye, E.; Kleckner, N. SeqA: A negative modulator of replication initiation in E. coli. Cell 1994, 77, 413–426. [Google Scholar] [CrossRef]
- Von Freiesleben, U.; Rasmussen, K.V.; Schaechter, M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 1994, 14, 763–772. [Google Scholar] [PubMed]
- Campbell, J.L.; Kleckner, N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 1990, 62, 967–979. [Google Scholar] [CrossRef]
- Nievera, C.; Torgue, J.J.-C.; Grimwade, J.E.; Leonard, A.C. SeqA Blocking of DnaA-oriC Interactions Ensures Staged Assembly of the E. coli Pre-RC. Mol. Cell 2006, 24, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Kang, S.; Lee, H.; Kim, H.K.; Hwang, D.S. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher order complexes. J. Biol. Chem. 2003, 278, 34983–34989. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, H.; Han, J.S.; Hwang, D.S. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J. Biol. Chem. 1999, 274, 11463–11468. [Google Scholar] [CrossRef] [PubMed]
- Waldminghaus, T.; Skarstad, K. The Escherichia coli SeqA protein. Plasmid 2009, 61, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Helgesen, E.; Fossum-Raunehaug, S.; Saetre, F.; Schink, K.O.; Skarstad, K. Dynamic Escherichia coli SeqA complexes organize the newly replicated DNA at a considerable distance from the replisome. Nucleic Acids Res. 2015, 43, 2730–2743. [Google Scholar] [CrossRef] [PubMed]
- Zawilak-Pawlik, A.; Donczew, R.; Szafrański, S.; Mackiewicz, P.; Terradot, L.; Zakrzewska-Czerwińska, J. DiaA/HobA and DnaA: A pair of proteins co-evolved to cooperate during bacterial orisome assembly. J. Mol. Biol. 2011, 408, 238–2351. [Google Scholar] [CrossRef] [PubMed]
- Keyamura, K.; Fujikawa, N.; Ishida, T.; Ozaki, S.; Su, M.; Fujimitsu, K.; Kagawa, W.; Yokoyama, S.; Kurumizaka, H. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP—DnaA-specific initiation complexes. Genes Dev. 2007, 21, 2083–2099. [Google Scholar] [CrossRef] [PubMed]
- Natrajan, G.; Hall, D.R.; Thompson, A.C.; Gutsche, I.; Terradot, L. Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli. Mol. Microbiol. 2007, 65, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Terradot, L.; Zawilak-Pawlik, A. Structural insight into Helicobacter pylori DNA replication initiation. Gut Microbes 2010, 1, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhou, A.; Li, S.S.; Ni, J.; Tao, J.; Lu, J.; Wan, B.; Li, S.S.; Zhang, J.; Zhao, S.; et al. Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci. Rep. 2016, 6, 30837. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.C.; Grimwade, J.E. Regulating DnaA complex assembly: It is time to fill the gaps. Curr. Opin. Microbiol. 2010, 13, 766–772. [Google Scholar] [CrossRef] [PubMed]

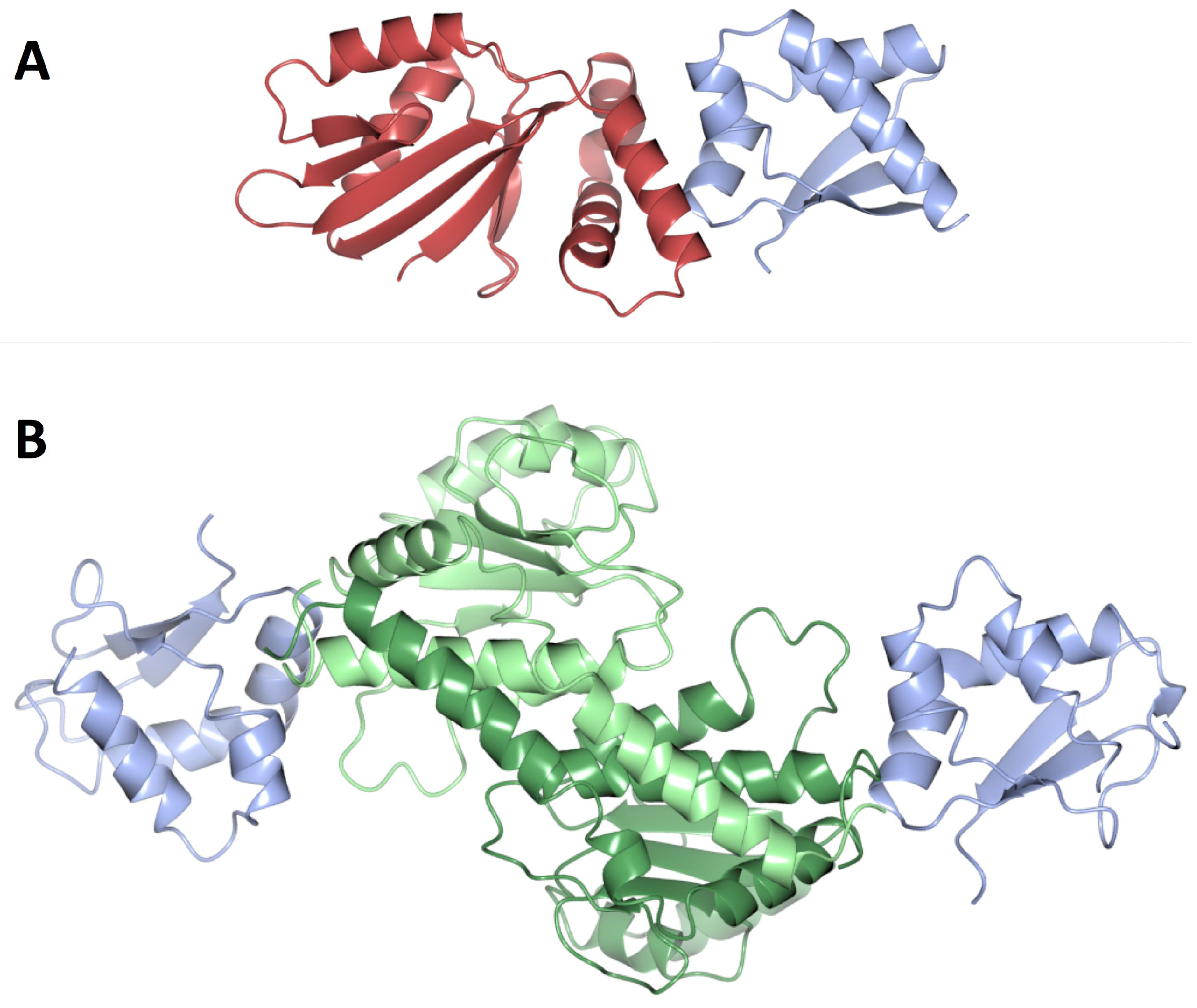
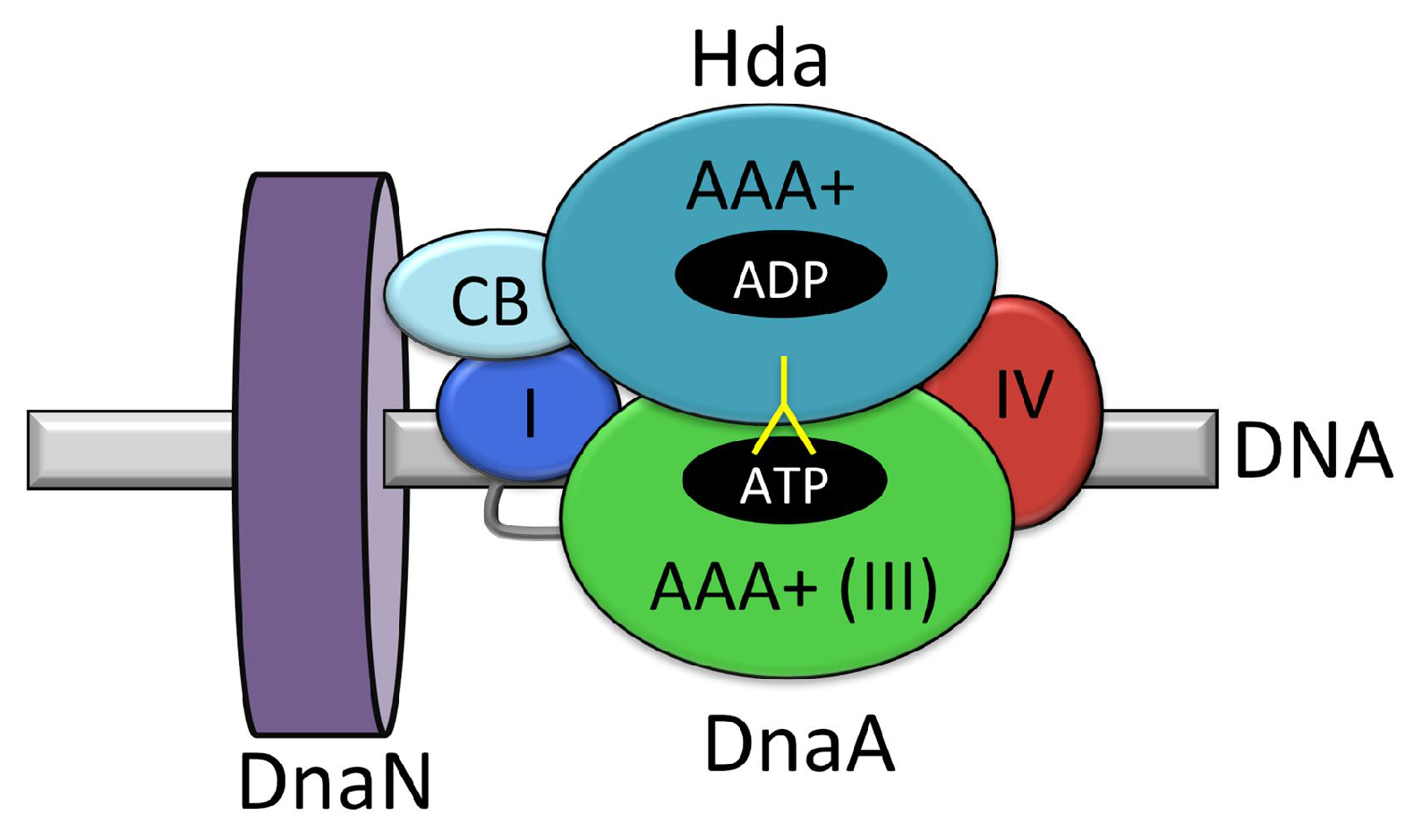
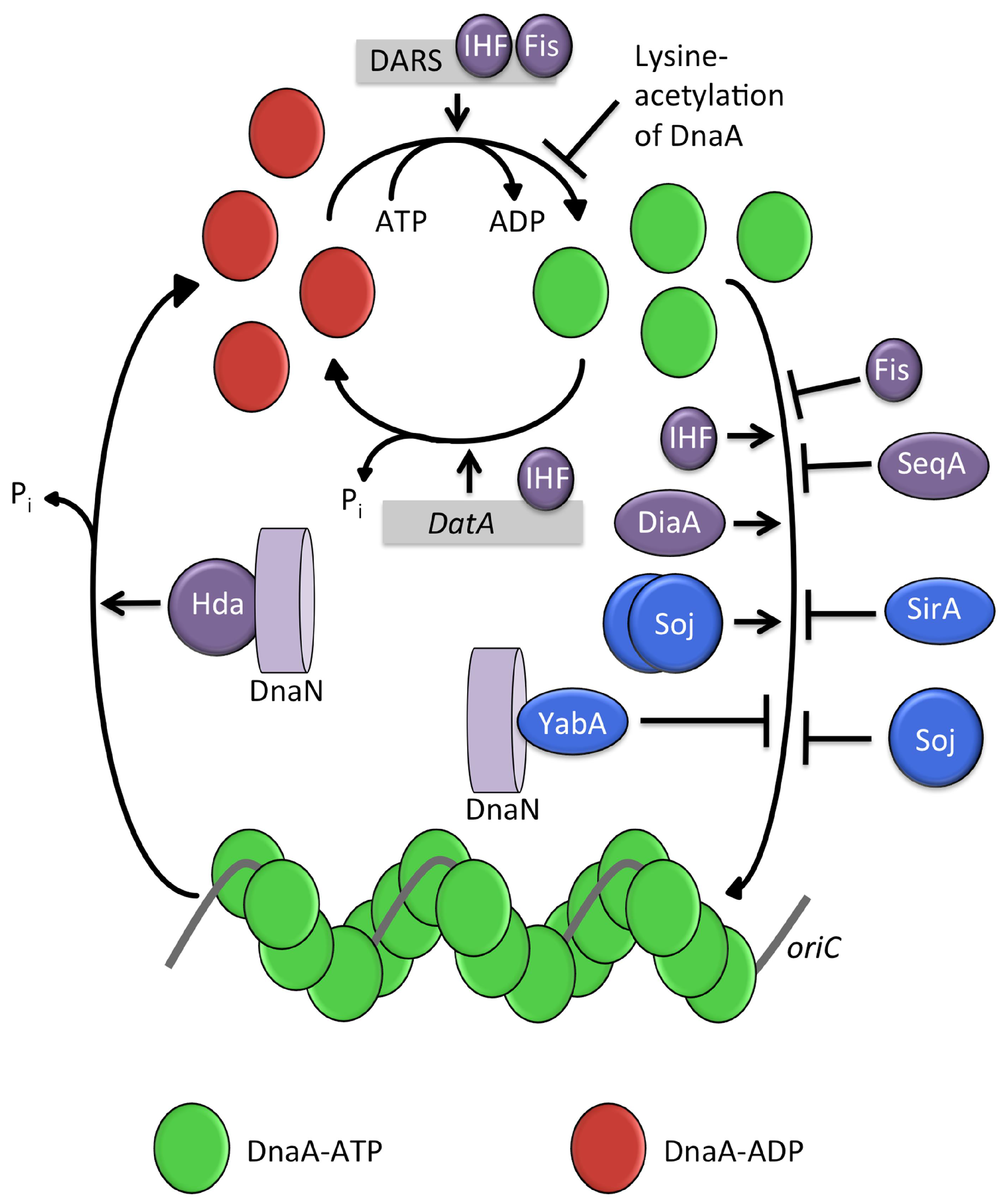
| Role in DNA Replication Initiation | B. subtilis | E. coli |
|---|---|---|
| Initiator | DnaA | DnaA |
| Helicase | DnaC | DnaB |
| DNA Remodelling | DnaB, DnaD | _ |
| Helicase Loader | DnaI | DnaC |
| Primase | DnaG | DnaG |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jameson, K.H.; Wilkinson, A.J. Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes 2017, 8, 22. https://doi.org/10.3390/genes8010022
Jameson KH, Wilkinson AJ. Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes. 2017; 8(1):22. https://doi.org/10.3390/genes8010022
Chicago/Turabian StyleJameson, Katie H., and Anthony J. Wilkinson. 2017. "Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli" Genes 8, no. 1: 22. https://doi.org/10.3390/genes8010022
APA StyleJameson, K. H., & Wilkinson, A. J. (2017). Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes, 8(1), 22. https://doi.org/10.3390/genes8010022






