Effect of River Ecological Restoration on Biofilm Microbial Community Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Habitat Survey and Physico-Chemical Parameters of Stream Water
2.3. Biofilm Sampling Procedure
2.4. DNA Extraction and Analysis of Bacterial Community Composition
2.5. Statistical Analysis
3. Results
3.1. Habitat Characteristics
3.2. Effects of Habitat Restoration on Physico-Chemical Properties of Stream Water
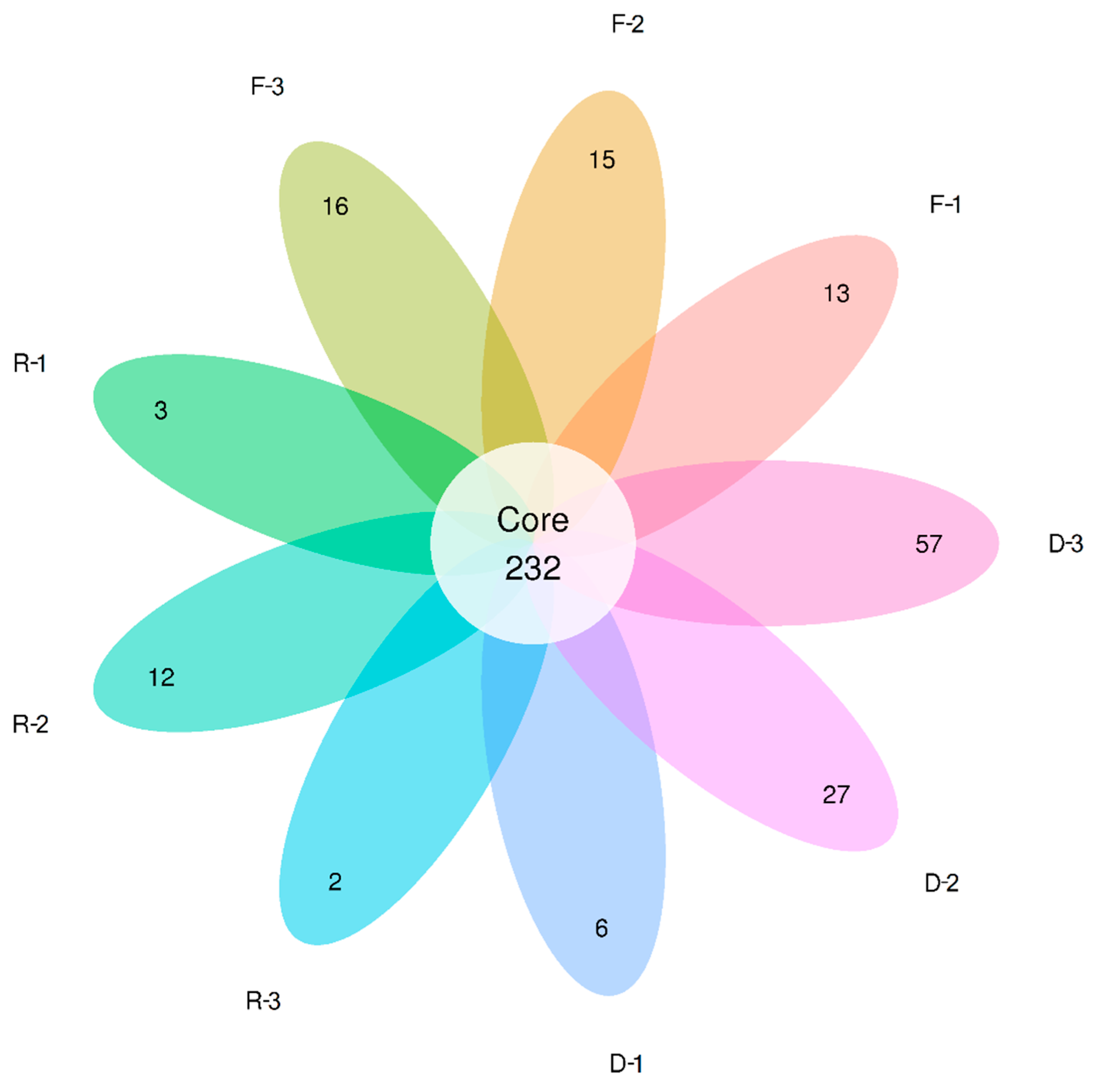
3.3. Effects of Habitat Restoration on Bacterial Community Composition
3.4. Correlation between Bacterial Community Composition and Environmental Variables
4. Discussion
4.1. Habitat Restoration Impact on Physico-Chemical Properties of Stream Water
4.2. Impact of Habitat Restoration on the Bacterial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bernhardt, E.S.; Sudduth, E.B.; Palmer, M.A.; Allan, J.D.; Meyer, J.L.; Alexander, G.; Follastad-Shah, J.; Hassett, B.; Jenkinson, R.; Lave, R.; et al. Restoring rivers one reach at a time: Results from a survey of US river restoration practitioners. Restor. Ecol. 2007, 15, 482–493. [Google Scholar] [CrossRef]
- Malmqvist, B.; Rundle, S. Threats to the running water ecosystems of the world. Environ. Conserv. 2002, 29, 134–153. [Google Scholar] [CrossRef]
- Naiman, R.J.; Bunn, S.E.; Nilsson, C.; Petts, G.E.; Pinay, G.; Thompson, L.C. Legitimizing fluvial ecosystems as users of water: An overview. Environ. Manag. 2002, 30, 455–467. [Google Scholar] [CrossRef]
- Laasonen, P.; Muotka, T.; Kivijarvi, I. Recovery of macroinvertebrate communities from stream habitat restoration. Aquat. Conserv.-Mar. Freshw. Ecosyst. 1998, 8, 101–113. [Google Scholar] [CrossRef]
- Lepori, F.; Palm, D.; Brannas, E.; Malmqvist, B. Does restoration of structural heterogeneity in streams enhance fish and macroinvertebrate diversity? Ecol. Appl. 2005, 15, 2060–2071. [Google Scholar] [CrossRef]
- Miller, S.W.; Budy, P.; Schmidt, J.C. Quantifying macroinvertebrate responses to in-stream habitat restoration: Applications of meta-analysis to river restoration. Restor. Ecol. 2010, 18, 8–19. [Google Scholar] [CrossRef]
- Palmer, M.A.; Filoso, S.; Fanelli, R.M. From ecosystems to ecosystem services: Stream restoration as ecological engineering. Ecol. Eng. 2014, 65, 62–70. [Google Scholar] [CrossRef]
- Kail, J.; Brabec, K.; Poppe, M.; Januschke, K. The effect of river restoration on fish, macro-invertebrates and aquatic macrophytes: A meta-analysis. Ecol. Indic. 2015, 58, 311–321. [Google Scholar] [CrossRef]
- Douglas, M.; Lake, P.S. Species richness of stream stones—An investigation of the mechanisms generating the species-area relationship. Oikos 1994, 69, 387–396. [Google Scholar] [CrossRef]
- Palmer, M.A.; Allan, J.D.; Butman, C.A. Dispersal as a regional process affecting the local dynamics of marine and stream benthic invertebrates. Trends Ecol. Evol. 1996, 11, 322–326. [Google Scholar] [CrossRef]
- Lake, P.S. Disturbance, patchiness, and diversity in streams. J. N. Am. Benthol. Soc. 2000, 19, 573–592. [Google Scholar] [CrossRef] [Green Version]
- Louhi, P.; Mykra, H.; Paavola, R.; Huusko, A.; Vehanen, T.; Maki-Petays, A.; Muotka, T. Twenty years of stream restoration in Finland: Little response by benthic macroinvertebrate communities. Ecol. Appl. 2011, 21, 1950–1961. [Google Scholar] [CrossRef]
- Simaika, J.P.; Stoll, S.; Lorenz, A.W.; Thomas, G.; Sundermann, A.; Haase, P. Bundles of stream restoration measures and their effects on fish communities. Limnologica 2015, 55, 1–8. [Google Scholar] [CrossRef]
- Flores, L.; Giorgi, A.; Gonzalez, J.M.; Larranaga, A.; Diez, J.R.; Elosegi, A. Effects of wood addition on stream benthic invertebrates differed among seasons at both habitat and reach scales. Ecol. Eng. 2017, 106, 116–123. [Google Scholar] [CrossRef]
- Nuttle, T.; Logan, M.N.; Parise, D.J.; Foltz, D.A.; Silvis, J.M.; Haibach, M.R. Restoration of macro-invertebrates, fish, and habitats in streams following mining subsidence: Replicated analysis across 18 mitigation sites. Restor. Ecol. 2017, 25, 820–831. [Google Scholar] [CrossRef]
- Lear, G.; Ancion, P.Y.; Harding, J.; Lewis, G.D. Use of bacterial communities to assess the ecological health of a recently restored stream. N. Z. J. Mar. Freshw. Res. 2012, 46, 291–301. [Google Scholar] [CrossRef]
- Gantzer, C.J.; Rittmann, B.E.; Herricks, E.E. Effect of long-term water velocity changes on streambed biofilm activity. Water Res. 1991, 25, 15–20. [Google Scholar] [CrossRef]
- Lawrence, J.R.; Chenier, M.R.; Roy, R.; Beaumier, D.; Fortin, N.; Swerhone, G.D.W.; Neu, T.R.; Greer, C.W. Microscale and molecular assessment of impacts of nickel, nutrients, and oxygen level on structure and function of river biofilm communities. Appl. Environ. Microbiol. 2004, 70, 4326–4339. [Google Scholar] [CrossRef]
- Lear, G.; Anderson, M.J.; Smith, J.P.; Boxen, K.; Lewis, G.D. Spatial and temporal heterogeneity of the bacterial communities in stream epilithic biofilms. FEMS Microbiol. Ecol. 2008, 65, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romani, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Fischer, H.; Sukhodolov, A.; Wilczek, S.; Engelhardt, C. Effects of flow dynamics and sediment movement on microbial activity in a lowland river. River Res. Appl. 2003, 19, 473–482. [Google Scholar] [CrossRef]
- Sheldon, F.; Walker, K.F. Changes in biofilms induced by flow regulation could explain extinctions of aquatic snails in the lower River Murray, Australia. Hydrobiologia 1997, 347, 97–108. [Google Scholar] [CrossRef]
- Cotner, J.B.; Biddanda, B.A. Small players, large role: Microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 2002, 5, 105–121. [Google Scholar] [CrossRef]
- Battin, T.J.; Kaplan, L.A.; Newbold, J.D.; Cheng, X.H.; Hansen, C. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 2003, 69, 5443–5452. [Google Scholar] [CrossRef]
- Zeglin, L.H. Stream microbial diversity in response to environmental changes: Review and synthesis of existing research. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Valett, H.M.; Thomas, S.A.; Mulholland, P.J.; Webster, J.R.; Dahm, C.N.; Fellows, C.S.; Crenshaw, C.L.; Peterson, C.G. Endogenous and exogenous control of ecosystem function: N cycling in headwater streams. Ecology 2008, 89, 3515–3527. [Google Scholar] [CrossRef]
- Mulholland, P.J.; Helton, A.M.; Poole, G.C.; Hall, R.O.; Hamilton, S.K.; Peterson, B.J.; Tank, J.L.; Ashkenas, L.R.; Cooper, L.W.; Dahm, C.N. Stream denitrification across biomes and its response to anthropogenic nitrate loading. Nature 2008, 452, 202. [Google Scholar] [CrossRef]
- Violin, C.R.; Cada, P.; Sudduth, E.B.; Hassett, B.A.; Bernhardt, P.E.S. Effects of urbanization and urban stream restoration on the physical and biological structure of stream ecosystems. Ecol. Appl. 2011, 21, 1932–1949. [Google Scholar] [CrossRef]
- Kondolf, G.M. Application of the pebble count: Notes on purpose, method, and variants. J. Am. Water Resour. Assoc. 1997, 33, 79–87. [Google Scholar] [CrossRef]
- Shannon, C.E. The mathematical theory of communication (Reprinted). M D Comput. 1997, 14, 306–317. [Google Scholar]
- Elizabeth, J.; Arar, G.B.C. Method 445.0 In Vitro Determination of Chlorophyll a and Pheophytin a in Marine and Freshwater Algae by Fluorescence; United States Environmental Protection Agency, Office of Research and Development, National Exposure Research Laboratory: Washington, DC, USA, 1997.
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Microbiol. Biotechnol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Torres-Mellado, G.A.; Escobar, I.; Palfner, G.; Casanova-Katny, M.A. Mycotrophy in Gilliesieae, a threatened and poorly known tribe of Alliaceae from central Chile. Rev. Chil. Hist. Nat. 2012, 85, 179–186. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Wang, Y.J.; Zhang, H.; Zhu, L.; Xu, Y.L.; Liu, N.; Sun, X.M.; Hu, L.P.; Huang, H.; Wei, K.; Zhu, R.L. Dynamic distribution of gut microbiota in goats at different ages and health states. Front. Microbiol. 2018, 9, 2509. [Google Scholar] [CrossRef]
- Oksanen, J.F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.; Hara, R.; Simpson, G.; Solymos, P.; et al. Vegan, Community Ecology Package. R Package Version 2.5-2. 2018. Available online: https://CRAN.R-project.org/package=vegan (accessed on 25 May 2019).
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F. CANOCO—An extension of decorana to analyze species-environment relationships. Vegetatio 1988, 75, 159–160. [Google Scholar] [CrossRef]
- Fernandez-Gomez, B.; Richter, M.; Schueler, M.; Pinhassi, J.; Acinas, S.G.; Gonzalez, J.M.; Pedros-Alio, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef]
- Geist, J.; Hawkins, S.J. Habitat recovery and restoration in aquatic ecosystems: Current progress and future challenges. Aquat. Conserv.-Mar. Freshw. Ecosyst. 2016, 26, 942–962. [Google Scholar] [CrossRef]
- Shrestha, S.; Farrelly, J.; Eggleton, M.; Chen, Y.S. Effects of conservation wetlands on stream habitat, water quality and fish communities in agricultural watersheds of the lower Mississippi River Basin. Ecol. Eng. 2017, 107, 99–109. [Google Scholar] [CrossRef]
- Fisher, S.G. Stream ecology—Structure and function of running waters—Allan, Jd. Science 1995, 270, 1858. [Google Scholar]
- Lear, G.; Washington, V.; Neale, M.; Case, B.; Buckley, H.; Lewis, G. The biogeography of stream bacteria. Glob. Ecol. Biogeogr. 2013, 22, 544–554. [Google Scholar] [CrossRef]
- Fierer, N.; Morse, J.L.; Berthrong, S.T.; Bernhardt, E.S.; Jackson, R.B. Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 2007, 88, 2162–2173. [Google Scholar] [CrossRef]
- Vannote, R.L.; Minshall, G.W.; Cummins, K.W.; Sedell, J.R.; Cushing, C.E. The river continuum concept. Can. J. Fish. Aquat. Sci. 1980, 37, 130–137. [Google Scholar] [CrossRef]
- Atashgahi, S.; Aydin, R.; Dimitrov, M.R.; Sipkema, D.; Hamonts, K.; Lahti, L.; Maphosa, F.; Kruse, T.; Saccenti, E.; Springael, D.; et al. Impact of a wastewater treatment plant on microbial community composition and function in a hyporheic zone of a eutrophic river. Sci. Rep. 2015, 5, 17284. [Google Scholar] [CrossRef]
- Levi, P.S.; Starnawski, P.; Poulsen, B.; Baattrup-Pedersen, A.; Schramm, A.; Riis, T. Microbial community diversity and composition varies with habitat characteristics and biofilm function in macrophyte-rich streams. Oikos 2016. [Google Scholar] [CrossRef]
- Drury, B.; Rosi-Marshall, E.; Kelly, J.J. Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl. Environ. Microbiol. 2013, 79, 1897–1905. [Google Scholar] [CrossRef]
- Lu, X.M.; Lu, P.Z. Characterization of bacterial communities in sediments receiving various wastewater effluents with high-throughput sequencing analysis. Microb. Ecol. 2014, 67, 612. [Google Scholar] [CrossRef]
- Ponsatí, L.; Corcoll, N.; Petrović, M.; Picó, Y.; Ginebreda, A.; Tornés, E.; Guasch, H.; Barcelo, D.; Sabater, S. Multiple-stressor effects on river biofilms under different hydrological conditions. Freshw. Biol. 2016, 61, 2102–2115. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.G.; Xu, H.; He, Y.; Zhao, F.; Huang, M.S. Remediation of urban river water by pontederia cordata combined with artificial aeration: Organic matter and nutrients removal and root-adhered bacterial communities. Int. J. Phytoremediat. 2015, 17, 1105–1114. [Google Scholar] [CrossRef]
- Olapade, O.A.; Leff, L.G. Seasonal response of stream biofilm communities to dissolved organic matter and nutrient enrichments. Appl. Environ. Microbiol. 2005, 71, 2278–2287. [Google Scholar] [CrossRef]
- Rier, S.T.; Stevenson, R.J. Effects of light, dissolved organic carbon, and inorganic nutrients on the relationship between algae and heterotrophic bacteria in stream periphyton. Hydrobiologia 2002, 489, 179–184. [Google Scholar] [CrossRef]
- Tank, J.L.; Webster, J.R. Interaction of substrate and nutrient availability on wood biofilm processes in streams. Ecology 1998, 79, 2168–2179. [Google Scholar] [CrossRef]
- Hempel, M.; Grossart, H.P.; Gross, E.M. Community composition of bacterial biofilms on two submerged macrophytes and an artificial substrate in a pre-alpine Lake. Aquat. Microb. Ecol. 2010, 58, 79–94. [Google Scholar] [CrossRef]
- Verma, D.K.; Rathore, G. New host record of five Flavobacterium species associated with tropical fresh water farmed fishes from North India. BJM 2015, 46, 969–976. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Wang, H.; Cao, Y.J.; Li, X.Y.; Wang, G.J. High quality draft genomic sequence of Arenimonas donghaensis DSM 18148(T). Stand. Genom. Sci. 2015, 10, 59. [Google Scholar] [CrossRef]
- Garcia-Garcera, M.; Touchon, M.; Brisse, S.; Rocha, E. Metagenomic assessment of the interplay between the environment and the genetic diversification of Acinetobacter. Environ. Microbiol. 2017, 19, 5010–5024. [Google Scholar] [CrossRef]
- Vorobev, A.; Beck, D.A.; Kalyuzhnaya, M.G.; Lidstrom, M.E.; Chistoserdova, L. Comparative transcriptomics in three Methylophilaceae species uncover different strategies for environmental adaptation. PeerJ 2013, 1, e115. [Google Scholar] [CrossRef]
- Risso, C.; Sun, J.; Zhuang, K.; Mahadevan, R.; DeBoy, R.; Ismail, W.; Shrivastava, S.; Huot, H.; Kothari, S.; Daugherty, S. Genome-scale comparison and constraint-based metabolic reconstruction of the facultative anaerobic Fe(III)-reducer Rhodoferax ferrireducens. BMC Genom. 2009, 10, 447. [Google Scholar] [CrossRef]
- Zhao, D.; Cao, X.; Huang, R.; Zeng, J.; Wu, Q.L. Variation of bacterial communities in water and sediments during the decomposition of Microcystis biomass. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Kämpfer, P.; Wellner, S.; Lohse, K.; Martin, K.; Lodders, N. Duganella phyllosphaerae sp. nov. isolated from the leaf surface of trifolium repens and proposal to reclassify duganella violaceinigra into a novel genus as pseudoduganella violceinigra gen. nov. comb. nov. Syst. Appl. Microbiol. 2012, 35, 19–23. [Google Scholar] [CrossRef]





| River Type | Width (m) | Mean Depth (cm) | Dissolved Oxygen (mg/L) | pH | Turbidity | NH4-N (mg/L) | NO3-N (mg/L) | TN (mg/L) | TP (mg/L) | Chemical Oxygen Demand (mg/L) | Total Organic C (mg/L) | Chlorophyll a (mg/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Forest | 8.83 ± 1.64 | 35.87 ± 7.97 | 14.16 ± 0.80 | 7.33 ± 0.11 | 0.62 ± 0.14 | 0.02 ± 0.01 | 1.06 ± 0.13 | 1.99 ± 0.21 | 0.18 ± 0.02 | 2.44 ± 0.15 | 0.48 ± 0.16 | 0.61 ± 0.23 |
| Restored | 13.17 ± 3.09 | 28.13 ± 7.22 | 13.14 ± 0.65 | 7.64 ± 0.14 | 3.52 ± 0.85 | 0.08 ± 0.02 | 1.13 ± 0.40 | 2.74 ± 0.77 | 0.17 ± 0.02 | 3.35 ± 0.76 | 2.81 ± 0.32 | 1.22 ± 0.19 |
| Degraded | 11.57 ± 5.72 | 22.87 ± 3.86 | 7.91 ± 1.52 | 7.38 ± 0.11 | 22.81 ± 14.93 | 1.37 ± 1.19 | 0.79 ± 0.40 | 4.01 ± 0.76 | 0.18 ± 0.05 | 8.82 ± 3.40 | 6.70 ± 2.21 | 0.20 ± 0.09 |
| River Type | Observed OTUs | Unique OTUs | Diversity Indices | |
|---|---|---|---|---|
| Chao 1 Value | Shannon-Weiner Index | |||
| Forest | 604.11 ± 38.87 | 14.67 ± 0.88 | 715.45 ± 36.27 | 6.42 ± 0.12 |
| Restored | 585.00 ± 19.86 | 5.67 ± 3.18 | 708.84 ± 21.18 | 5.89 ± 0.15 |
| Degraded | 666.89 ± 69.17 | 30.00 ± 14.80 | 769.73 ± 72.81 | 6.98 ± 0.17 |
| River-Type Comparison | ANOSIM | |
|---|---|---|
| R | p | |
| Forest vs. Degraded | 0.645 | 0.001 |
| Forest vs. Restored | 0.733 | 0.001 |
| Restored vs. Degraded | 0.256 | 0.008 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Q.; Sekar, R.; Marrs, R.; Zhang, Y. Effect of River Ecological Restoration on Biofilm Microbial Community Composition. Water 2019, 11, 1244. https://doi.org/10.3390/w11061244
Lin Q, Sekar R, Marrs R, Zhang Y. Effect of River Ecological Restoration on Biofilm Microbial Community Composition. Water. 2019; 11(6):1244. https://doi.org/10.3390/w11061244
Chicago/Turabian StyleLin, Qiaoyan, Raju Sekar, Rob Marrs, and Yixin Zhang. 2019. "Effect of River Ecological Restoration on Biofilm Microbial Community Composition" Water 11, no. 6: 1244. https://doi.org/10.3390/w11061244
APA StyleLin, Q., Sekar, R., Marrs, R., & Zhang, Y. (2019). Effect of River Ecological Restoration on Biofilm Microbial Community Composition. Water, 11(6), 1244. https://doi.org/10.3390/w11061244







