Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Bacterial Strains and Culture Conditions
2.3. AGO5 Down-Regulation by RNA Interference
2.4. Treatments
2.5. Gene Expression Analysis
2.6. AGO5 Protein Accumulation in Response to Rhizobia
2.7. Root Hair Deformation Analysis
2.8. Nodulation Assay
2.9. Histology of Nodules by Light Microscopy
2.10. Sequence Collection and Phylogenetic Analysis
2.11. Statistical Analyses
3. Results
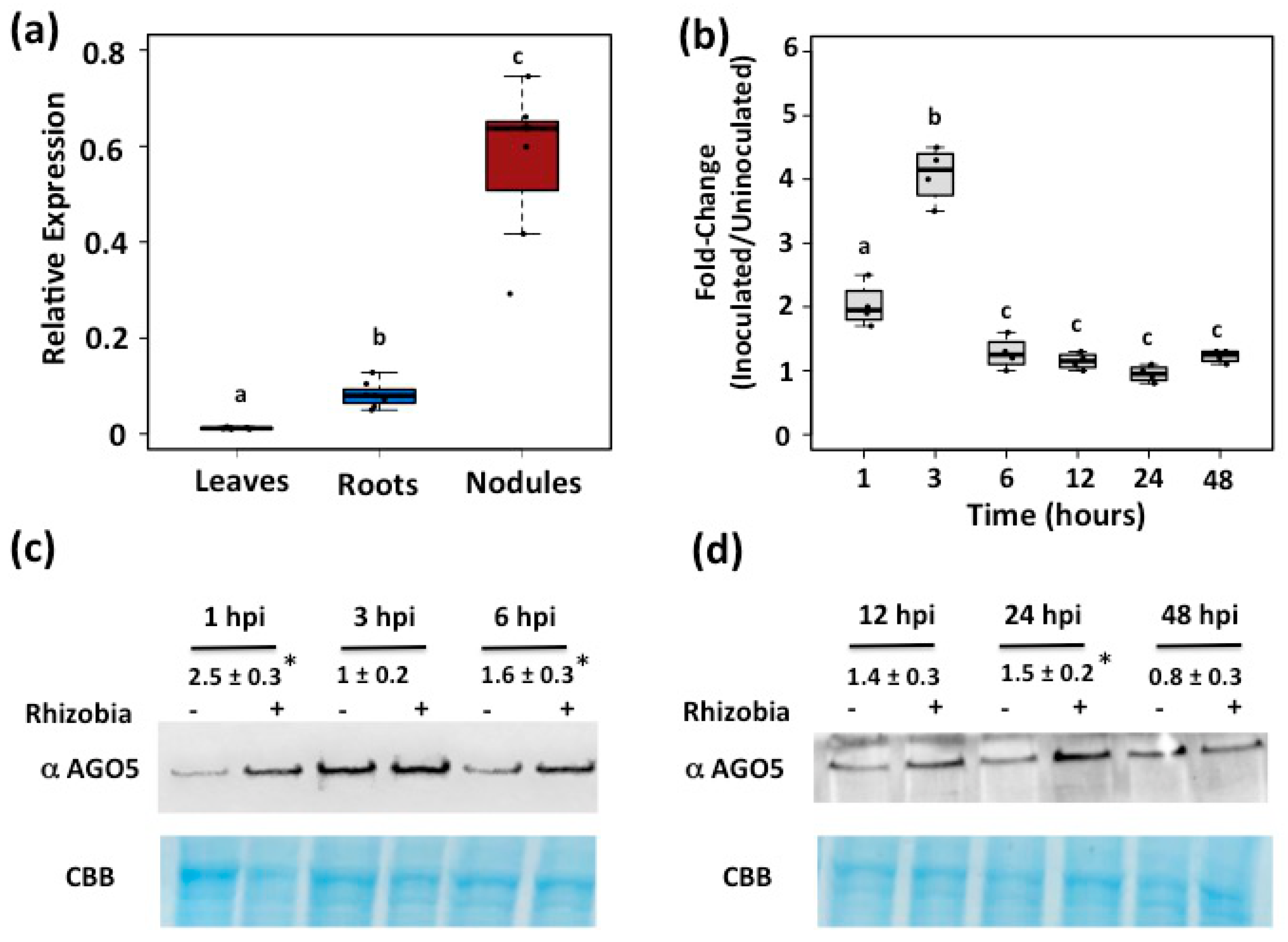
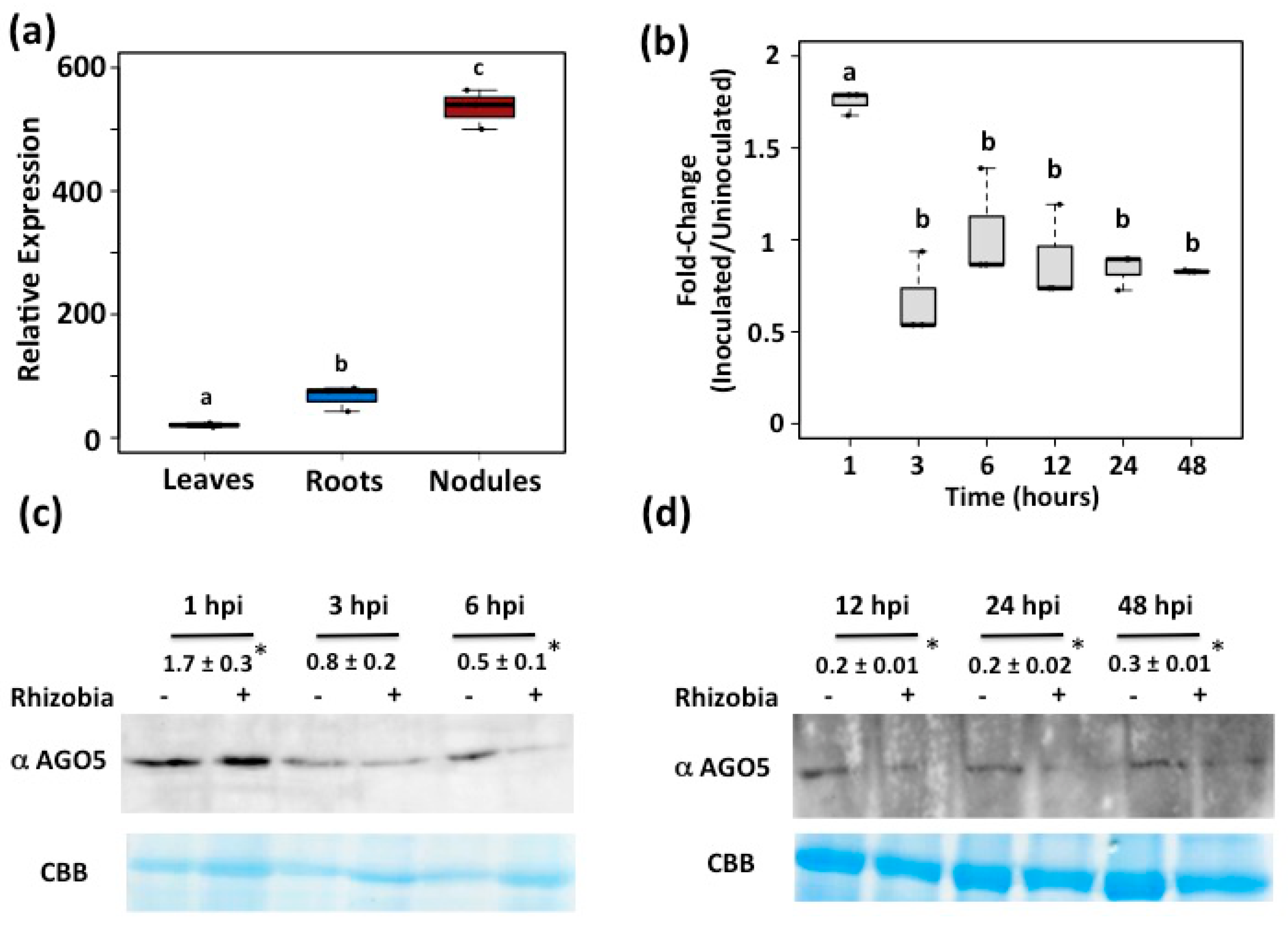
3.1. AGO5 Is Preferentially Expressed in Roots and Nodules of Common Bean Plants
3.2. PvAGO5 Expression Is Induced in Response to Rhizobia
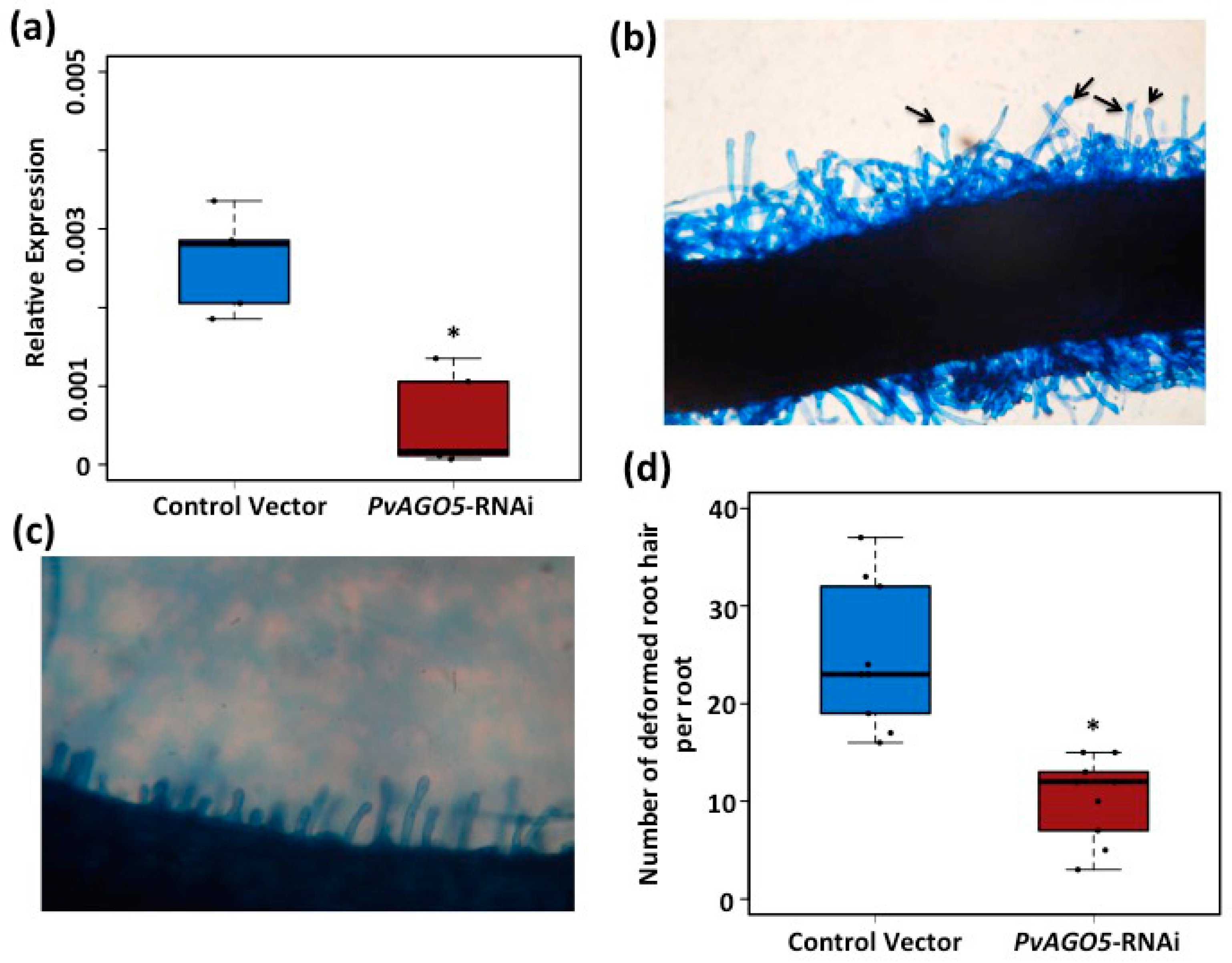
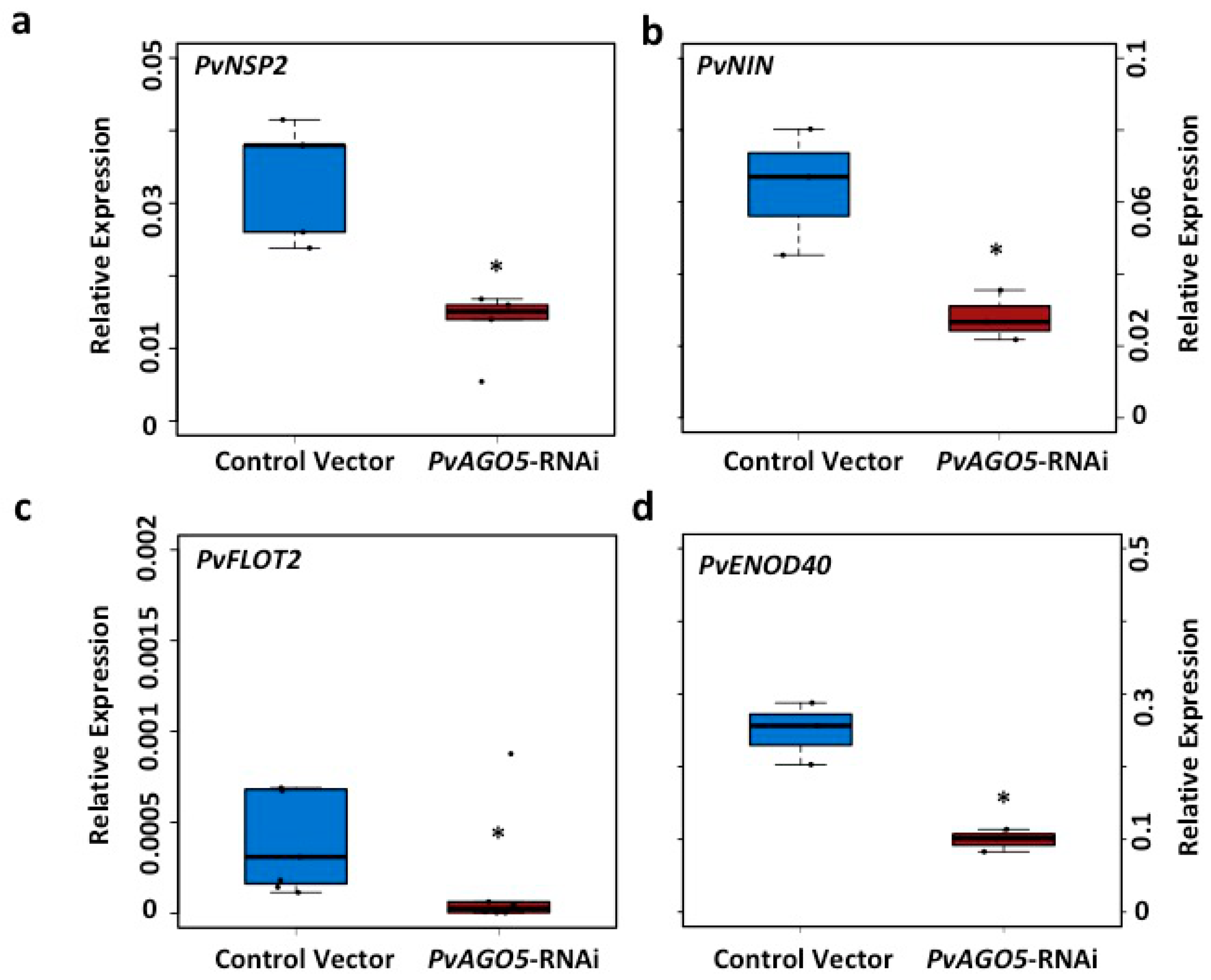
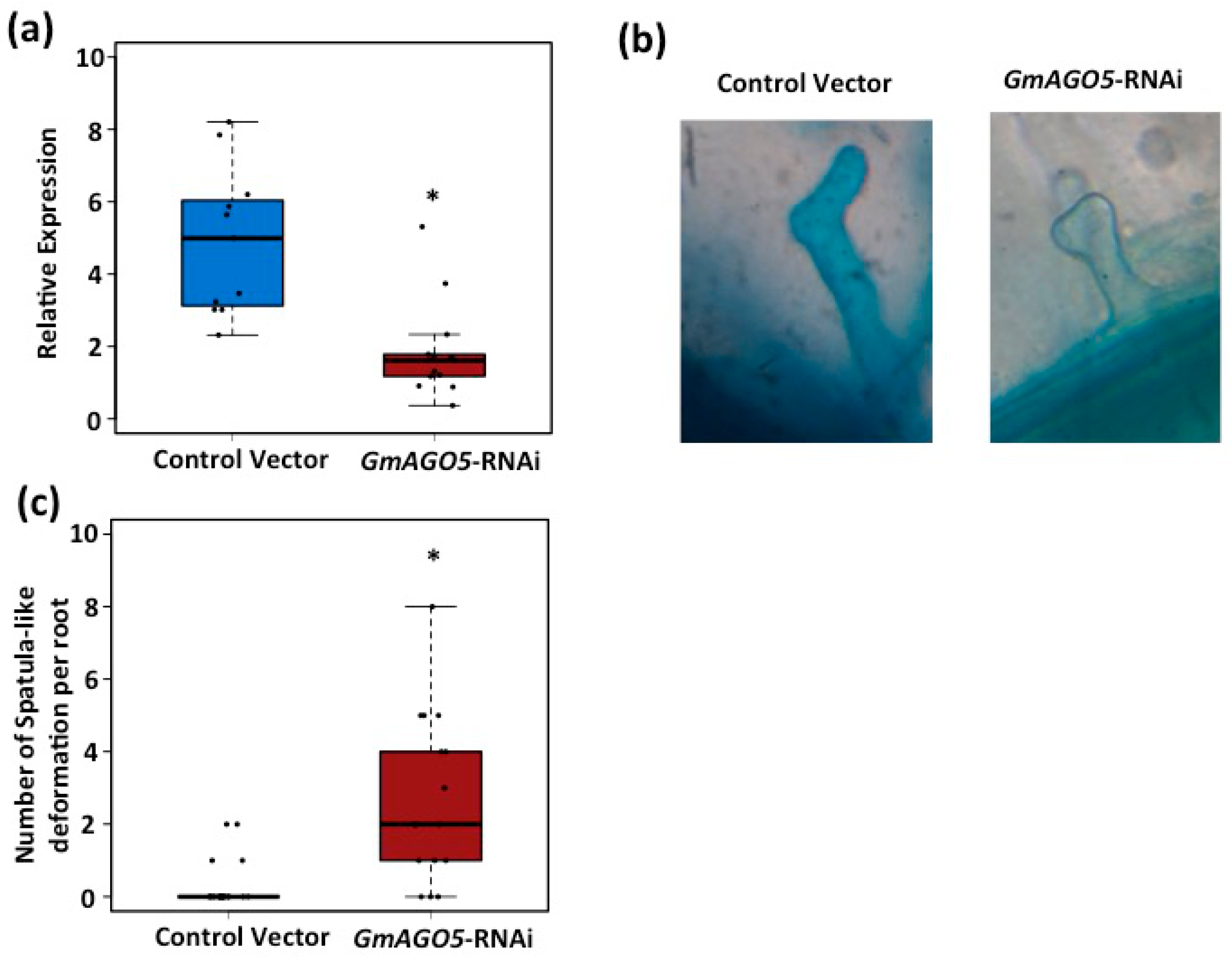
3.3. AGO5 Is Required for Rhizobia-Induced Root Hair Deformation and the Activation of Symbiosis-Specific Genes
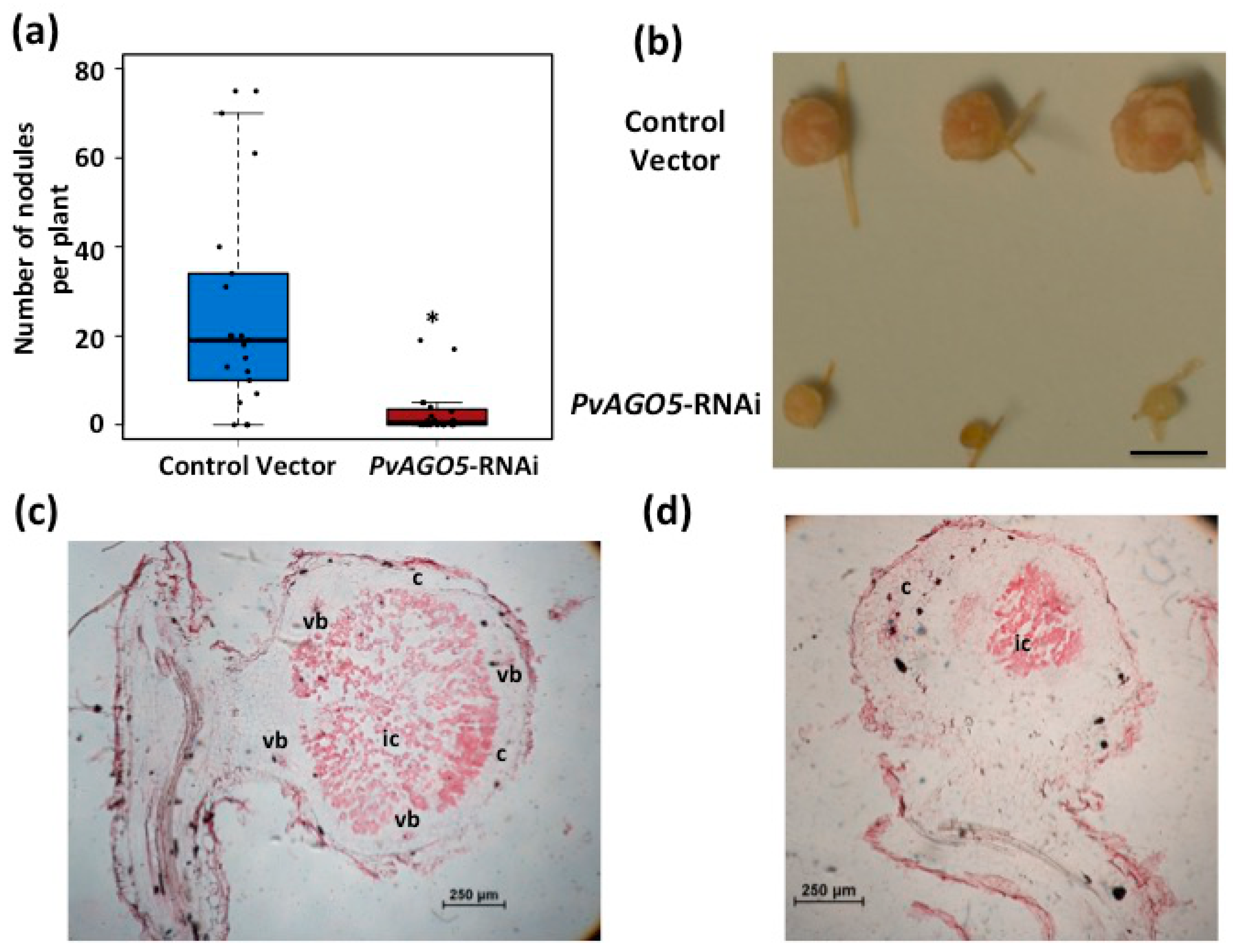
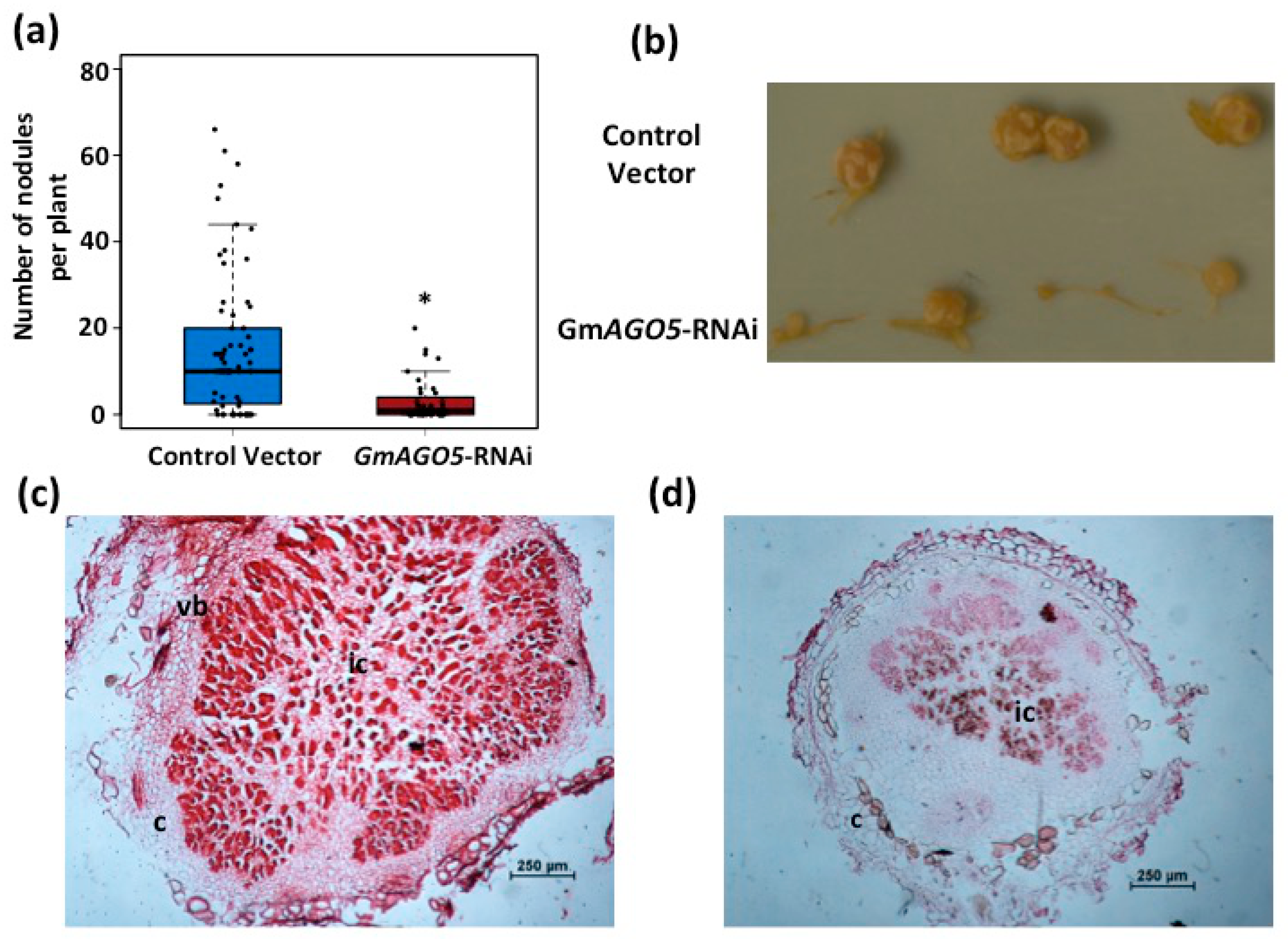
3.4. Down-Regulation of PvAGO5 Affects Nodule Development in Common Bean
3.5. AGO5 Is Also Required in Soybean to Establish Symbiosis with B. japonicum
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gresshoff, P.M.; Hayashi, S.; Biswas, B.; Mirzaei, S.; Indrasumunar, A.; Reid, D.; Samuel, S.; Tollenaere, A.; van Hameren, B.; Hastwell, A.; et al. The value of biodiversity in legume symbiotic nitrogen fixation and nodulation for biofuel and food production. J. Plant Physiol. 2015, 172, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Castro-Guerrero, N.A.; Isidra-Arellano, M.C.; Mendoza-Cozatl, D.M.; Valdés-López, O. Common bean: A legume model on the rise for unraveling adaptations to iron, zinc and phosphate deficiencies. Front. Plant Sci. 2016, 7, 600. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Russo, G. Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 2016, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Venkateshwaran, M.; Volkening, J.D.; Sussman, M.R.; Ané, J.M. Symbiosis and the social network of higher plants. Curr. Opin. Plant Biol. 2013, 16, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D. Speak, friend, and enter: Signaling system that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.D. Invasion by invitation: Rhizobial infection in legumes. Mol. Plant Microbe Interact. 2011, 24, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Nievel, F.; Barker, D. Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.T.; Schilderink, S.; Moling, S.; Deinum, E.E.; Kondorosi, E.; Franssen, H.; Kulikova, O.; Niebel, A.; Bisseling, T. Fate map of Medicago truncatula root nodules. Development 2014, 141, 3517–3528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Parniske, M. Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 2012, 15, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Ané, J.M.; Kiss, G.B.; Riely, B.K.; Penmetsa, R.V.; Oldroyd, G.E.; Ayax, C.; Lévy, J.; Debellé, F.; Baek, J.M.; Kalo, P.; et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Sciences 2004, 303, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, N.; Madsen, L.H.; Radutoiu, S.; Frantescu, M.; Quistgaard, E.M.; Miwa, H.; Downie, J.A.; Jame, E.K.; Felle, H.H.; Haaning, L.L.; et al. A nucleoporin is required for induction of calcium spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Sun, J.; Heckmann, A.B.; Venkateshwaran, M.; Riely, B.K.; Otegui, M.S.; Edwards, A.; Freshour, G.; Hahn, M.G.; Cook, D.R.; et al. The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 2007, 145, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yoshikawa, M.; Yano, K.; Miwa, H.; Uchida, H.; Asamizu, E.; Sato, S.; Tabata, S.; Imaizumi-Anraku, H.; Umehara, Y.; et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 2007, 19, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Takeda, N.; Perry, J.; Uchida, H.; Dräxl, S.; Brachmann, A.; Sato, S.; Tabata, S.; Kawaguchi, M. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhizal fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 2010, 22, 2059–2526. [Google Scholar] [CrossRef] [PubMed]
- Capoen, W.; Sun, J.; Wysham, D.; Otegui, M.S.; Venkateshwaran, M.; Hirsch, S.; Miwa, H.; Downie, J.A.; Morris, R.J.; Ané, J.M.; et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 2011, 108, 14348–14353. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Sun, J.; Vaz Martins, T.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Véry, A.A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Venkateshwaran, M.; Jayaraman, D.; Chabaud, M.; Genre, A.; Ballon, A.J.; Maeda, J.; Forshey, K.; den Os, D.; Kwiecien, N.W.; Coon, J.J.; et al. A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 9781–9786. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Borneman, S.; Morris, R.J.; Oldroyd, G.E. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPs, a DNA-binding transcriptional activator orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Zogli, P.; Libault, M. Plant response to biotic stress: Is there a common epigenetic response during plant-pathogenic and symbiotic interactions? Plant Sci. 2017, 263, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Satgé, C.; Moreau, S.; Sallet, E.; Lefort, G.; Auriac, M.C.; Rembliere, C.; Cottret, L.; Gallardo, K.; Noirot, C.; Jardinaud, M.F.; et al. Reprograming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plant. 2016, 16166. [Google Scholar] [CrossRef] [PubMed]
- Nagymihály, M.; Veluchamy, A.; Györgypál, Z.; Ariel, F.; Jégu, T.; Benchamed, M.; Szücs, A.; Margaert, P.; Kondorosi, E. Ploidy-dependent changes in the epigenome of symbiotic cells correlate with specific patterns of gene expression. Proc. Natl. Acad. Sci. USA 2017, 114, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Ichida, H.; Yoneyama, K.; Koba, T.; Abe, T. Epigenetic modification of rhizobial genome is essential for efficient nodulation. Biochem. Biophys. Res. Commun. 2009, 389, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Qi, Y. RNAi in plants: An Argonaute-centered view. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xia, R.; Meyers, B.C.; Walbot, V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 2015, 27, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.A.; Meyers, B.C.; Sherrier, D.J. MicroRNAs in the rhizobia legume symbiosis. Plant Physiol. 2009, 151, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Pereira, P.A.; Slotkin, R.K.; Martienssen, R.A.; Becker, J.D. MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 2011, 62, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Okada, T.; Hu, Y.; Scholefield, A.; Taylor, J.M.; Koltunow, A.M. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 2012, 139, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Ville-Calzada, J.P. Control of female gamete formation by small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Brechenmacher, L.; Aldrich, J.T.; Clauss, T.R.; Gritsenko, M.A.; Hixson, K.K.; Libault, M.; Tanaka, K.; Yao, F. Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrrhizobium japonicum. Mol. Cell. Proteom. 2012, 11, 1140–1155. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.M.; Venkateshwaran, M.; Volkening, J.D.; Grimsrud, P.A.; Maeda, J.; Bailey, D.J.; Park, K.; Howes-Podoll, M.; den Os, D.; Yeun, L.H.; et al. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteom. 2012, 11, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-Garcia, N.; Yu, H.J.; Hwang, H.J.; Oh, M.; Kim, G.B.; Surrendrarrao, A.K.; Chasman, D.; et al. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Iñiguez, L.P.; Fu, F.; Bucciarelli, B.; Miller, S.S.; Jackson, S.A.; McClean, P.E.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq based gene expression atlas of the common bean. BMC Genom. 2014, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Patil, K.; Fitzpatrick, M.R.; Frnaklin, L.D.; Yao, Q.; Cook, J.R.; Zhen, W.; Libault, M.; Brechenmacher, L.; Valliyodan, B.; et al. Soybean knowledge (SoyKB): A web resource for soybean translational genomics. BMC Genom. 2012, 1, S15. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Fitzpatrick, M.R.; Chen, S.; Liu, Y.; Zhan, H.; Endacott, R.Z.; Gaudiello, E.C.; Stacey, G.; Nguyen, H.T.; Xu, D. Soybean knowledge base: (SoyKB): A web resource for interaction of soybean translational genomics and molecular breeding. Nucleic Acid. Res. 2014, 42, D1245–D1252. [Google Scholar] [CrossRef] [PubMed]
- Catoira, R.; Galera, C.; de Billy, F.; Penmetsa, R.V.; Journet, E.P.; Maillet, F.; Rosenberg, C.; Cood, D.; Gough, C.; Dénarié, J. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 2000, 12, 1647–1665. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, O.; Arenas-Huertero, C.; Ramírez, M.; Girard, L.; Sánchez, F.; Vance, C.P.; Reyes, J.L.; Hernández, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signaling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Navarrete, G.; Alvarado-Affantrager, X.; Olivares, J.E.; Guillén, G.; Díaz-Camino, C.; Campos, F.; Gresshoff, P.M.; Sanchez, F. Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat. Protoc. 2007, 2, 1819–1824. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of four soybean reference genes for gene expression normalization. Plant Genome 2008, 1, 44–54. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assuption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosc. Lett. 2003, 13, 62–66. [Google Scholar] [CrossRef]
- ImageJ. Available online: https://imagej.net (accessed on 10 October 2016).
- InterPro: Protein Sequence Analysis & Classification. Available online: http://www.ebi.ac.uk/interpro/ (accessed on 10 October 2016).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Dénarié, S.R.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod Factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Mondo, S.J.; Topp, S.; Starker, C.G.; Gantt, S.; Harrison, M. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrizal symbiosis. Plant J. 2010, 61, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Zhang, X.C.; Govindarajulu, M.; Qiu, J.; Ong, Y.T.; Brechenmacher, L.; Berg, R.H.; Hurley-Sommer, A.; Taylor, C.G.; Stacey, G. A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J. 2010, 62, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Mbengue, M.; Camut, S.; de Carvalho-Niebel, F.; Deslandes, L.; Froidure, S.; Klaus-Heisen, D.K.; Moreau, S.; Rivas, S.; Timmers, T.; Hervé, C. The Medicago truncatula E3 Ubiquitin ligase PUB1 interacts with LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 2010, 22, 3474–3488. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.D.; Muni, R.R.D.; Torres-Jerez, I.; Tang, Y.; Allen, S.; Andriankaja, M.; Li, G.; Laxmi, A.; Cheng, X.; Wen, J.; et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011, 65, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Liu, C.; Li, N.; Zhang, S. Global identification and expression analysis of stress-responsive genes in the Argonaute family in apple. Mol. Genet. Genom. 2016, 291, 2015–2030. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Moffett, P. Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 2015, 27, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Jones, S.I.; Vodkin, L.O. Mutation in Argonaute5 illuminate epistatic interactions of the K1 and I loci leading to saddle seed color patterns in Glycine max. Plant Cell 2017, 29, 708–725. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.R.; Frances, L.; Kelner, A.; Fournier, J.; Middleton, P.H.; Auriac, M.C.; Mysore, K.S.; Wen, J.; Erard, M.; Barker, D.G.; et al. The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol. 2016, 171, 1037–1054. [Google Scholar] [CrossRef] [PubMed]
- Arrighi, J.F.; Godfroy, O.; de Billy, F.; Saurat, O.; Jauneau, A.; Gough, C. The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. USA 2008, 105, 9817–9822. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.; Schultze, M.; Ratet, P.; Oldroyd, G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Hontelez, J.; Lillo, A.; Guarnerio, C.; van de Peut, D.; Fedorova, E.; Bisseling, T.; Franssen, H. Medicago truncatula ENOD40-1 and ENOD40-2 are both involved in nodule initiation and bacteroid development. J. Exp. Bot. 2007, 58, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Kim, J.; Muñoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E.D. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Haney, C.H.; Long, S.R. Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Long, S.R. Identification and characterization of Nodulation-Signaling Pathway 2, a gene of Medicago truncatula involved in Nod Factor signaling. Plant Physiol. 2003, 131, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K.I. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014, 78, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Nova-Franco, B.; Íñiguez, L.P.; Valdés-López, O.; Alvarado-Affantranger, X.; Leija, A.; Fuentes, S.I.; Ramírez, M.; Paul, S.; Reyes, J.L.; Girard, L.; et al. The Micro-RNA171c-APETALA2–1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol. 2015, 168, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 2015, 168, 101–116. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyero-Saavedra, M.D.R.; Qiao, Z.; Sánchez-Correa, M.D.S.; Díaz-Pineda, M.E.; Reyes, J.L.; Covarrubias, A.A.; Libault, M.; Valdés-López, O. Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis. Genes 2017, 8, 352. https://doi.org/10.3390/genes8120352
Reyero-Saavedra MDR, Qiao Z, Sánchez-Correa MDS, Díaz-Pineda ME, Reyes JL, Covarrubias AA, Libault M, Valdés-López O. Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis. Genes. 2017; 8(12):352. https://doi.org/10.3390/genes8120352
Chicago/Turabian StyleReyero-Saavedra, María Del Rocio, Zhenzhen Qiao, María Del Socorro Sánchez-Correa, M. Enrique Díaz-Pineda, Jose L. Reyes, Alejandra A. Covarrubias, Marc Libault, and Oswaldo Valdés-López. 2017. "Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis" Genes 8, no. 12: 352. https://doi.org/10.3390/genes8120352
APA StyleReyero-Saavedra, M. D. R., Qiao, Z., Sánchez-Correa, M. D. S., Díaz-Pineda, M. E., Reyes, J. L., Covarrubias, A. A., Libault, M., & Valdés-López, O. (2017). Gene Silencing of Argonaute5 Negatively Affects the Establishment of the Legume-Rhizobia Symbiosis. Genes, 8(12), 352. https://doi.org/10.3390/genes8120352




