Genome-Wide Quantification of the Effect of Gene Overexpression on Escherichia coli Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Plasmid Construction
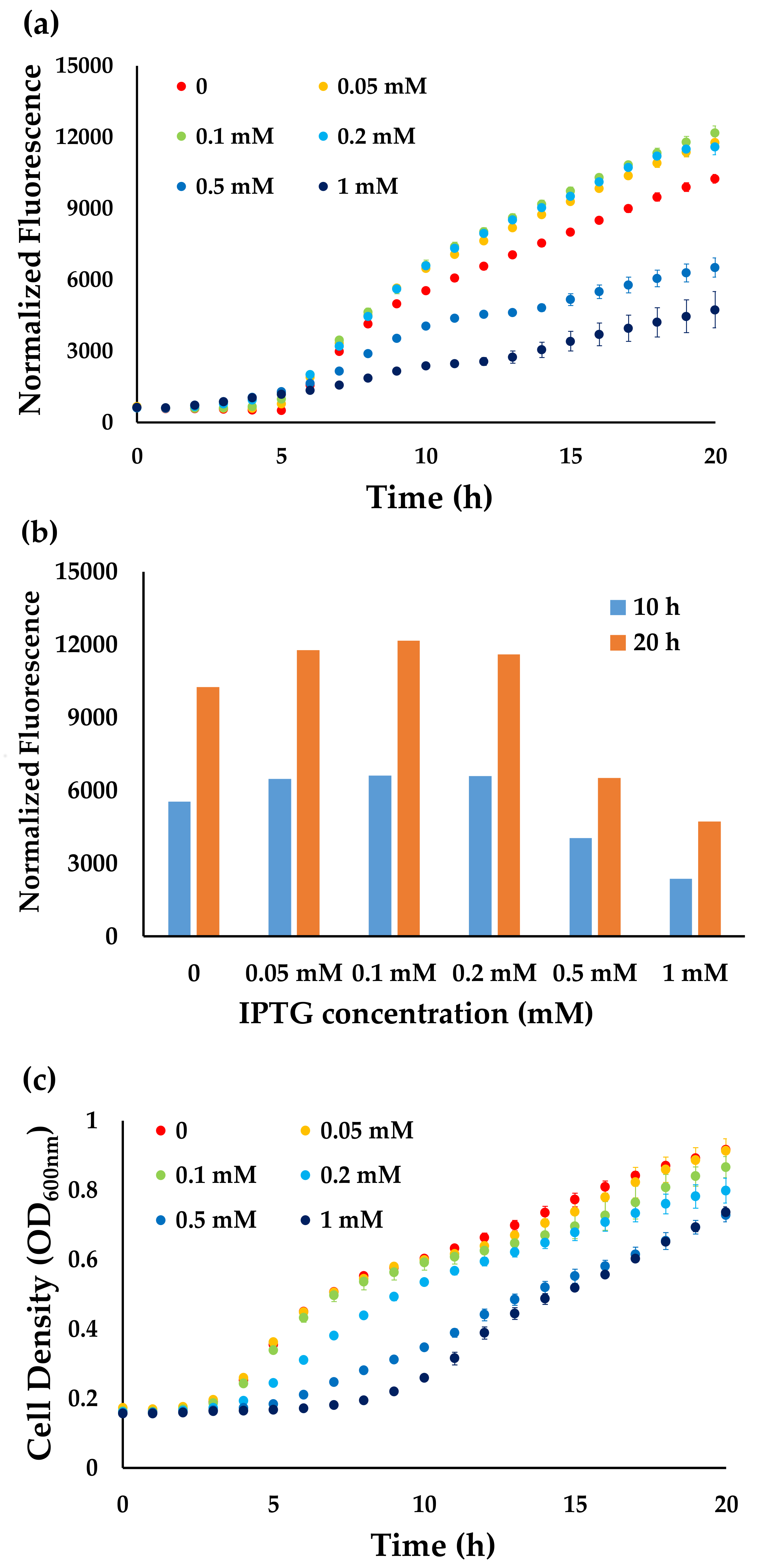
2.2. Cell Growth Experiments
2.3. Bioinformatical Analyses
3. Results and Discussion
3.1. Growth Condition Selection
3.2. High-Throughput Growth Tests of the ASKA Collection
3.3. Bioinformatical Analyses of Factors Affecting Cell Growth
3.3.1. The Effect of Protein Length on Cell Growth
3.3.2. The Effect of Amino Acid Compositions on Cell Growth
3.3.3. The Effect of Codon Bias on Cell Growth
3.3.4. Gene Ontology Analyses
3.3.5. Protein Functional Interaction Network Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Bentley, W.E.; Kompala, D.S. Optimal induction of protein synthesis in recombinant bacterial cultures. Ann. N. Y. Acad. Sci. 1990, 589, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Rinas, U. Stress induced by recombinant protein production in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 2004, 89, 73–92. [Google Scholar] [PubMed]
- Dong, H.; Nilsson, L.; Kurland, C.G. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 1995, 177, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Dumon-Seignovert, L.; Cariot, G.; Vuillard, L. The toxicity of recombinant proteins in Escherichia coli: A comparison of overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr. Purif. 2004, 37, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Trepod, C.M.; Mott, J.E. A spontaneous runaway vector for production-scale expression of bovine somatotropin from Escherichia coli. Appl. Microbiol. Biotechnol. 2002, 58, 84–88. [Google Scholar] [PubMed]
- Chang, D.E.; Smalley, D.J.; Conway, T. Gene expression profiling of Escherichia coli growth transitions: An expanded stringent response model. Mol. Microbiol. 2002, 45, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Venkat, S.; Nannapaneni, D.T.; Gregory, C.; Gan, Q.; McIntosh, M.; Fan, C. Genetically encoding thioacetyl-lysine as a non-deacetylatable analog of lysine acetylation in Escherichia coli. FEBS Open Biol. 2017, 7, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Venkat, S.; Sturges, J.; Stahman, A.; Gregory, C.; Gan, Q.; Fan, C. Genetically incorporating two distinct post-translational modifications into one protein simultaneously. ACS Synth. Biol. 2018, 7, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Tsuchisaka, A.; Yukawa, H. Branched-chain amino acids. Adv. Biochem. Eng. Biotechnol. 2017, 159, 103–128. [Google Scholar] [PubMed]
- Massey, L.K.; Sokatch, J.R.; Conrad, R.S. Branched-chain amino acid catabolism in bacteria. Bacteriol. Rev. 1976, 40, 42–54. [Google Scholar] [PubMed]
- Kurland, C.; Gallant, J. Errors of heterologous protein expression. Curr. Opin. Biotechnol. 1996, 7, 489–493. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Kane, J.F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 1995, 6, 494–500. [Google Scholar] [CrossRef]
- Li, G.W.; Oh, E.; Weissman, J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 2012, 484, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984, 3, 2895–2898. [Google Scholar] [PubMed]
- Sorensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Jornvall, H.; Berndt, K.D.; Oppermann, U. Codon optimization reveals critical factors for high level expression of two rare codon genes in Escherichia coli: RNA stability and secondary structure but not tRNA abundance. Biochem. Biophys. Res. Commun. 2004, 313, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb. Cell Fact. 2009, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Kazana, E.; Bellanger, N.; Singh, T.; Tuite, M.F.; von der Haar, T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014, 33, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Oresic, M.; Shalloway, D. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol. 1998, 281, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Sauna, Z.E.; Kimchi-Sarfaty, C.; Ambudkar, S.V.; Gottesman, M.M.; Nussinov, R. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 2008, 383, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Huntley, R.P.; Sawford, T.; Mutowo-Meullenet, P.; Shypitsyna, A.; Bonilla, C.; Martin, M.J.; O’Donovan, C. The GOA database: Gene ontology annotation updates for 2015. Nucleic Acids Res. 2015, 43, D1057–D1063. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; McCandlish, A.C.; Gronenberg, L.S.; Chng, S.S.; Silhavy, T.J.; Kahne, D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 11754–11759. [Google Scholar] [CrossRef] [PubMed]
- Daley, D.O.; Rapp, M.; Granseth, E.; Melen, K.; Drew, D.; von Heijne, G. Global topology analysis of the Escherichia coli inner membrane proteome. Science 2005, 308, 1321–1323. [Google Scholar] [CrossRef] [PubMed]
- Narita, S.; Tokuda, H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009, 583, 2160–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abouhamad, W.N.; Manson, M.D. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 1994, 14, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Fixen, K.R.; Janakiraman, A.; Garrity, S.; Slade, D.J.; Gray, A.N.; Karahan, N.; Hochschild, A.; Goldberg, M.B. Genetic reporter system for positioning of proteins at the bacterial pole. MBio 2012, 3, e00251-11. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Gagnon, M.; Sans, D.; Michnick, S.; Brakier-Gingras, L. Mapping of the RNA recognition site of Escherichia coli ribosomal protein S7. RNA 2000, 6, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, P.; Woodson, S.A. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20. J. Mol. Biol. 2009, 392, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Metspalu, E.; Maimets, T.; Ustav, M.; Villems, R. A quaternary complex consisting of two molecules of tRNA and ribosomal proteins L2 and L17. FEBS Lett. 1981, 132, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Robertson, H.D.; Dunn, J.J. Ribonucleic acid processing activity of Escherichia coli ribonuclease III. J. Biol. Chem. 1975, 250, 3050–3056. [Google Scholar] [PubMed]
- Ling, J.; Roy, H.; Ibba, M. Mechanism of tRNA-dependent editing in translational quality control. Proc. Natl. Acad. Sci. USA 2007, 104, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Venkat, S.; Gregory, C.; Gan, Q.; Fan, C. Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli. Chembiochem 2017, 18, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.; Rak, B.; Benz, R. The gene bglH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of β-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol. Microbiol. 1999, 31, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Yamamoto, K.; Ishihama, A. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 2009, 191, 4562–4571. [Google Scholar] [CrossRef] [PubMed]
- Skretas, G.; Georgiou, G. Simple genetic selection protocol for isolation of overexpressed genes that enhance accumulation of membrane-integrated human G protein-coupled receptors in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 5852–5859. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.J.; Price, N.D. Genetic co-occurrence network across sequenced microbes. PLoS Comput. Biol. 2011, 7, e1002340. [Google Scholar] [CrossRef] [PubMed]





| Growth Effects 1 | Delay Factors 2 | Numbers of Strains | Percentage |
|---|---|---|---|
| No or moderate | <2 | 921 | 22.6% |
| Significant | 2 to 7 | 3049 | 74.9% |
| Severe | >7 | 101 | 2.5% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Venkat, S.; Wilson, J.; McGuire, P.; Chang, A.L.; Gan, Q.; Fan, C. Genome-Wide Quantification of the Effect of Gene Overexpression on Escherichia coli Growth. Genes 2018, 9, 414. https://doi.org/10.3390/genes9080414
Chen H, Venkat S, Wilson J, McGuire P, Chang AL, Gan Q, Fan C. Genome-Wide Quantification of the Effect of Gene Overexpression on Escherichia coli Growth. Genes. 2018; 9(8):414. https://doi.org/10.3390/genes9080414
Chicago/Turabian StyleChen, Hao, Sumana Venkat, Jessica Wilson, Paige McGuire, Abigail L. Chang, Qinglei Gan, and Chenguang Fan. 2018. "Genome-Wide Quantification of the Effect of Gene Overexpression on Escherichia coli Growth" Genes 9, no. 8: 414. https://doi.org/10.3390/genes9080414
APA StyleChen, H., Venkat, S., Wilson, J., McGuire, P., Chang, A. L., Gan, Q., & Fan, C. (2018). Genome-Wide Quantification of the Effect of Gene Overexpression on Escherichia coli Growth. Genes, 9(8), 414. https://doi.org/10.3390/genes9080414







