Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of the CmaGSTs
2.2. Phylogenetic Analysis
2.3. Chromosome Localization and Gene Duplication Analysis
2.4. Microsynteny of the GST Gene Family
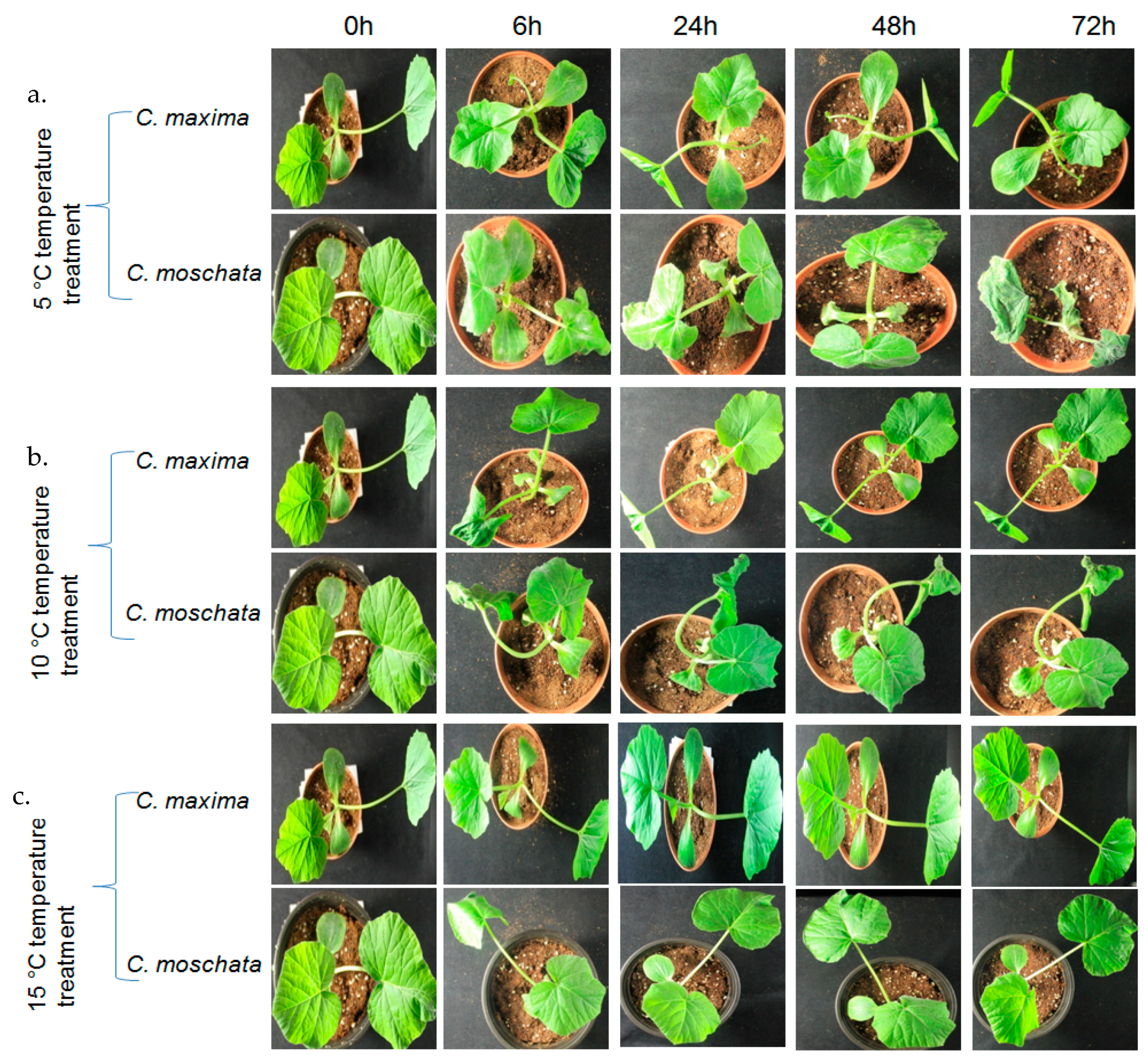
2.5. Plant Materials, Growth, and Treatments
2.6. RNA Extraction and cDNA Synthesis
2.7. Qualitative and Quantitative PCR Expression Analyses
2.8. Statistical Analysis
3. Results
3.1. Identification of GST Genes in C. maxima
3.2. Phylogenetic Analysis
3.3. Chromosomal Locations and Gene Duplications of the CmaGSTs
3.4. Structural and Motif Analyses of the CmaGSTs
3.5. Microsynteny Relationships and Analysis of CmaGST Protein Interactions
3.6. Expression Profiles of CmaGST Genes in Various Organs
3.7. Expression Profiling of the CmaGST Genes during Cold Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Cummins, I.; Cole, D.J.; Edwards, R. Glutathione-mediated detoxification systems in plants. Curr. Opin. Plant Biol. 1998, 1, 258–266. [Google Scholar] [CrossRef]
- Chronopoulou, E.; Kontouri, K.; Chantzikonstantinou, M.; Pouliou, F.; Perperopoulou, F.; Voulgari, G.; Bosmali, E.; Axarli, I.; Nianiou-Obeidat, I.; Madesis, P. Plant glutathione transferases: Structure, antioxidant catalytic function and in planta protective role in biotic and abiotic stress. Curr. Chem. Biol. 2014, 8, 58–75. [Google Scholar] [CrossRef]
- Esmaeili, M.; Shahrtash, M.; Moosavi, F.; Mohsenzadeh, S.; Mohabatkar, H. Plant glutathione S-transferase function. In Proceedings of the 6th National Biotechnology Congress of Iran, Tehran, Iran, 13 August 2009. [Google Scholar]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 1998, 10, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-W.; Liu, M.-J.; Chen, C.; Huang, C.-H.; Chao, L.-Y.; Hsieh, H.-L. A glutathione S-transferase regulated by light and hormones participates in the modulation of arabidopsis seedling development. Plant Physiol. 2010, 154, 1646–1658. [Google Scholar] [CrossRef] [PubMed]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Edwards, R. Glutathione transferases. Arabidopsis Book 2010, e0131. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, I.; Cicero, L.L.; Lo Piero, A.R. The glutathione S-transferase gene superfamily: An in silico approach to study the post translational regulation. Biodegradation 2013, 24, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Kilili, K.G.; Atanassova, N.; Vardanyan, A.; Clatot, N.; Al-Sabarna, K.; Kanellopoulos, P.N.; Makris, A.M.; Kampranis, S.C. Differential roles of tau class glutathione S-transferases in oxidative stress. J. Biol. Chem. 2004, 279, 24540–24551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, H. Expression of Suaeda salsa glutathione S-transferase in transgenic rice resulted in a different level of abiotic stress resistance. J. Agric. Sci. 2006, 144, 547–554. [Google Scholar] [CrossRef]
- Ji, W.; Zhu, Y.; Li, Y.; Yang, L.; Zhao, X.; Cai, H.; Bai, X. Over-expression of a glutathione S-transferase gene, GSGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol. Lett. 2010, 32, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Kampranis, S.C.; Damianova, R.; Atallah, M.; Toby, G.; Kondi, G.; Tsichlis, P.N.; Makris, A.M. A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J. Bio. Chem. 2000, 275, 29207–29216. [Google Scholar] [CrossRef] [PubMed]
- Frova, C. The plant glutathione transferase gene family: Genomic structure, functions, expression and evolution. Physiol. Plant. 2003, 119, 469–479. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Mercurio, V.; Puglisi, I.; Petrone, G. Different roles of functional residues in the hydrophobic binding site of two sweet orange tau glutathione S-transferases. FEBS J. 2010, 277, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Cicero, L.L.; Madesis, P.; Tsaftaris, A.; Lo Piero, A.R. Tobacco plants over-expressing the sweet orange tau glutathione transferases (CsGSTUs) acquire tolerance to the diphenyl ether herbicide fluorodifen and to salt and drought stresses. Phytochemistry 2015, 116, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Hawkins, T.; Hussey, P.J.; Edwards, R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot 2009, 60, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Yang, Z.-L.; Yang, X.; Liu, Y.-J.; Wang, X.-R.; Zeng, Q.-Y. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 2009, 21, 3749–3766. [Google Scholar] [CrossRef] [PubMed]
- Moons, A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam. Horm. 2005, 72, 155–202. [Google Scholar] [PubMed]
- Lallement, P.-A.; Brouwer, B.; Keech, O.; Hecker, A.; Rouhier, N. The still mysterious roles of cysteine-containing glutathione transferases in plants. Front. Pharmacol. 2014, 5, 192. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Cole, D.J.; Edwards, R. Characterisation of a zeta class glutathione transferase from Arabidopsis thaliana with a putative role in tyrosine catabolism. Arch. Biochem. Biophys 2000, 384, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Davis, D.G. Abiotic stress alters transcript profiles and activity of glutathione S-transferase, glutathione peroxidase, and glutathione reductase in Euphorbia esula. Physiol. Plant. 2004, 120, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Edwards, R. Roles for stress-inducible lambda glutathione transferases in flavonoid metabolism in plants as identified by ligand fishing. J. Biol. Chem. 2010, 285, 36322–36329. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Davis, B.G.; Edwards, R. Functional divergence in the glutathione transferase superfamily in plants identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 30859–30869. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K. Cold stress tolerance mechanisms in plants. A review. Agron. Sustain. Dev. 2010, 30, 515–527. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Ann. rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-h.; Lee, H.; Xiong, L.; Zhu, J.-K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Li, Q.-B.; Haskell, D.W.; Guy, C.L. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994, 104, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Thamilarasan, S.K.; Park, J.-I.; Jung, H.-J.; Nou, I.-S. Genome-wide analysis of the distribution of Ap2/Erf transcription factors reveals duplication and cbfs genes elucidate their potential function in Brassica oleracea. BMC Genom. 2014, 15, 422. [Google Scholar] [CrossRef] [PubMed]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation–what is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Kousik, C.S.; Levi, A.; Wehner, T.C.; Maynard, D.N. Cucurbitaceae (vine crops). eLS 2015. [Google Scholar] [CrossRef]
- Lee, J.-M.; Kubota, C.; Tsao, S.; Bie, Z.; Echevarria, P.H.; Morra, L.; Oda, M. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 2010, 127, 93–105. [Google Scholar] [CrossRef]
- Cucurbita maxima (Rimu) Genome. Cucurbit Genomics Database. Available online: http://cucurbitgenomics.org/search/genome/8 (accessed on 20 November 2017).
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASsy server. In The Proteomics Protocols Handbook; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Information Resource (TAIR). Available online: https:// www.arabidopsis.org (accessed on 18 November 2017).
- Rice Genome Annotation Project. Available online: http://rice.plantbiology.msu.edu (accessed on 22 November 2017).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Heredity 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Baum, M.; Grando, S.; Ceccarelli, S.; Bai, G.; Li, R.; Von Korff, M.; Varshney, R.K.; Graner, A.; Valkoun, J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 2009, 60, 3531–3544. [Google Scholar] [CrossRef] [PubMed]
- Cluster 3.0 Open source clustering software. Available online: http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv (accessed on 4 February 2018).
- Vijayakumar, H.; Thamilarasan, S.K.; Shanmugam, A.; Natarajan, S.; Jung, H.-J.; Park, J.-I.; Kim, H.; Chung, M.-Y.; Nou, I.-S. Glutathione transferases superfamily: Cold-inducible expression of distinct GST genes in Brassica oleracea. Int. J. Mol. Sci. 2016, 17, 1211. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.; Bowers, J.; Chapman, B. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef] [PubMed]
- Oakley, A.J. Glutathione transferases: New functions. Curr. Opin. Struct. Biol. 2005, 15, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2010, 71, 338–350. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, B.; Keeler, S.J.; Lau, S.-M.C.; Koeppe, M.K.; O'Keefe, D.P. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000, 124, 1105–1120. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, C.; D’Agostino, N.; Traini, A.; Recupero, G.R.; Frusciante, L.; Chiusano, M.L. Characterization of the glutathione S-transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L.) osbeck. BMC Plant Biol. 2014, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Sappl, P.G.; Oñate-Sánchez, L.; Singh, K.B.; Millar, A.H. Proteomic analysis of glutathione S-transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Mol. Biol. 2004, 54, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Raval, G.; Rawal, R.; Patel, J.; Sainger, R.; Patel, M.; Shah, M.; Patel, D.; Patel, P. Serum glutathione-S-transferase and glutathione reductase activity in head and neck cancer patients. Neoplasma 2002, 49, 260–266. [Google Scholar] [PubMed]
- Dalton, D.A.; Boniface, C.; Turner, Z.; Lindahl, A.; Kim, H.J.; Jelinek, L.; Govindarajulu, M.; Finger, R.E.; Taylor, C.G. Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol. 2009, 150, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.-K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Plant Stress Toler. Methods and Protocols 2010, 39–55. [Google Scholar]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Ann. Rev. Plant Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]






| Sl. No. | Gene Name | Accession No. | Chromosome | Start Location | End Location | Strand | pI | MW(kDa) | AA | Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| 01 | CmaGSTU1 | CmaCh13G011300 | 13 | 8381645 | 8382352 | + | 5.74 | 27.04 | 235 | Cytoplasm |
| 02 | CmaGSTU2 | CmaCh11G002990 | 11 | 1483599 | 1484338 | + | 5.17 | 25.08 | 219 | Cytoplasm |
| 03 | CmaGSTU3 | CmaCh14G019610 | 14 | 13801828 | 13805918 | + | 6.31 | 44.50 | 387 | Cytoplasm |
| 04 | CmaGSTU4 | CmaCh06G011550 | 06 | 7777316 | 7778827 | − | 5.93 | 26.16 | 227 | Cytoplasm |
| 05 | CmaGSTU5 | CmaCh00G006890 | 00 | 47643604 | 47645087 | − | 6.98 | 27.96 | 242 | Cytoplasm |
| 06 | CmaGSTU6 | CmaCh06G012770 | 06 | 8499592 | 8500393 | − | 6.02 | 26.29 | 226 | Cytoplasm |
| 07 | CmaGSTU7 | CmaCh14G018620 | 14 | 13214607 | 13217686 | − | 5.98 | 41.95 | 370 | Cytoplasm |
| 08 | CmaGSTU8 | CmaCh06G012780 | 06 | 8501349 | 8502406 | + | 6.62 | 25.61 | 222 | Cytoplasm |
| 09 | CmaGSTU9 | CmaCh14G018610 | 14 | 13213235 | 13214322 | − | 5.28 | 25.41 | 222 | Cytoplasm |
| 10 | CmaGSTU10 | CmaCh03G003840 | 03 | 4366133 | 4366923 | + | 5.94 | 24.72 | 217 | Cytoplasm |
| 11 | CmaGSTU11 | CmaCh07G010310 | 07 | 5378533 | 5380070 | − | 6.20 | 25.66 | 220 | Cytoplasm |
| 12 | CmaGSTU12 | CmaCh16G004650 | 16 | 2333498 | 2334374 | + | 6.14 | 24.95 | 219 | Cytoplasm |
| 13 | CmaGSTU13 | CmaCh04G006390 | 04 | 3269916 | 3271375 | − | 8.63 | 33.08 | 288 | Cytoplasm |
| 14 | CmaGSTU14 | CmaCh16G004640 | 16 | 2326120 | 2328293 | + | 7.66 | 23.19 | 202 | Cytoplasm |
| 15 | CmaGSTU15 | CmaCh04G006400 | 04 | 3271577 | 3272321 | − | 5.27 | 24.41 | 212 | Cytoplasm |
| 16 | CmaGSTU16 | CmaCh04G006410 | 04 | 3273122 | 3286431 | − | 9.02 | 91.29 | 810 | Nucleus |
| 17 | CmaGSTU17 | CmaCh04G022700 | 04 | 15857294 | 15858045 | − | 5.58 | 25.47 | 224 | Cytoplasm |
| 18 | CmaGSTU18 | CmaCh04G006380 | 04 | 3268691 | 3269778 | − | 5.65 | 25.65 | 224 | Cytoplasm |
| 19 | CmaGSTF1 | CmaCh08G004090 | 08 | 2344840 | 2346569 | + | 6.38 | 24.14 | 217 | Cytoplasm |
| 20 | CmaGSTF2 | CmaCh08G004100 | 08 | 2346863 | 2348162 | − | 5.75 | 24.12 | 214 | Cytoplasm |
| 21 | CmaGSTF3 | CmaCh12G003850 | 12 | 1988809 | 1990016 | − | 5.51 | 25.09 | 220 | Cytoplasm |
| 22 | CmaEF1G1 | CmaCh16G007670. | 16 | 4206363 | 4213994 | + | 5.54 | 61.96 | 547 | Cytoplasm |
| 23 | CmaEF1G2 | CmaCh04G008920 | 04 | 4645271 | 4652681 | + | 5.65 | 60.31 | 531 | Cytoplasm |
| 24 | CmaEF1G3 | CmaCh06G008870 | 06 | 5603706 | 5606718 | − | 6.01 | 47.14 | 415 | Cytoplasm |
| 25 | CmaGSTT1 | CmaCh16G005900 | 16 | 3054792 | 3058033 | − | 9.38 | 27.09 | 238 | Cytoplasm |
| 26 | CmaGSTT2 | CmaCh04G007350 | 00 | 47643604 | 47645087 | + | 9.34 | 26.96 | 238 | Cytoplasm |
| 27 | CmaGSTZ1 | CmaCh02G006430 | 02 | 3895323 | 3899120 | + | 5.85 | 24.89 | 222 | Cytoplasm |
| 28 | CmaGSTZ2 | CmaCh02G006410 | 02 | 3887013 | 3892301 | + | 5.17 | 24.99 | 223 | Cytoplasm |
| 29 | CmaGSTZ3 | CmaCh06G011350 | 06 | 7652425 | 7656514 | + | 5.20 | 25.63 | 228 | Cytoplasm |
| 30 | CmaGSTL1 | CmaCh06G003420 | 06 | 1588222 | 1591488 | − | 5.25 | 29.35 | 257 | Cytoplasm |
| 31 | CmaGHR1 | CmaCh18G005590 | 18 | 3671672 | 3674403 | − | 7.62 | 40.83 | 355 | Chloroplast |
| 32 | CmaGHR2 | CmaCh19G001140 | 19 | 681203 | 684746 | + | 8.79 | 45.58 | 415 | Cytoplasm |
| Duplicated Gene Pairs | Ks | Ka | Ka/Ks | Duplication Type | Type of Selection | Divergence Time (Mya) | ||
|---|---|---|---|---|---|---|---|---|
| CmaGSTU3 | vs. | CmaGSTU4 | 0.215 | 0.127 | 0.593 | Segmental | Purifying | 7.18 |
| CmaGSTU4 | vs. | CmaGSTU5 | 0.139 | 0.056 | 0.405 | Segmental | Purifying | 4.66 |
| CmaGSTU12 | vs. | CmaGSTU14 | 0.038 | 0.017 | 0.452 | Tandem | Purifying | 1.30 |
| CmaGSTU12 | vs. | CmaGSTU13 | 0.412 | 0.142 | 0.346 | Segmental | Purifying | 13.75 |
| CmaGSTU12 | vs. | CmaGSTU15 | 0.455 | 0.137 | 0.302 | Segmental | Purifying | 15.19 |
| CmaGSTU12 | vs. | CmaGSTU16 | 0.533 | 0.229 | 0.430 | Segmental | Purifying | 17.78 |
| CmaGSTU13 | vs. | CmaGSTU14 | 0.387 | 0.142 | 0.367 | Segmental | Purifying | 12.91 |
| CmaGSTU13 | vs. | CmaGSTU15 | 0.405 | 0.172 | 0.424 | Tandem | Purifying | 13.52 |
| CmaGSTU13 | vs. | CmaGSTU16 | 0.309 | 0.242 | 0.784 | Tandem | Purifying | 10.32 |
| CmaGSTU14 | vs. | CmaGSTU15 | 0.398 | 0.151 | 0.379 | Segmental | Purifying | 13.27 |
| CmaGSTU15 | vs. | CmaGSTU16 | 0.308 | 0.226 | 0.735 | Tandem | Purifying | 10.28 |
| CmaGSTZ1 | vs. | CmaGSTZ2 | 0.382 | 0.220 | 0.575 | Tandem | Purifying | 12.76 |
| CmaEF1G1 | vs. | CmaEF1G2 | 0.080 | 0.119 | 1.483 | Segmental | Positive | 2.68 |
| CmaEF1G1 | vs. | CmaEF1G3 | 0.536 | 0.293 | 0.547 | Segmental | Purifying | 17.89 |
| CmaEF1G2 | vs. | CmaEF1G3 | 0.566 | 0.279 | 0.493 | Segmental | Purifying | 18.87 |
| CmaGSTT1 | vs. | CmaGSTT2 | 0.201 | 0.130 | 0.646 | Segmental | Purifying | 6.72 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Kayum, M.; Nath, U.K.; Park, J.-I.; Biswas, M.K.; Choi, E.K.; Song, J.-Y.; Kim, H.-T.; Nou, I.-S. Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance. Genes 2018, 9, 84. https://doi.org/10.3390/genes9020084
Abdul Kayum M, Nath UK, Park J-I, Biswas MK, Choi EK, Song J-Y, Kim H-T, Nou I-S. Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance. Genes. 2018; 9(2):84. https://doi.org/10.3390/genes9020084
Chicago/Turabian StyleAbdul Kayum, Md., Ujjal Kumar Nath, Jong-In Park, Manosh Kumar Biswas, Eung Kyoo Choi, Jae-Young Song, Hoy-Taek Kim, and Ill-Sup Nou. 2018. "Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance" Genes 9, no. 2: 84. https://doi.org/10.3390/genes9020084
APA StyleAbdul Kayum, M., Nath, U. K., Park, J.-I., Biswas, M. K., Choi, E. K., Song, J.-Y., Kim, H.-T., & Nou, I.-S. (2018). Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance. Genes, 9(2), 84. https://doi.org/10.3390/genes9020084






