Comparative Analysis of Airborne Bacterial and Fungal Communities in South-Eastern Italy and in Albania Using the Compositional Analysis of 16S and ITS rRNA Gene Sequencing Datasets
Abstract
:1. Introduction
2. Materials and Methods
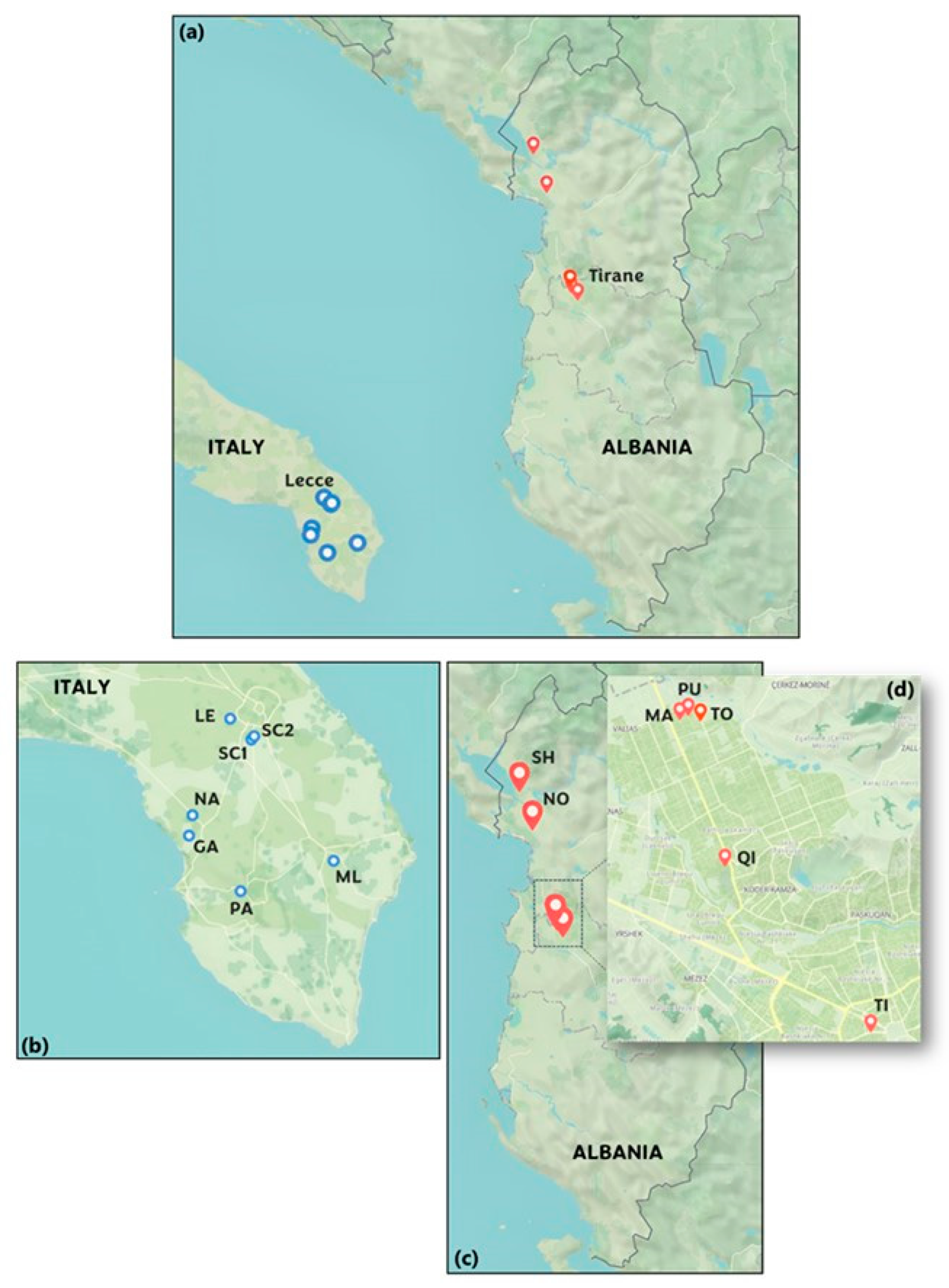
2.1. Description of the Sites Selected for the Monitoring Campaign
- -
- Two urban sites: one at the Department of Mathematics and Physics of the University of Salento in Lecce (~95,000 inhabitants, denoted as LE) and another in the city center of Nardò (~31,500 inhabitants, denoted as NA). Both are significantly influenced by vehicular traffic.
- -
- Three small-town sites: in the Lecce province, these included Parabita (~9000 inhabitants, PA), San Cesario di Lecce (~8000 inhabitants, SC1), and Muro Leccese (~5000 inhabitants, ML). These sites were selected for their varied, yet relatively lower, levels of pollution.
- -
- Two rural sites: in the Lecce province, located in San Cesario di Lecce (~8000 inhabitants, SC2) and Galatone (~15,500 inhabitants, GA). These sites were chosen to explore the biogenic component of atmospheric aerosols, given their minimal vehicular traffic and limited sources of pollution.
- -
- Two urban sites: located in the city center of Tirana, the first one in the Rruga Myslym Shyri Tiranë area (denoted as TI), and the second one in the area of the University College Qiriazi (denoted as QI).
- -
- Three suburban sites: located in the suburban area of Tirana, in the zone of Kamëz, and identified as MA (Marashi), PU (Pusi), and TO (Tokat).
- -
- Two rural sites: in northern Albania, at Shkodër (~135,000 inhabitants, denoted as SH), located between Lake Shkodër and the Albanian Alps, and in the Lezhë Municipality (~65,000 inhabitants, denoted as NO), spanning from the Plain of Zadrima to the Adriatic Sea. Shkodër and Lezhë represent rural conditions with minimal anthropogenic impact. More information on the climate characteristics of Northern Albania can be found in Dervishi et al. [47].
2.2. Description of the Monitoring Campaign and the Methodology Used for Aerosol Detection
2.3. Methodology for DNA Extraction and 16S/ITS rRNA Gene Metabarcoding
2.4. Description of the Methodology for Compositional Data Analysis
3. Results
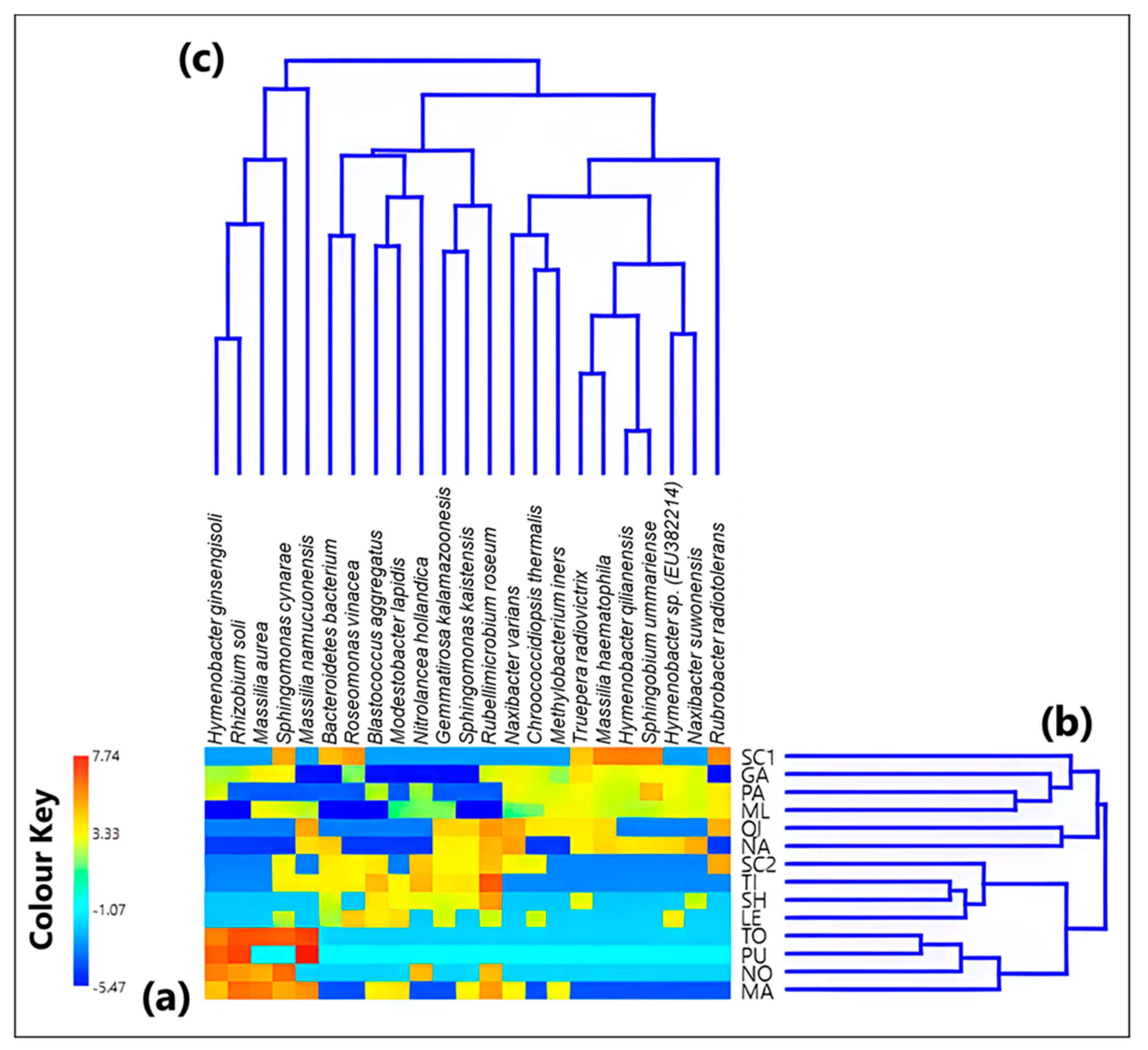
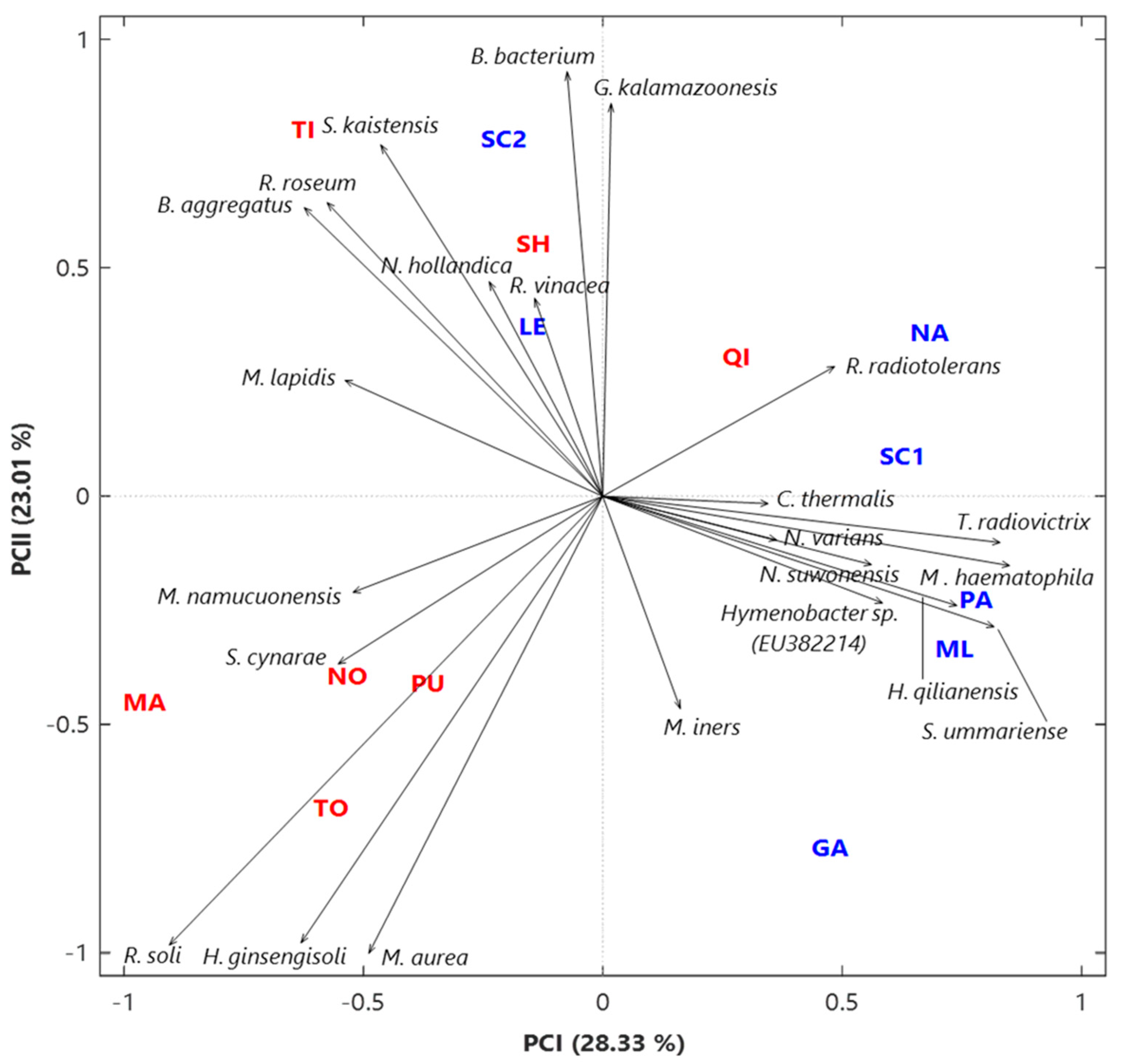
3.1. Analysis of the Bacterial Communities at the Species Level
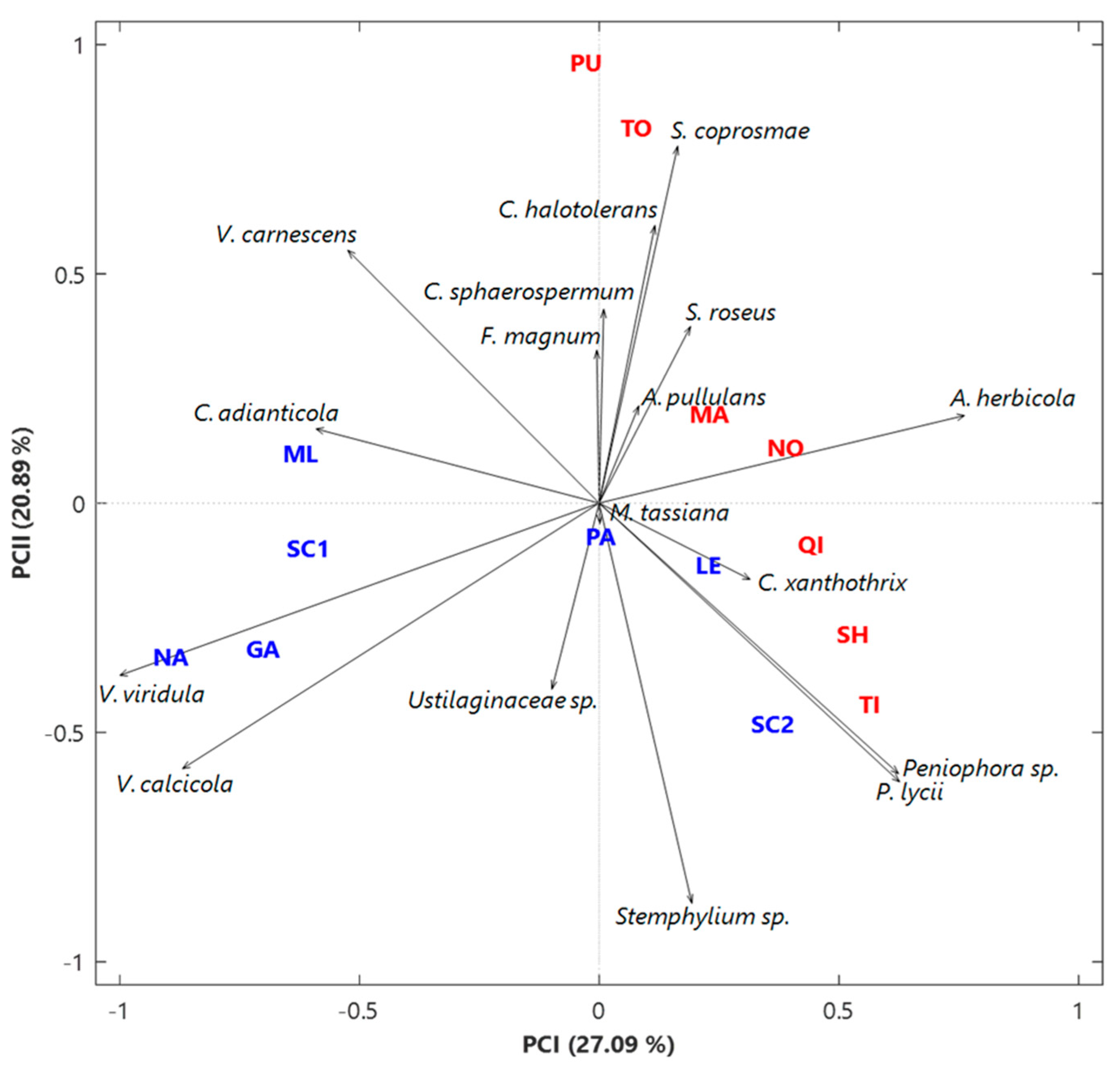
3.2. Analysis of the Fungal Communities at the Species Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary Biological Aerosol Particles in the Atmosphere: A Review. Tellus B Chem. Phys. Meteorol. 2012, 64, 15598. [Google Scholar] [CrossRef]
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the Earth System: Climate, Health, and Ecosystem Interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kabir, E.; Jahan, S.A. Airborne Bioaerosols and Their Impact on Human Health. J. Environ. Sci. 2018, 67, 23–35. [Google Scholar] [CrossRef]
- Tang, K.; Huang, Z.; Huang, J.; Maki, T.; Zhang, S.; Shimizu, A.; Ma, X.; Shi, J.; Bi, J.; Zhou, T.; et al. Characterization of Atmospheric Bioaerosols along the Transport Pathway of Asian Dust during the Dust-Bioaerosol 2016 Campaign. Atmos. Chem. Phys. 2018, 18, 7131–7148. [Google Scholar] [CrossRef]
- Xie, W.; Li, Y.; Bai, W.; Hou, J.; Ma, T.; Zeng, X.; Zhang, L.; An, T. The Source and Transport of Bioaerosols in the Air: A Review. Front. Environ. Sci. Eng. 2021, 15, 44. [Google Scholar] [CrossRef]
- Yassin, M.F.; Almouqatea, S. Assessment of Airborne Bacteria and Fungi in an Indoor and Outdoor Environment. Int. J. Environ. Sci. Technol. 2010, 7, 535–544. [Google Scholar] [CrossRef]
- Kowalski, M.; Wolany, J.; Pastuszka, J.S.; Płaza, G.; Wlazło, A.; Ulfig, K.; Malina, A. Characteristics of Airborne Bacteria and Fungi in Some Polish Wastewater Treatment Plants. Int. J. Environ. Sci. Technol. 2017, 14, 2181–2192. [Google Scholar] [CrossRef]
- Reanprayoon, P.; Yoonaiwong, W. Airborne Concentrations of Bacteria and Fungi in Thailand Border Market. Aerobiologia 2012, 28, 49–60. [Google Scholar] [CrossRef]
- Park, D.; Ryu, S.; Kim, S.; Byun, H.; Yoon, C.; Lee, K. Airborne Bacteria and Fungi Associated with Waste-Handling Work. Int. J. Occup. Environ. Health 2013, 19, 311–318. [Google Scholar] [CrossRef]
- Rdzanek, M.; Pusz, W.; Gębarowska, E.; Pląskowska, E. Airborne Bacteria and Fungi in a Coal Mine in Poland. J. Caves. Karst Stud. 2015, 77, 177–182. [Google Scholar] [CrossRef]
- Jacob, J.H.; Irshaid, F.I.; Alhalib, M.A. Estimation and Identification of Airborne Bacteria and Fungi in the Outdoor Atmosphere of Al-Mafraq Area—Jordan. Jordan J. Biol. Sci. 2016, 9, 3–10. [Google Scholar] [CrossRef]
- Gębarowska, E.; Pusz, W.; Kucińska, J.; Kita, W. Comparative Analysis of Airborne Bacteria and Fungi in Two Salt Mines in Poland. Aerobiologia 2018, 34, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.D.; Ma, M.; Xia, Y.; Wang, T.; Shu, B.; Lednicky, J.A.; Ma, M.-J.; Lu, J.; Gray, G.C. Bioaerosol Sampling in Modern Agriculture: A Novel Approach for Emerging Pathogen Surveillance? J. Infect. Dis. 2016, 214, 537–545. [Google Scholar] [CrossRef]
- Mubareka, S.; Groulx, N.; Savory, E.; Cutts, T.; Theriault, S.; Scott, J.A.; Roy, C.J.; Turgeon, N.; Bryce, E.; Astrakianakis, G.; et al. Bioaerosols and Transmission, a Diverse and Growing Community of Practice. Front. Public Health 2019, 7, 23. [Google Scholar] [CrossRef]
- Mbareche, H.; Morawska, L.; Duchaine, C. On the Interpretation of Bioaerosol Exposure Measurements and Impacts on Health. J. Air Waste Manag. Assoc. 2019, 69, 789–804. [Google Scholar] [CrossRef]
- Shammi, M.; Rahman, M.M.; Tareq, S.M. Distribution of Bioaerosols in Association with Particulate Matter: A Review on Emerging Public Health Threat in Asian Megacities. Front. Environ. Sci. 2021, 9, 698215. [Google Scholar] [CrossRef]
- Maki, T.; Sano, I.; Shimamoto, S.; Miyata, H.; Fujiwara, Y.; Watanabe, K.; Kurosaki, Y.; Aoki, K.; Matsuki, A.; Noda, J.; et al. Long-range transport of airborne microorganisms by Asian dust events: Bioaerosols relating to human health. In Proceedings of the EGU General Assembly Conference Abstracts 2023, Vienna, Australia, 24–28 April 2023. [Google Scholar] [CrossRef]
- Innocente, E.; Squizzato, S.; Visin, F.; Facca, C.; Rampazzo, G.; Bertolini, V.; Gandolfi, I.; Franzetti, A.; Ambrosini, R.; Bestetti, G. Influence of Seasonality, Air Mass Origin and Particulate Matter Chemical Composition on Airborne Bacterial Community Structure in the Po Valley, Italy. Sci. Total Environ. 2017, 593–594, 677–687. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, S.; Zheng, D.; Li, J.; Tian, H.; Wang, Y. Effects of Haze Pollution on Microbial Community Changes and Correlation with Chemical Components in Atmospheric Particulate Matter. Sci. Total Environ. 2018, 637–638, 507–516. [Google Scholar] [CrossRef]
- Romano, S.; Di Salvo, M.; Rispoli, G.; Alifano, P.; Perrone, M.R.; Talà, A. Airborne Bacteria in the Central Mediterranean: Structure and Role of Meteorology and Air Mass Transport. Sci. Total Environ. 2019, 697, 134020. [Google Scholar] [CrossRef]
- Calderón-Ezquerro, M.C.; Serrano-Silva, N.; Brunner-Mendoza, C. Metagenomic Characterisation of Bioaerosols during the Dry Season in Mexico City. Aerobiologia 2020, 36, 493–505. [Google Scholar] [CrossRef]
- Kang, S.; Cho, K.-S. Microbial Metagenome of Airborne Particulate Matter: Methodology, Characteristics, and Influencing Parameters. Korean J. Microbiol. Biotechnol. 2022, 50, 165–192. [Google Scholar] [CrossRef]
- Erkorkmaz, B.A.; Gat, D.; Rudich, Y. Aerial transport of bacteria by dust plumes in the Eastern Mediterranean revealed by complementary rRNA/rRNA-gene sequencing. Commun. Earth Environ. 2023, 4, 24. [Google Scholar] [CrossRef]
- Robinson, C.K.; Brotman, R.M.; Ravel, J. Intricacies of Assessing the Human Microbiome in Epidemiologic Studies. Ann. Epidemiol. 2016, 26, 311–321. [Google Scholar] [CrossRef]
- Xia, Y. Correlation and Association Analyses in Microbiome Study Integrating Multiomics in Health and Disease. Prog. Mol. Biol. Transl. Sci. 2020, 171, 309–491. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Fragola, M.; Romano, S.; Peccarrisi, D.; Talà, A.; Alifano, P.; Buccolieri, A.; Quarta, G.; Calcagnile, L. Characterization of the Airborne Microbiome in Different Indoor and Outdoor Locations of a University Building Using an Innovative Compositional Data Analysis Approach. Atmosphere 2023, 14, 1529. [Google Scholar] [CrossRef]
- Romano, S.; Catanzaro, V.; Paladini, F. Impacts of the COVID-19 Lockdown Measures on the 2020 Columnar and Surface Air Pollution Parameters over South-Eastern Italy. Atmosphere 2021, 12, 1366. [Google Scholar] [CrossRef]
- Romano, S.; Pichierri, S.; Fragola, M.; Buccolieri, A.; Quarta, G.; Calcagnile, L. Characterization of the PM2.5 aerosol fraction monitored at a suburban site in south-eastern Italy by integrating isotopic techniques and ion beam analysis. Front. Environ. Sci. 2022, 10, 971204. [Google Scholar] [CrossRef]
- Perrone, M.R.; Paladini, F.; Becagli, S.; Amore, A.; Romano, S. Daytime and Nighttime Chemical and Optical Properties of Fine and Coarse Particles at a Central Mediterranean Coastal Site. Environ. Sci. Pollut. Res. Int. 2022, 29, 43401–43420. [Google Scholar] [CrossRef]
- Romano, S.; Becagli, S.; Lucarelli, F.; Rispoli, G.; Perrone, M.R. Airborne Bacteria Structure and Chemical Composition Relationships in Winter and Spring PM10 Samples over Southeastern Italy. Sci. Total Environ. 2020, 730, 138899. [Google Scholar] [CrossRef]
- Fragola, M.; Perrone, M.R.; Alifano, P.; Talà, A.; Romano, S. Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean. Toxins 2021, 13, 518. [Google Scholar] [CrossRef] [PubMed]
- Fragola, M.; Arsieni, A.; Carelli, N.; Dattoli, S.; Maiellaro, S.; Perrone, M.R.; Romano, S. Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights. Int. J. Environ. Res. Public Health 2022, 19, 2624. [Google Scholar] [CrossRef] [PubMed]
- Mico, S.; Deda, A.; Alushllari, M. Particulate Matter Concentrations: Urban versus Suburban Areas of Vlora, Albania. In Proceedings of the AIP Conference Proceedings, Birmingham, UK, 16–20 July 2017. [Google Scholar] [CrossRef]
- Liti, A.; Cara, O.; Mercury, J.; Cara, M. The monitoring of PM2.5 and PM10 in the air in Durres, Albania. J. Int. Sci. Publ. Ecol. Saf. 2020, 14, 75–81. [Google Scholar]
- Hysenaj, M.; Duraj, S. Spatiotemporal Variations of Particulate Matter in Tirana. J. Ecol. Eng. 2021, 22, 209–214. [Google Scholar] [CrossRef]
- Hajderi, A.; Bozo, L. The Air Pollution from Vehicles and Health Risks in the City of Tirana. Adv. Eng. Days 2023, 7, 116–119. [Google Scholar]
- Kaçorri, D.; Totoni, R.; Prifti, L. Multinomial Logistic Regression to Estimate the Public Perception of Air Pollution and Its Impact on Health: A Case Study in Tirana. AS-Proceedings 2023, 1, 312–317. [Google Scholar]
- Troja, E.; Pinguli, L.; Troja, R.; Dhamo, E.; Muça, E. Microbiological Air Monitoring and Long-Term Evaluations of Selected Urban Areas in the City of Tirana. Environ. Sci. Proc. 2021, 9, 13. [Google Scholar] [CrossRef]
- Qarri, F.; Lazo, P.; Allajbeu, S.; Bekteshi, L.; Kane, S.; Stafilov, T. The Evaluation of Air Quality in Albania by Moss Biomonitoring and Metals Atmospheric Deposition. Arch. Environ. Contam. Toxicol. 2019, 76, 554–571. [Google Scholar] [CrossRef]
- Lazo, P.; Kika, A.; Qarri, F.; Bekteshi, L.; Allajbeu, S.; Stafilov, T. Air Quality Assessment by Moss Biomonitoring and Trace Metals Atmospheric Deposition. Aerosol Air Qual. Res. 2022, 22, 220008. [Google Scholar] [CrossRef]
- Romano, S.; Perrone, M.R.; Pavese, G.; Esposito, F.; Calvello, M. Optical Properties of PM2.5 Particles: Results from a Monitoring Campaign in Southeastern Italy. Atmos. Environ. 2019, 203, 35–47. [Google Scholar] [CrossRef]
- Perrone, M.R.; Romano, S.; Orza, J.A.G. Particle Optical Properties at a Central Mediterranean Site: Impact of Advection Routes and Local Meteorology. Atmos. Res. 2014, 145–146, 152–167. [Google Scholar] [CrossRef]
- Perrone, M.R.; Romano, S.; Orza, J.A.G. Columnar and Ground-Level Aerosol Optical Properties: Sensitivity to the Transboundary Pollution, Daily and Weekly Patterns, and Relationships. Environ. Sci. Pollut. Res. Int. 2015, 22, 16570–16589. [Google Scholar] [CrossRef] [PubMed]
- Banja, M.; Como, E.; Murtaj, B.; Zotaj, A. Mapping Air Pollution in Urban Tirana Area Using GIS. In Proceedings of the International Conference SDI 2010, Skopje, North Macedonia, 15–17 September 2010. [Google Scholar]
- Kopali, A.; Libohova, Z.; Teqja, Z.; Owens, P.R. Bioclimatic Suitability for Wine Vineyards in Mediterranean Climate-Tirana Region, Albania. Eur. J. Hortic. Sci. 2021, 86, 179–188. [Google Scholar] [CrossRef]
- Dervishi, S.; Pashako, F.; Dushaj, X.; Dervishi, I.O. Energy Performance Optimization of Traditional Housing in Mediterranean Climate. J. Build. Eng. 2022, 45, 103423. [Google Scholar] [CrossRef]
- Robert, D.; Pirro, I. A Comparative Study of Ambient Air Quality Status in Major Cities of Albania. Ann. “VALAHIA” Uni Targoviste 2014, 8. [Google Scholar]
- Shu, H.; Xiangchao, C.; Peng, L.; Hui, G. Study on Electret Technology of Air Filtration Material. IOP Conf. Ser. Earth Environ. Sci. 2017, 100, 012110. [Google Scholar] [CrossRef]
- Romay, F.J.; Liu, B.Y.H.; Chae, S.-J. Experimental Study of Electrostatic Capture Mechanisms in Commercial Electret Filters. Aerosol Sci. Technol. 1998, 28, 224–234. [Google Scholar] [CrossRef]
- King, P.; Pham, L.K.; Waltz, S.; Sphar, D.; Yamamoto, R.T.; Conrad, D.; Taplitz, R.; Torriani, F.; Forsyth, R.A. Longitudinal Metagenomic Analysis of Hospital Air Identifies Clinically Relevant Microbes. PLoS ONE 2016, 11, e0160124. [Google Scholar] [CrossRef]
- Perrone, M.R.; Romano, S.; De Maria, G.; Tundo, P.; Bruno, A.R.; Tagliaferro, L.; Maffia, M.; Fragola, M. Compositional Data Analysis of 16S rRNA Gene Sequencing Results from Hospital Airborne Microbiome Samples. Int. J. Environ. Res. Public Health 2022, 19, 10107. [Google Scholar] [CrossRef]
- Perrone, M.R.; Romano, S.; De Maria, G.; Tundo, P.; Bruno, A.R.; Tagliaferro, L.; Maffia, M.; Fragola, M. Simultaneous Monitoring of SARS-CoV-2 and Bacterial Profiles from the Air of Hospital Environments with COVID-19 Affected Patients. Aerobiologia 2022, 38, 391–412. [Google Scholar] [CrossRef]
- Jaing, C.; Thissen, J.; Morrison, M.; Dillon, M.B.; Waters, S.M.; Graham, G.T.; Be, N.A.; Nicoll, P.; Verma, S.; Caro, T.; et al. Sierra Nevada Sweep: Metagenomic Measurements of Bioaerosols Vertically Distributed across the Troposphere. Sci. Rep. 2020, 10, 12399. [Google Scholar] [CrossRef] [PubMed]
- Ginn, O.; Rocha-Melogno, L.; Bivins, A.; Lowry, S.; Cardelino, M.; Nichols, D.; Tripathi, S.N.; Soria, F.; Andrade, M.; Bergin, M.; et al. Detection and Quantification of Enteric Pathogens in Aerosols near Open Wastewater Canals in Cities with Poor Sanitation. Environ. Sci. Technol. 2021, 55, 14758–14771. [Google Scholar] [CrossRef] [PubMed]
- Ginn, O.; Berendes, D.; Wood, A.; Bivins, A.; Rocha-Melogno, L.; Deshusses, M.A.; Tripathi, S.N.; Bergin, M.H.; Brown, J. Open Waste Canals as Potential Sources of Antimicrobial Resistance Genes in Aerosols in Urban Kanpur, India. Am. J. Trop. Med. Hyg. 2021, 104, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Paytuví, A.; Battista, E.; Scippacercola, F.; Aiese Cigliano, R.; Sanseverino, W. GAIA: An integrated metagenomics suite. bioRxiv 2019, bioRxiv:804690. [Google Scholar] [CrossRef]
- Pollegioni, P.; Mattioni, C.; Ristorini, M.; Occhiuto, D.; Canepari, S.; Korneykova, M.V.; Gavrichkova, O. Diversity and Source of Airborne Microbial Communities at Differential Polluted Sites of Rome. Atmosphere 2022, 13, 224. [Google Scholar] [CrossRef]
- Pawlowsky-Glahn, V.; Egozcue, J.J.; Tolosana-Delgado, R. Modeling and Analysis of Compositional Data, 1st ed.; John Wiley & Sons: Nashville, TN, USA, 2015. [Google Scholar]
- Martín-Fernández, J.A.; Barceló-Vidal, C.; Pawlowsky-Glahn, V. Dealing with zeros and missing values in compositional data sets using nonparametric imputation. Math. Geol. 2003, 35, 253–278. [Google Scholar] [CrossRef]
- Bian, G.; Gloor, G.B.; Gong, A.; Jia, C.; Zhang, W.; Hu, J.; Zhang, H.; Zhang, Y.; Zhou, Z.; Zhang, J.; et al. The Gut Microbiota of Healthy Aged Chinese Is Similar to That of the Healthy Young. mSphere 2017, 2, 10–1128. [Google Scholar] [CrossRef]
- Khaledi, E.; Moghadam, J.N.; Abdollahzadeh, J.; Amini, J. Fungi Associated with Grapevine Trunk Diseases (GTDs) with Emphasize on Pestalotioid Species in Kurdistan Province, Iran. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Kaur, H.; Gupta, P.; Ahmad, H.; Shankarnarayan, S.A.; Srivastava, S.; Sahu, S.; Karuna, T.; Narang, T.; Gupta, S.; Ghosh, A.; et al. Cladosporium Halotolerans: Exploring an Unheeded Human Pathogen. Mycopathologia 2023, 188, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Batra, N.; Kaur, H.; Mohindra, S.; Singh, S.; Shamanth, A.S.; Rudramurthy, S.M. Cladosporium Sphaerospermum Causing Brain Abscess, a Saprophyte Turning Pathogen: Case and Review of Published Reports. J. Mycol. Med. 2019, 29, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, J. The Statistical Analysis of Compositional Data; Chapman and Hall: London, UK, 1986; 416p. [Google Scholar] [CrossRef]
- Quinn, T.P.; Erb, I.; Gloor, G.; Notredame, C.; Richardson, M.F.; Crowley, T.M. A field guide for the compositional analysis of any-omics data. GigaScience 2019, 8, 9. [Google Scholar] [CrossRef]
- Zhen, Q.; Deng, Y.; Wang, Y.; Wang, X.; Zhang, H.; Sun, X.; Ouyang, Z. Meteorological factors had more impact on airborne bacterial communities than air pollutants. Sci. Total Environ. 2017, 601–602, 703–712. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Barberán, A.; Ramirez, K.S.; Leff, J.W.; Bradford, M.A.; Wall, D.H.; Fierer, N. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 2014, 17, 794–802. [Google Scholar] [CrossRef]
- Fröhlich-Nowoisky, J.; Ruzene Nespoli, C.; Pickersgill, D.A.; Galand, P.E.; Müller-Germann, I.; Nunes, T.; Gomes Cardoso, J.; Almeida, S.M.; Pio, C.; Andreae, M.O.; et al. Diversity and seasonal dynamics of airborne archaea. Biogeosciences 2014, 11, 6067–6079. [Google Scholar] [CrossRef]
- Sousa, A.C.A.; Almeida, J.R.S.L.; Pereira, C.C.; Ramiro Pastorinho, M.; Pereira, Â.M.C.; Nogueira, A.J.A.; Taborda-Barata, L.; Teixeira, J.P.; Correia, A.C.M.; Alves, A. Characterization of Fungal Communities in House Dust Samples Collected From Central Portugal—A Preliminary Survey. J. Toxicol. Environ. Health A 2014, 77, 972–982. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Bertolini, V.; Gandolfi, I.; Ambrosini, R.; Bestetti, G.; Innocente, E.; Rampazzo, G.; Franzetti, A. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 2013, 97, 6561–6570. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Park, E.H.; Lee, S.; Ko, G.; Honda, Y.; Hashizume, M.; Deng, F.; Yi, S.M.; Kim, H. Airborne Bacterial Communities in Three East Asian Cities of China, South Korea, and Japan. Sci. Rep. 2017, 7, 5545. [Google Scholar] [CrossRef] [PubMed]
- Peccia, J.; Hernandez, M. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: A review. Atmos. Environ. 2006, 40, 3941–3961. [Google Scholar] [CrossRef]
- Chawla, H.; Anand, P.; Garg, K.; Bhagat, N.; Varmani, S.G.; Bansal, T.; McBain, A.J.; Gulati Marwah, R. A comprehensive review of microbial contamination in the indoor environment: Sources, sampling, health risks, and mitigation strategies. Front. Public Health 2023, 11, 1285393. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, S.; Pepkolaj, L.; Fragola, M.; Peccarrisi, D.; Dhimitri, J.; Buccolieri, A.; Talà, A.; Alifano, P.; Quarta, G.; Calcagnile, L. Comparative Analysis of Airborne Bacterial and Fungal Communities in South-Eastern Italy and in Albania Using the Compositional Analysis of 16S and ITS rRNA Gene Sequencing Datasets. Atmosphere 2024, 15, 1155. https://doi.org/10.3390/atmos15101155
Romano S, Pepkolaj L, Fragola M, Peccarrisi D, Dhimitri J, Buccolieri A, Talà A, Alifano P, Quarta G, Calcagnile L. Comparative Analysis of Airborne Bacterial and Fungal Communities in South-Eastern Italy and in Albania Using the Compositional Analysis of 16S and ITS rRNA Gene Sequencing Datasets. Atmosphere. 2024; 15(10):1155. https://doi.org/10.3390/atmos15101155
Chicago/Turabian StyleRomano, Salvatore, Lekë Pepkolaj, Mattia Fragola, Dalila Peccarrisi, Jostina Dhimitri, Alessandro Buccolieri, Adelfia Talà, Pietro Alifano, Gianluca Quarta, and Lucio Calcagnile. 2024. "Comparative Analysis of Airborne Bacterial and Fungal Communities in South-Eastern Italy and in Albania Using the Compositional Analysis of 16S and ITS rRNA Gene Sequencing Datasets" Atmosphere 15, no. 10: 1155. https://doi.org/10.3390/atmos15101155









