Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers
Abstract
:1. Introduction
1.1. PBAP Detection Methods
1.2. UV-LIF Classification Methods
1.3. Aims and Objectives
- To assess the efficiency and effectiveness of gradient boosting ensemble decision trees to accurately classify UV-LIF data into broad PBAP classes.
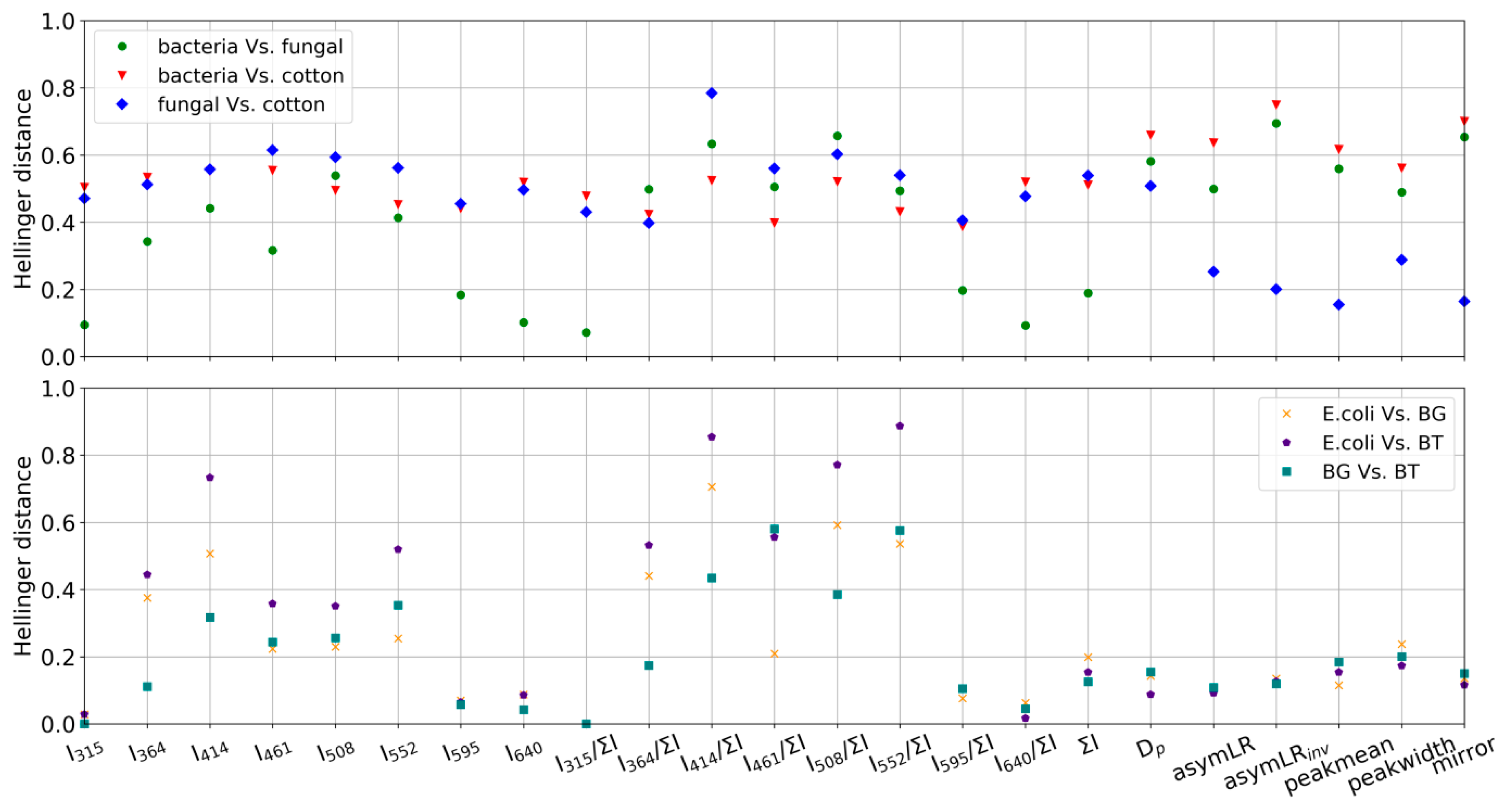
- To develop a framework for the UV-LIF machine learning community to assess how training data may be conflated independently of the choice of classification model and to also appraise the applicability of a training dataset to generate a classification model to represent a given ambient dataset. This is achieved using the Hellinger distance metric to quantify the similarity of parameter probability distributions between training data and model outputs for each class.
- To demonstrate real-world use of the above to quantify airborne concentrations of broad PBAP classes in a busy, multi-functional indoor environment.
2. Methods
2.1. The Multiparameter Bioaerosol Spectrometer
- Peakwidth: An estimate of the mean width of the array peak, defined as the mid-point between the mean and peak values.
- Peakmean: The ratio of the peak to mean parameters. This is a simple method of differentiating various particle morphologies, especially those of an elongated nature such as fibres or rod-shaped from round or irregular particles.
- Mirror: A measure of the scattering symmetry between the top and bottom half of each array, where the two halves are subtracted in an element by element fashion from the centre of the array and the resultant modulus is summed. Spherical particles produce values approaching zero and non-spherical particles yield larger values.
- AsymLR: Variant of mirror. A measure of the symmetry between the left and right arrays.
- AsymLRinv: As AsymLR but the right hand array is inverted.
2.2. Data Preparation
2.3. Gradient Boosting Ensemble Decision Trees
2.4. Laboratory Experimental Arrangement and Ambient Monitoring Site
2.4.1. Aerosol Challenge Simulator
2.4.2. University Place Indoor Ambient Sampling
3. Results
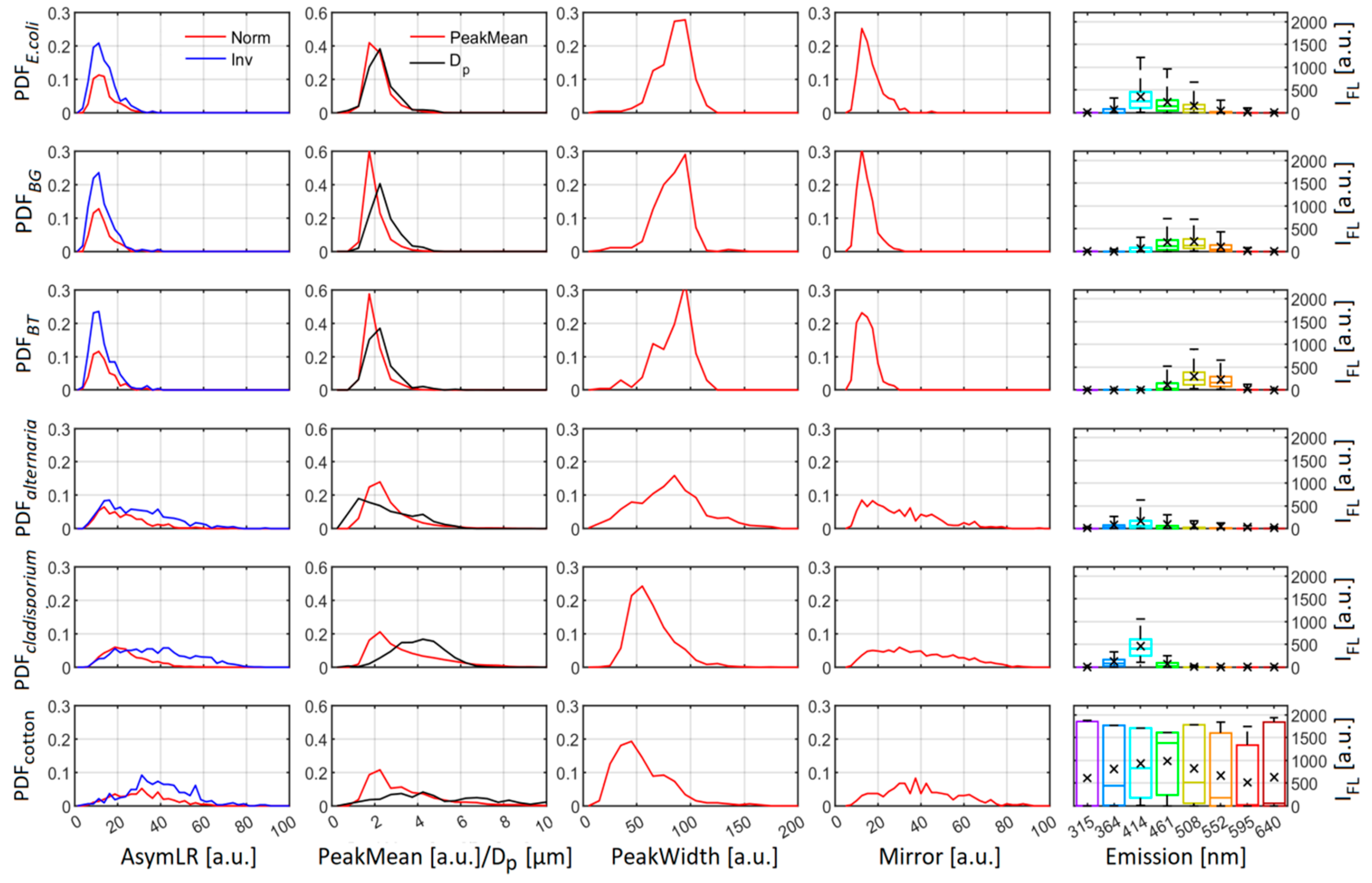
3.1. ACS Laboratory Data
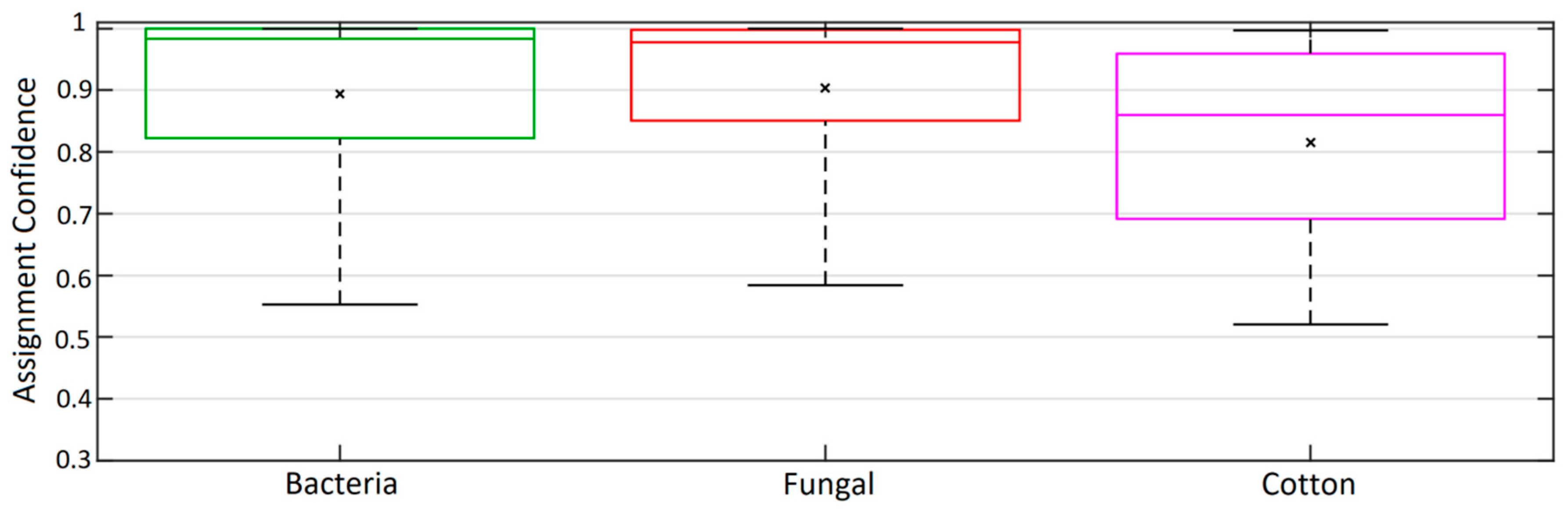
3.2. GBA Classification
- The training data are not representative of ambient fungal spore fluorescence due to how they are produced and aerosolized. As noted earlier, the fungal material used in this study is intended for allergenic testing use and has undergone chemical processing by the manufacturer. This may impact their fluorescent and morphological characteristics.
- That ambient fungal fluorescence is significantly altered by external factors.
- That we observed a fluorescent particle type with similar morphological properties to the ACS fungal material particles which are not fully representative of building mycology resulting in conflation/misattribution. The training dataset used in this study does not contain all of the most commonly observed fungal particles in building mycology studies (e.g., Aspergillius and Penicillium species [15]) which may exhibit different autofluorescent characteristics to the training samples.
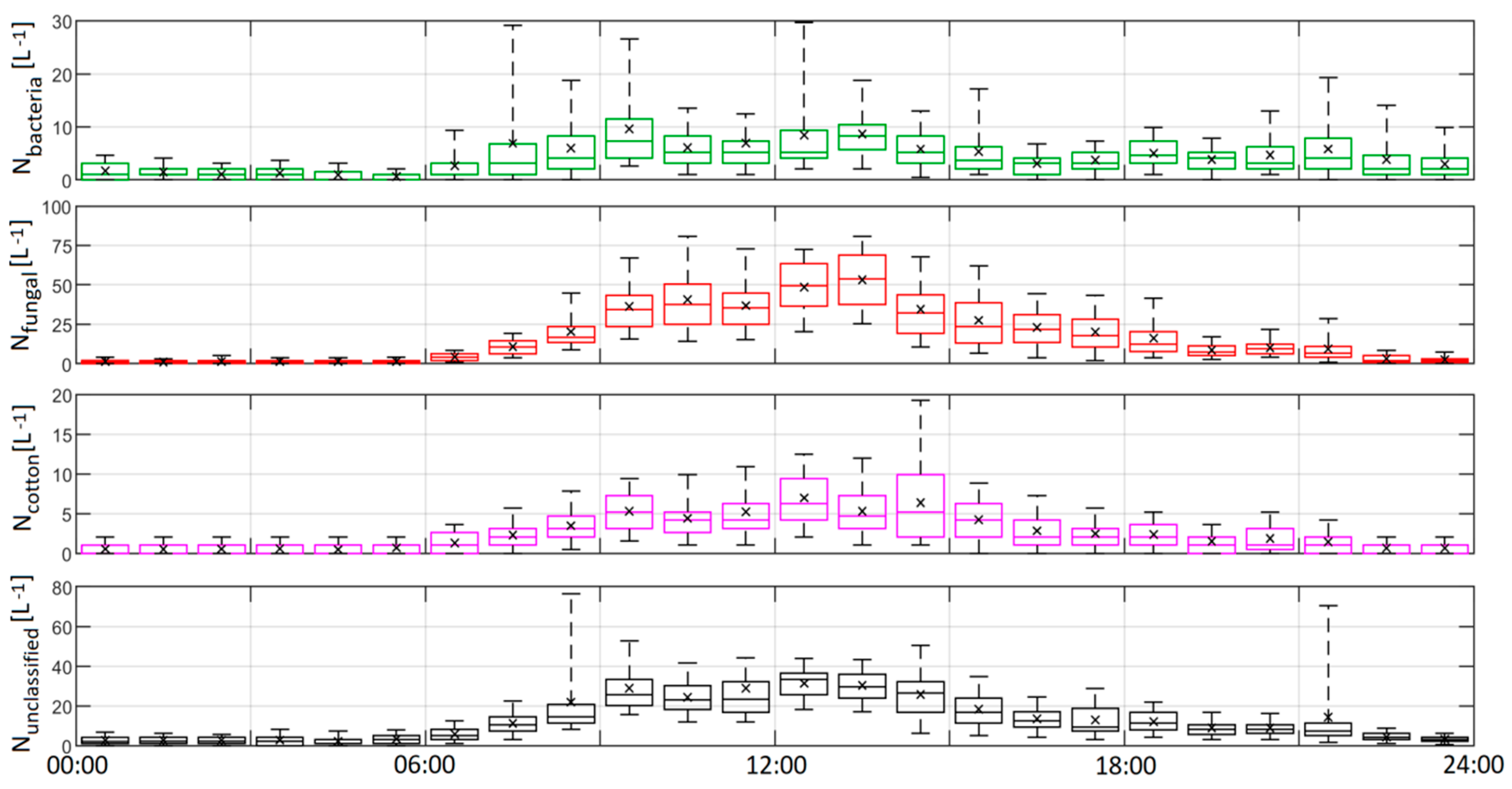
3.3. Ambient Indoor Air Time Series Product Analysis
4. Conclusions
- We demonstrate that the GBA classification model can accurately classify the training data into broad PBAP classes.
- The advanced CMOS shape information was demonstrated to be useful for minimising conflation between particle types with similar fluorescent characteristics but differing morphologies (e.g., E. coli bacteria and fungi).
- The Hellinger distance metric framework displays a high level of utility for assessing both the likelihood of training data conflations (e.g., bacteria samples display similarity) and the applicability of the training data to generate an appropriate model for a given ambient dataset.
- Some deficiencies in the fungal training samples were found using the above framework. They may arise due to either characteristic changes introduced by processing during manufacture or because the samples did not adequately represent the building mycology. This highlights the need to appraise the applicability of training data used to generate a classification model to build confidence in data outputs.
- The application of the model to ambient indoor data yielded illuminating results about PBAP within the building investigated; bacteria-like aerosol were well captured by the training data and they exhibited a strong, yet episodic and complex response to human activity within the building; fungal-like aerosol were observed to display a strong diurnal response to human activity with maximum concentrations at midday, correlating to a maximum in footfall. Interestingly large, rapidly decaying spikes in concentration were observed around the hour, corresponding with a high flux of people through the building. Concentrations of all classes fell to baseline minimums when the building was closed.
- High time resolution UV-LIF spectrometers can potentially reveal trends and mechanisms which may be obfuscated by offline methods that require long sample collections times.
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACS | Aerosol challenge simulator |
| BG | Bacillus atrophaeus |
| BT | Bacillus thuringensis |
| CMOS | Complementary metal-oxide-semiconductor |
| Dstl | Defence science and technologyl |
| FT | Forced trigger |
| GBA | Gradient boosting ensemble decision trees |
| HAC | Hierarchical agglomerative clustering |
| MBS | Multiparameter Bioaerosol Spectrometer |
| PBAP | Primary biological aerosol particle |
| UV-APS | Ultraviolet aerodynamic particle sizer |
| UV-LIF | Ultraviolet light-induced fluorescence |
| WIBS | Wideband integrated bioaerosol spectrometer |
References
- Bauer, H.; Kasper-Giebl, A.; Löflund, M.; Giebl, H.; Hitzenberger, R.; Zibuschka, F.; Puxbaum, H. The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols. Atmos. Res. 2002, 64, 109–119. [Google Scholar] [CrossRef]
- Bauer, H.; Schueller, E.; Weinke, G.; Berger, A.; Hitzenberger, R.; Marr, I.L.; Puxbaum, H. Significant contributions of fungal spores to the organic carbon and to the aerosol mass balance of the urban atmospheric aerosol. Atmos. Environ. 2008, 42, 5542–5549. [Google Scholar] [CrossRef]
- Möhler, O.; Georgakopoulos, D.G.; Morris, C.E.; Benz, S.; Ebert, V.; Hunsmann, S.; Saathoff, H.; Schnaiter, M.; Wagner, R. Heterogeneous ice nucleation activity of bacteria: New laboratory experiments at simulated cloud conditions. Biogeosciences 2008, 5, 1425–1435. [Google Scholar] [CrossRef] [Green Version]
- Crawford, I.; Bower, K.N.; Choularton, T.W.; Dearden, C.; Crosier, J.; Westbrook, C.; Capes, G.; Coe, H.; Connolly, P.J.; Dorsey, J.R.; et al. Ice formation and development in aged, wintertime cumulus over the UK: Observations and modelling. Atmos. Chem. Phys. 2012, 12, 4963–4985. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.E.; Conen, F.; Alex Huffman, J.; Phillips, V.; Pöschl, U.; Sands, D.C. Bioprecipitation: A feedback cycle linking earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob. Chang. Biol. 2014, 20, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Huffman, J.A.; Prenni, A.J.; DeMott, P.J.; Pöhlker, C.; Mason, R.H.; Robinson, N.H.; Fröhlich-Nowoisky, J.; Tobo, Y.; Després, V.R.; Garcia, E.; et al. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos. Chem. Phys. 2013, 13, 6151–6164. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.E.; Flagan, R.C.; Valenta, R.; Glovsky, M.M. Release of allergens as respirable aerosols: A link between grass pollen and asthma. J. Allergy Clin. Immunol. 2002, 109, 51–56. [Google Scholar] [CrossRef]
- Polymenakou, P.N.; Mandalakis, M.; Stephanou, E.G.; Tselepides, A. Particle Size Distribution of Airborne Microorganisms and Pathogens during an Intense African Dust Event in the Eastern Mediterranean. Environ. Health Perspect. 2008, 116, 292–296. [Google Scholar] [CrossRef] [Green Version]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Ebbehoj, N.E.; Hansen, M.O.; Sigsgaard, T.; Larsen, L. Building-related symptoms and molds: A two-step intervention study. Indoor Air 2002, 12, 273–277. [Google Scholar] [CrossRef]
- Zeliger, H.I. Toxic Effects of Chemical Mixtures. Arch. Environ. Health Int. J. 2003, 58, 23–29. [Google Scholar] [CrossRef]
- Nag, P.K. Sick Building Syndrome and Other Building-Related Illnesses. In Office Buildings; Springer: Singapore, 2019; pp. 53–103. [Google Scholar]
- Netuveli, G.; Hurwitz, B.; Levy, M.; Fletcher, M.; Barnes, G.; Durham, S.R.; Sheikh, A. Ethnic variations in UK asthma frequency, morbidity, and health-service use: A systematic review and meta-analysis. Lancet 2005, 365, 312–317. [Google Scholar] [CrossRef]
- Tackling the Allergy Crisis in Europe—Concerted Policy Action Needed. Available online: http://www.eaaci.org/documents/EAACI_Advocacy_Manifesto.pdf (accessed on 20 July 2020).
- Haleem Khan, A.A.; Mohan Karuppayil, S. Fungal pollution of indoor environments and its management. Saudi J. Biol. Sci. 2012, 19, 405–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sailer, M.F.; van Nieuwenhuijzen, E.J.; Knol, W. Forming of a functional biofilm on wood surfaces. Ecol. Eng. 2010, 36, 163–167. [Google Scholar] [CrossRef]
- Doherty, W.O.S.; Mousavioun, P.; Fellows, C.M. Value-adding to cellulosic ethanol: Lignin polymers. Ind. Crops Prod. 2011, 33, 259–276. [Google Scholar] [CrossRef] [Green Version]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. USA 2009, 106, 16393–16399. [Google Scholar] [CrossRef] [Green Version]
- Handorean, A.; Robertson, C.E.; Harris, J.K.; Frank, D.; Hull, N.; Kotter, C.; Stevens, M.J.; Baumgardner, D.; Pace, N.R.; Hernandez, M. Microbial aerosol liberation from soiled textiles isolated during routine residuals handling in a modern health care setting. Microbiome 2015, 3, 72. [Google Scholar] [CrossRef] [Green Version]
- Bhangar, S.; Adams, R.I.; Pasut, W.; Huffman, J.A.; Arens, E.A.; Taylor, J.W.; Bruns, T.D.; Nazaroff, W.W. Chamber bioaerosol study: Human emissions of size-resolved fluorescent biological aerosol particles. Indoor Air 2016, 26, 193–206. [Google Scholar] [CrossRef] [Green Version]
- Huffman, J.A.; Perring, A.E.; Savage, N.J.; Clot, B.; Crouzy, B.; Tummon, F.; Shoshanim, O.; Damit, B.; Schneider, J.; Sivaprakasam, V.; et al. Real-time sensing of bioaerosols: Review and current perspectives. Aerosol Sci. Technol. 2020, 54, 465–495. [Google Scholar] [CrossRef] [Green Version]
- Ruske, S.; Topping, D.O.; Foot, V.E.; Kaye, P.H.; Stanley, W.R.; Crawford, I.; Morse, A.P.; Gallagher, M.W. Evaluation of machine learning algorithms for classification of primary biological aerosol using a new UV-LIF spectrometer. Atmos. Meas. Tech. 2017, 10, 695–708. [Google Scholar] [CrossRef] [Green Version]
- Forde, E.; Gallagher, M.; Walker, M.; Foot, V.; Attwood, A.; Granger, G.; Sarda-Estève, R.; Stanley, W.; Kaye, P.; Topping, D. Intercomparison of Multiple UV-LIF Spectrometers Using the Aerosol Challenge Simulator. Atmosphere 2019, 10, 797. [Google Scholar] [CrossRef] [Green Version]
- Könemann, T.; Savage, N.; Klimach, T.; Walter, D.; Fröhlich-Nowoisky, J.; Su, H.; Pöschl, U.; Huffman, J.A.; Pöhlker, C. Spectral Intensity Bioaerosol Sensor (SIBS): An instrument for spectrally resolved fluorescence detection of single particles in real time. Atmos. Meas. Tech. 2019, 12, 1337–1363. [Google Scholar] [CrossRef] [Green Version]
- Šaulienė, I.; Šukienė, L.; Daunys, G.; Valiulis, G.; Vaitkevičius, L.; Matavulj, P.; Brdar, S.; Panic, M.; Sikoparija, B.; Clot, B.; et al. Automatic pollen recognition with the Rapid-E particle counter: The first-level procedure, experience and next steps. Atmos. Meas. Tech. 2019, 12, 3435–3452. [Google Scholar] [CrossRef] [Green Version]
- Huffman, J.A.; Treutlein, B.; Pöschl, U. Fluorescent biological aerosol particle concentrations and size distributions measured with an Ultraviolet Aerodynamic Particle Sizer (UV-APS) in Central Europe. Atmos. Chem. Phys. 2010, 10, 3215–3233. [Google Scholar] [CrossRef] [Green Version]
- Gabey, A.M.; Vaitilingom, M.; Freney, E.; Boulon, J.; Sellegri, K.; Gallagher, M.W.; Crawford, I.P.; Robinson, N.H.; Stanley, W.R.; Kaye, P.H. Observations of fluorescent and biological aerosol at a high-altitude site in central France. Atmos. Chem. Phys. 2013, 13, 7415–7428. [Google Scholar] [CrossRef] [Green Version]
- Crawford, I.; Ruske, S.; Topping, D.O.; Gallagher, M.W. Evaluation of hierarchical agglomerative cluster analysis methods for discrimination of primary biological aerosol. Atmos. Meas. Tech. Discuss. 2015, 8, 7303–7333. [Google Scholar] [CrossRef]
- Forde, E.; Gallagher, M.; Foot, V.; Sarda-Esteve, R.; Crawford, I.; Kaye, P.; Stanley, W.; Topping, D. Characterisation and source identification of biofluorescent aerosol emissions over winter and summer periods in the United Kingdom. Atmos. Chem. Phys. 2019, 19, 1665–1684. [Google Scholar] [CrossRef] [Green Version]
- Savage, N.J.; Huffman, J.A. Evaluation of a hierarchical agglomerative clustering method applied to WIBS laboratory data for improved discrimination of biological particles by comparing data preparation techniques. Atmos. Meas. Tech. 2018, 11, 4929–4942. [Google Scholar] [CrossRef] [Green Version]
- Gabey, A.M.; Gallagher, M.W.; Whitehead, J.; Dorsey, J.R.; Kaye, P.H.; Stanley, W.R. Measurements and comparison of primary biological aerosol above and below a tropical forest canopy using a dual channel fluorescence spectrometer. Atmos. Chem. Phys. 2010, 10, 4453–4466. [Google Scholar] [CrossRef] [Green Version]
- Toprak, E.; Schnaiter, M. Fluorescent biological aerosol particles measured with the Waveband Integrated Bioaerosol Sensor WIBS-4: Laboratory tests combined with a one year field study. Atmos. Chem. Phys. 2013, 13, 225–243. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, D.J.; Healy, D.A.; Hellebust, S.; Buters, J.T.M.; Sodeau, J.R. Using the WIBS-4 (Waveband Integrated Bioaerosol Sensor) Technique for the On-Line Detection of Pollen Grains. Aerosol Sci. Technol. 2014, 48, 341–349. [Google Scholar] [CrossRef]
- Crawford, I.; Robinson, N.H.; Flynn, M.J.; Foot, V.E.; Gallagher, M.W.; Huffman, J.A.; Stanley, W.R.; Kaye, P.H. Characterisation of bioaerosol emissions from a Colorado pine forest: Results from the BEACHON-RoMBAS experiment. Atmos. Chem. Phys. 2014, 14, 8559–8578. [Google Scholar] [CrossRef] [Green Version]
- Perring, A.E.; Schwarz, J.P.; Baumgardner, D.; Hernandez, M.T.; Spracklen, D.V.; Heald, C.L.; Gao, R.S.; Kok, G.; McMeeking, G.R.; McQuaid, J.B.; et al. Airborne observations of regional variation in fluorescent aerosol across the United States. J. Geophys. Res. Atmos. 2015, 120, 1153–1170. [Google Scholar] [CrossRef]
- Gosselin, M.I.; Rathnayake, C.M.; Crawford, I.; Pöhlker, C.; Fröhlich-Nowoisky, J.; Schmer, B.; Després, V.R.; Engling, G.; Gallagher, M.; Stone, E.; et al. Fluorescent bioaerosol particle, molecular tracer, and fungal spore concentrations during dry and rainy periods in a semi-arid forest. Atmos. Chem. Phys. 2016, 16, 15165–15184. [Google Scholar] [CrossRef] [Green Version]
- Kaye, P.H.; Hirst, E.; Greenaway, R.S.; Ulanowski, Z.; Hesse, E.; DeMott, P.J.; Saunders, C.; Connolly, P. Classifying atmospheric ice crystals by spatial light scattering. Opt. Lett. 2008, 33, 1545. [Google Scholar] [CrossRef] [PubMed]
- Savage, N.; Krentz, C.; Könemann, T.; Han, T.T.; Mainelis, G.; Pöhlker, C.; Huffman, J.A. Systematic Characterization and Fluorescence Threshold Strategies for the Wideband Integrated Bioaerosol Sensor (WIBS) Using Size-Resolved Biological and Interfering Particles. Atmos. Meas. Tech. Discuss. 2017, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Freund, Y.; Schapire, R.E. A desicion-theoretic generalization of on-line learning and an application to boosting. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1995; pp. 23–37. [Google Scholar]
- Hernandez, M.; Perring, A.E.; McCabe, K.; Kok, G.; Granger, G.; Baumgardner, D. Chamber catalogues of optical and fluorescent signatures distinguish bioaerosol classes. Atmos. Meas. Tech. 2016, 9, 3283–3292. [Google Scholar] [CrossRef] [Green Version]
- Pollen Count Averages for Northwest England. Available online: https://www.worcester.ac.uk/documents/Pollen-Count-Averages-for-Northwest-England.pdf (accessed on 30 June 2020).





| Sample | Origin | Processing | Storage | Dispersal | Size (µm) | Morphology |
|---|---|---|---|---|---|---|
| Escherichia coli (G−) | Dstl stock | Re-suspended in phosphate-buffered saline | >5 °C | Medical nebuliser | 1.3 ± 0.6 | rod-shaped |
| Bacillus atrophaeus (G+) | Dstl stock | Re-suspended in phosphate-buffered saline | >5 °C | Medical nebuliser | 1.4 ± 0.4 | rod-shaped |
| Bacillus thuringensis (G+) | Dstl stock | Re-suspended in phosphate-buffered saline | >5 °C | Medical nebuliser | 1.2 ± 0.6 | rod-shaped |
| Alternaria Alternaria | Stallergenes Greer Strain ATCC 11680 | acetone | <0 °C | compressed air | 1.9 ± 4.2 | fibrous |
| Cladosporium herbarum | Stallergenes Greer Strain ATCC 6506 | acetone | <0 °C | compressed air | 2.7 ± 3.0 | fibrous |
| Cotton | Black T-Shirt | none | N/A | mechanical agitation | N/A | - |
| Predicted Label | ||||
|---|---|---|---|---|
| Bacteria | Fungal | Cotton | ||
| Bacteria | 100% | 0% | 0% | |
| True label | Fungal | 0% | 100% | 0% |
| Cotton | 0% | 0% | 100% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crawford, I.; Topping, D.; Gallagher, M.; Forde, E.; Lloyd, J.R.; Foot, V.; Stopford, C.; Kaye, P. Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers. Atmosphere 2020, 11, 1039. https://doi.org/10.3390/atmos11101039
Crawford I, Topping D, Gallagher M, Forde E, Lloyd JR, Foot V, Stopford C, Kaye P. Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers. Atmosphere. 2020; 11(10):1039. https://doi.org/10.3390/atmos11101039
Chicago/Turabian StyleCrawford, Ian, David Topping, Martin Gallagher, Elizabeth Forde, Jonathan R. Lloyd, Virginia Foot, Chris Stopford, and Paul Kaye. 2020. "Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers" Atmosphere 11, no. 10: 1039. https://doi.org/10.3390/atmos11101039
APA StyleCrawford, I., Topping, D., Gallagher, M., Forde, E., Lloyd, J. R., Foot, V., Stopford, C., & Kaye, P. (2020). Detection of Airborne Biological Particles in Indoor Air Using a Real-Time Advanced Morphological Parameter UV-LIF Spectrometer and Gradient Boosting Ensemble Decision Tree Classifiers. Atmosphere, 11(10), 1039. https://doi.org/10.3390/atmos11101039








