Selecting Bacteria Candidates for the Bioaugmentation of Activated Sludge to Improve the Aerobic Treatment of Landfill Leachate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Isolating the Bacteria from the AS and Soil
2.3. Sample Collection
2.4. Evaluating Catabolic Traits of the Bacterial Strains
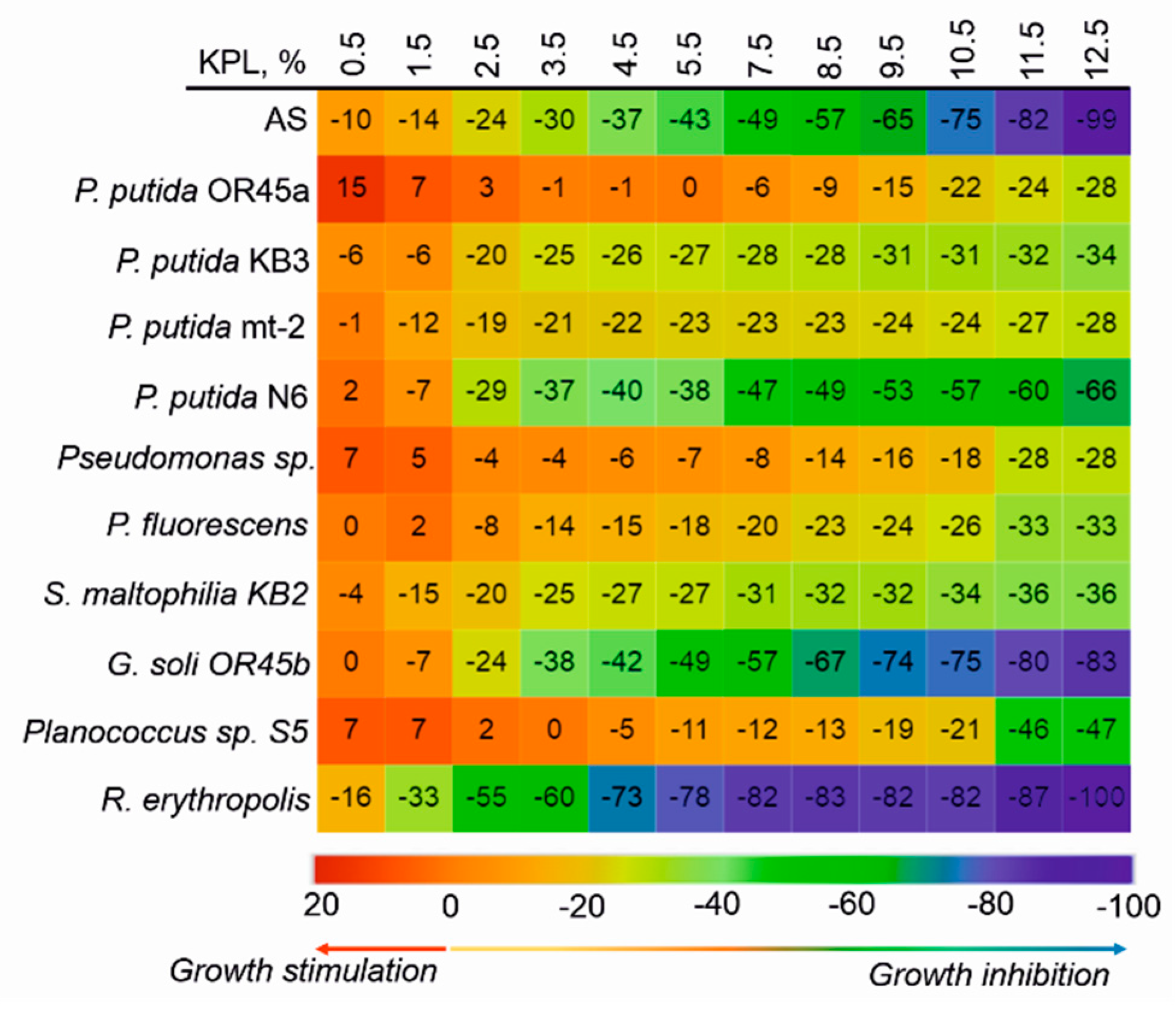
2.5. Acute Toxicity of the KPL
2.6. Evaluating the Auto-Aggregation and Co-Aggregation Ability of the Bacteria
2.7. Determining the Flocculation Activity Index (FAI)
2.8. Measuring the Hydrophobicity of the Bacterial Cell Surface and the AS
2.9. Qualitatively Assessing the N-Acyl Homoserine Lactone Signal Molecules and Exopolysaccharide Production
2.10. Detecting the Bacterial Motility and Chemotaxis
2.11. Qualitatively Screening the Biosurfactant Production and Determining the Emulsification Index
2.12. Qualitatively Assessing the Production of Siderophores
2.13. Determining the Non-Inhibitory Concentrations of Ions for the Bacteria
2.14. Whole Genome Sequencing of P. putida OR45a and P. putida KB3
2.15. Statistical Analysis
3. Results
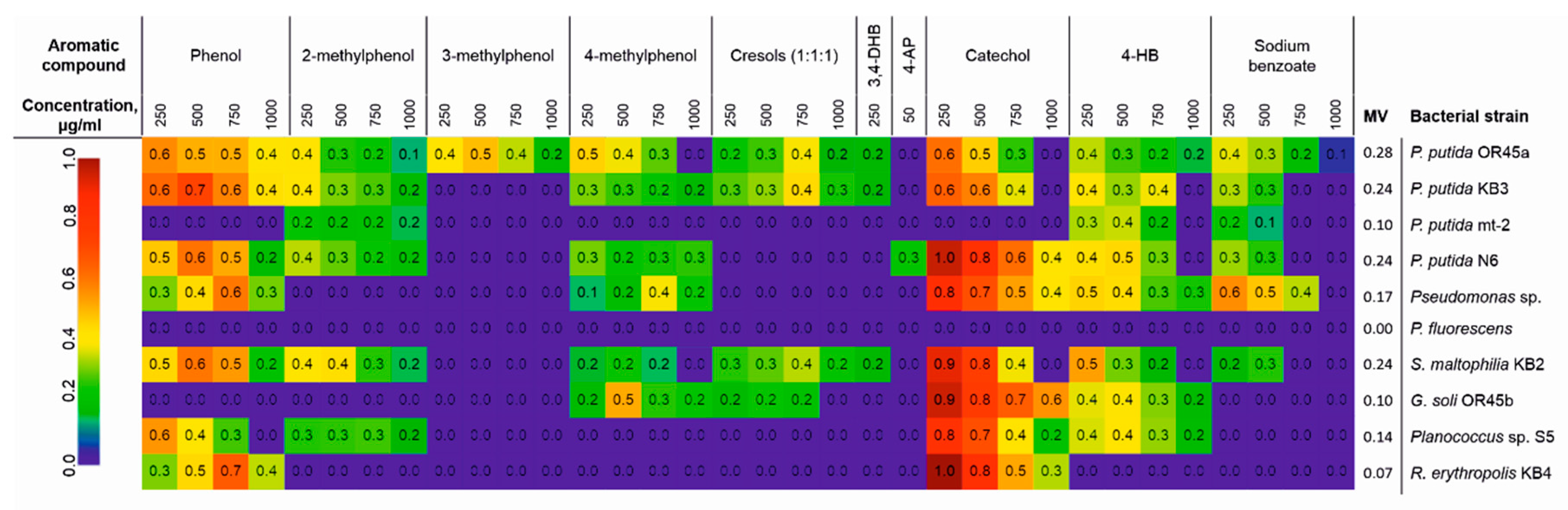
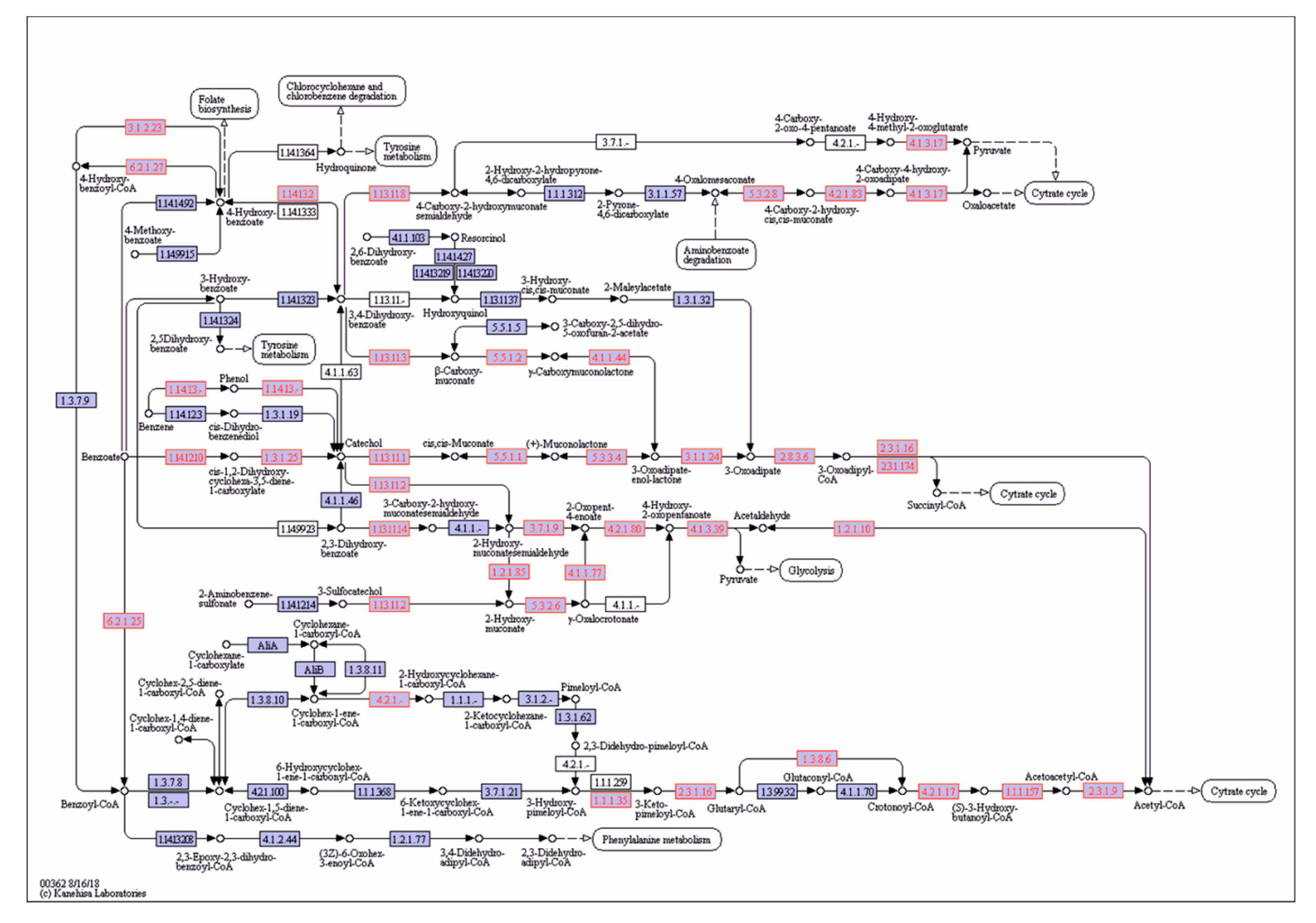
3.1. The Catabolic Potential of the Tested Bacteria and Their Ability to Grow in the Presence of the KPL
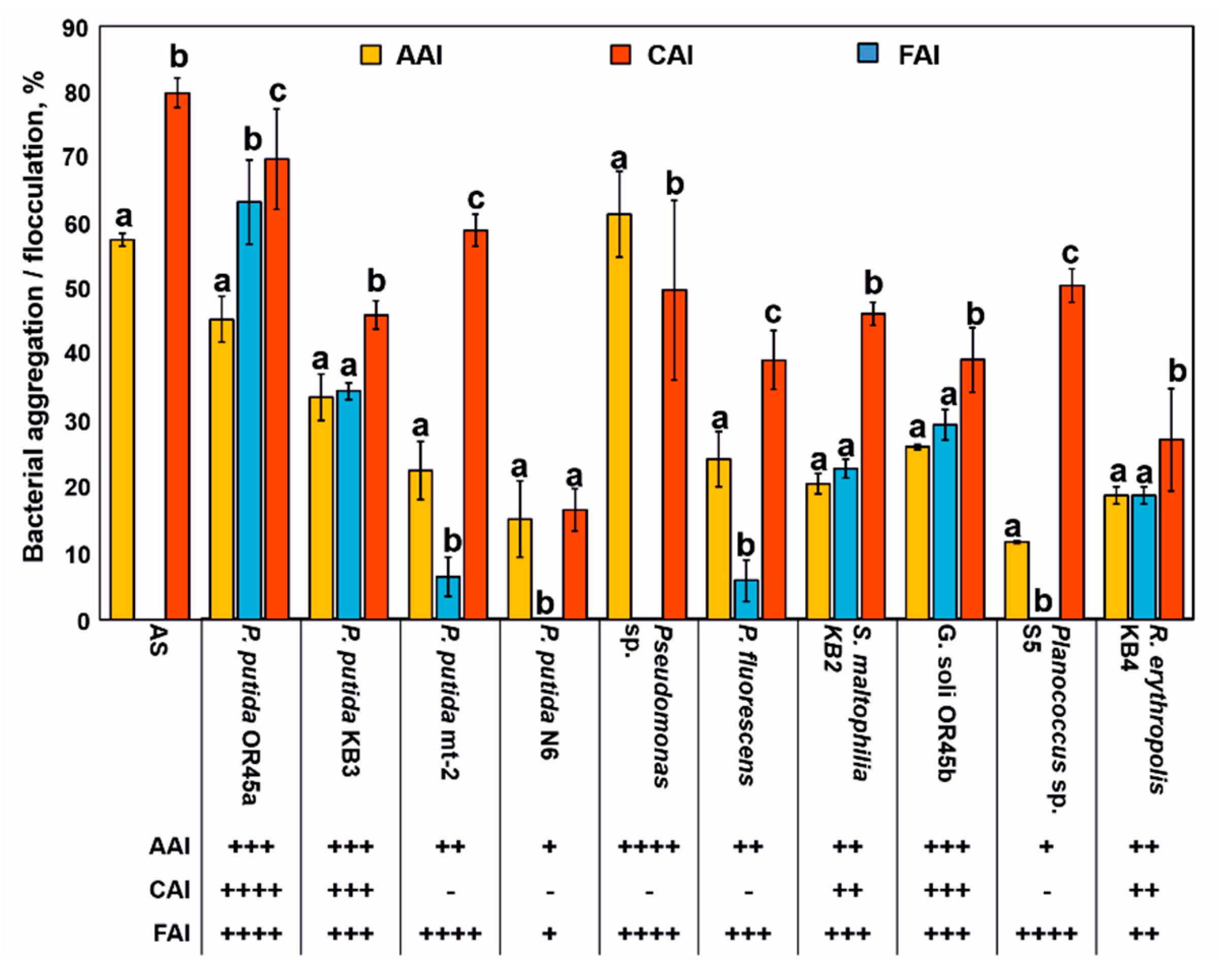
3.2. Ability of Bacterial Strains to Incorporate into the AS Structure
3.3. Biosurfactant and Siderophore Production by the Bacterial Strains
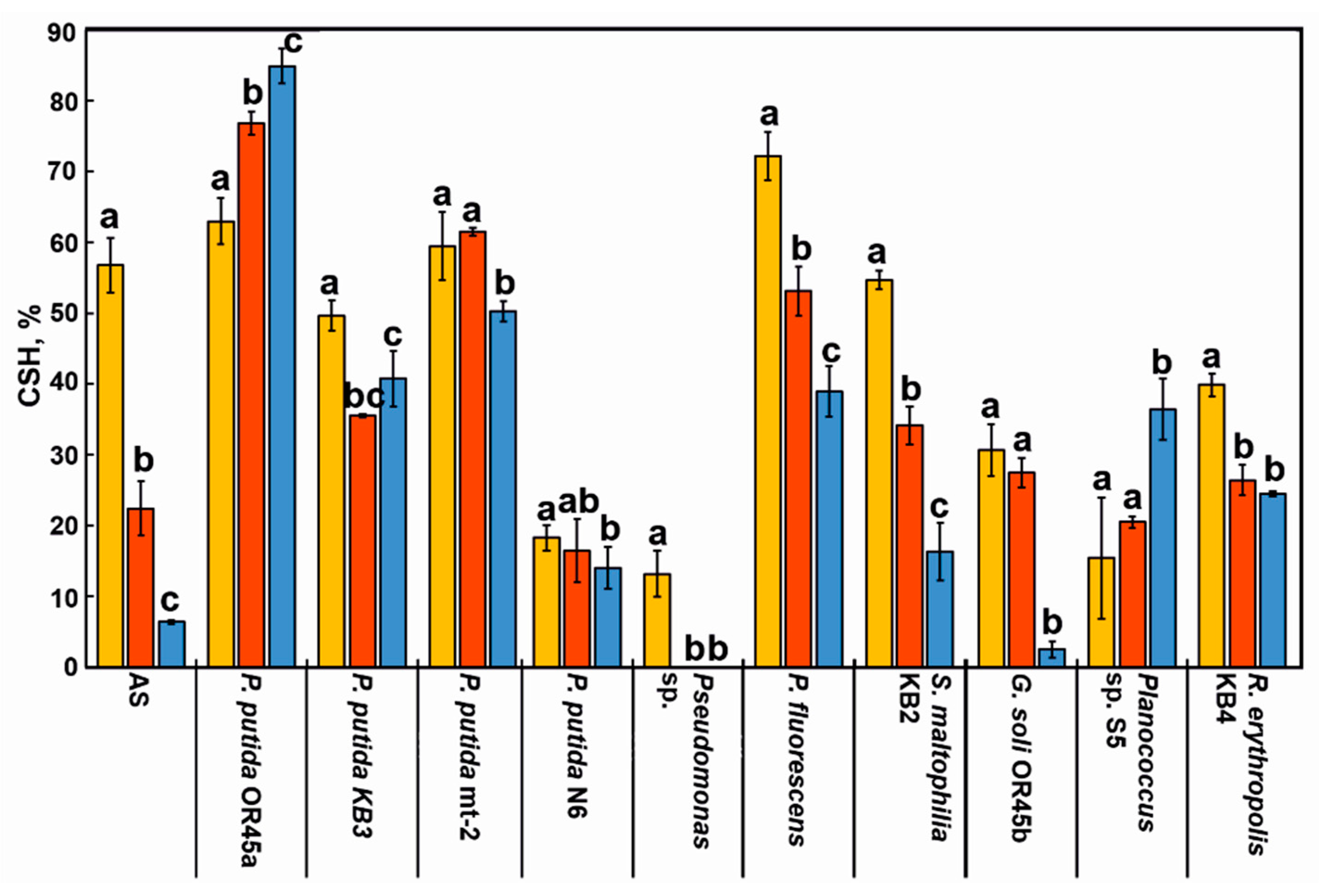
3.4. Cell Surface Hydrophobicity (CSH) of the Bacterial Strains and the AS
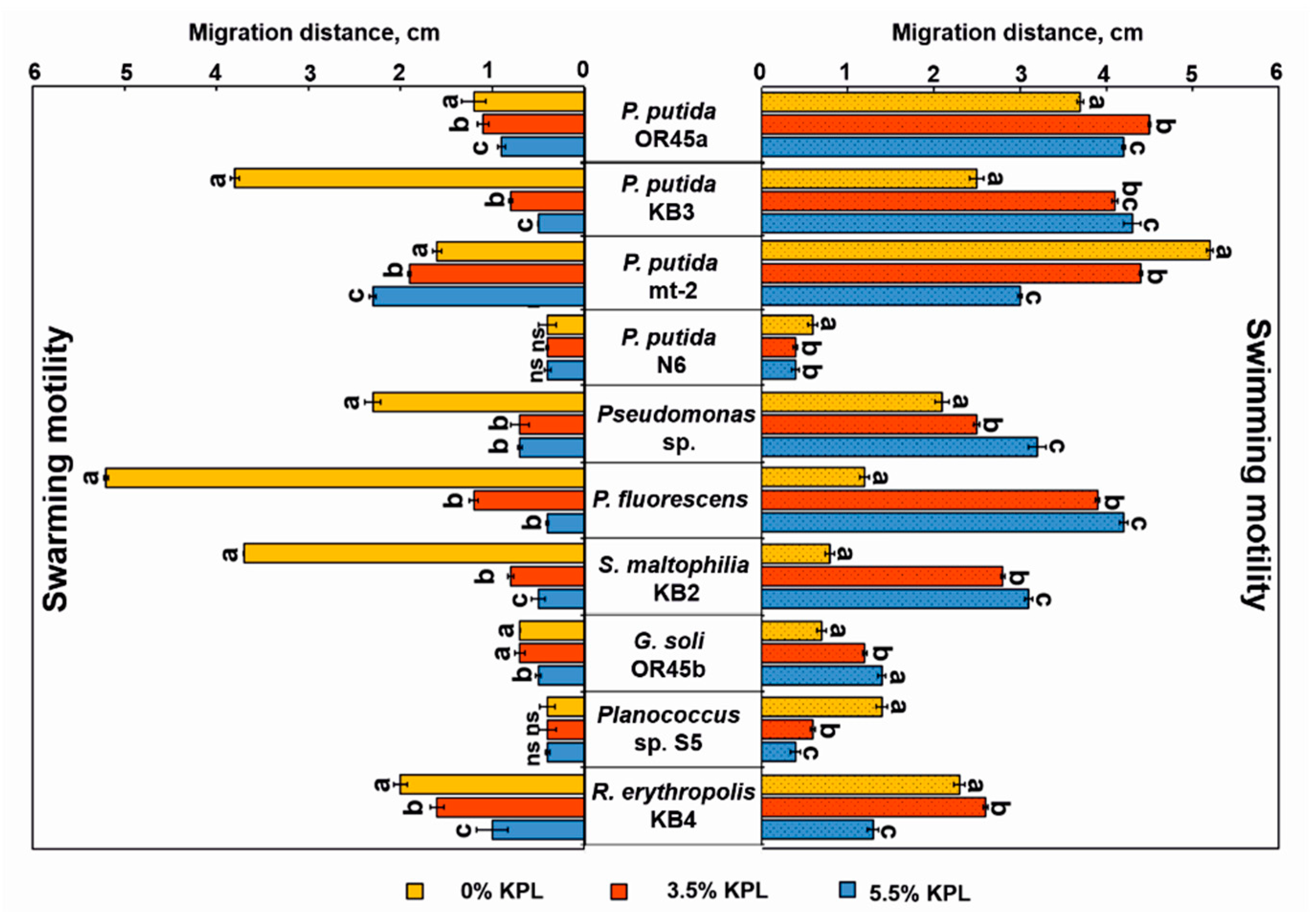
3.5. Bacterial Motility and Chemotaxis
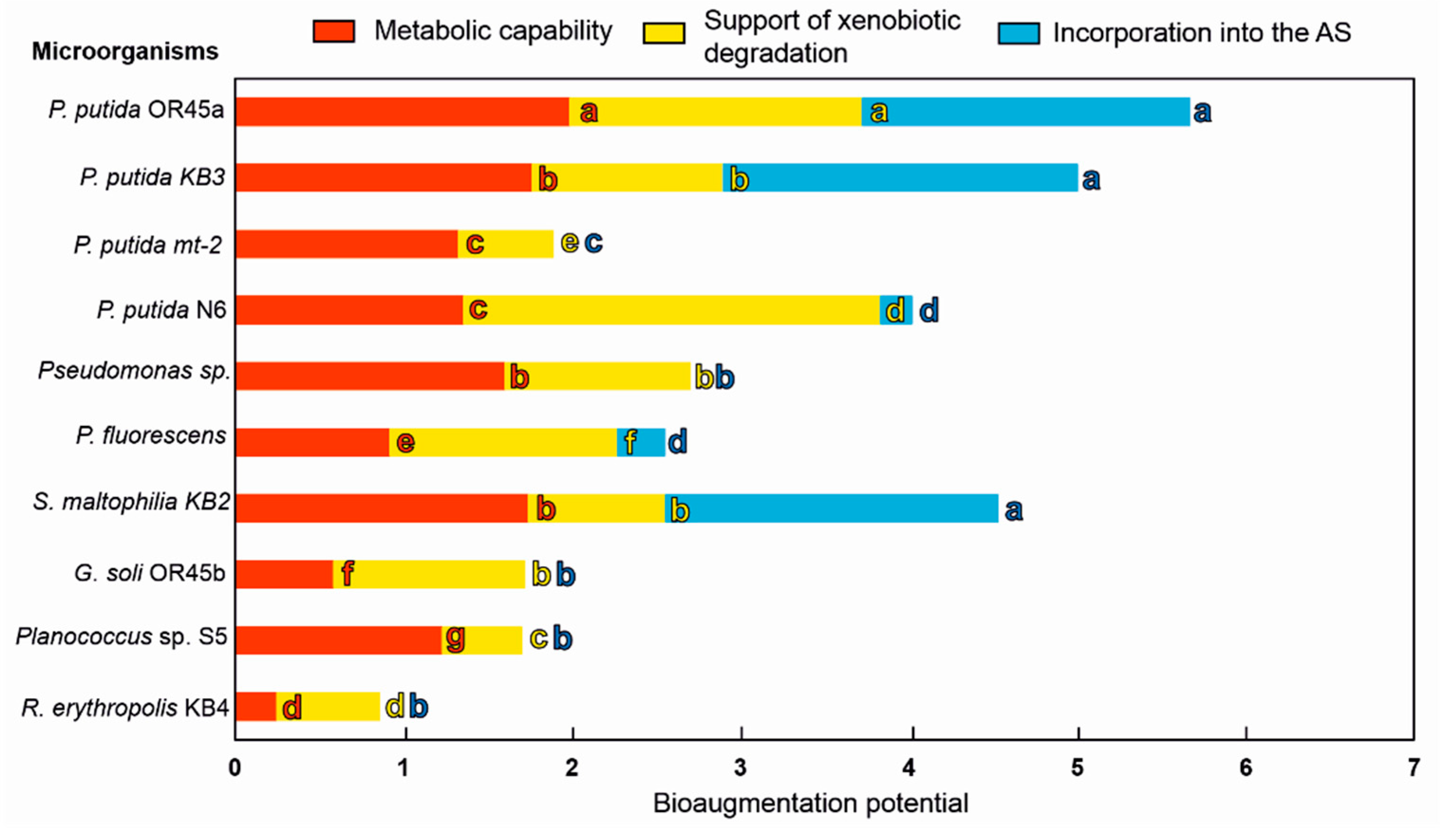
3.6. Selecting the Most Promising Candidates for Bioaugmentation
3.7. Non-Inhibitory Concentration (NIC) of the Selected Ions for P. putida OR45a and P. putida KB3
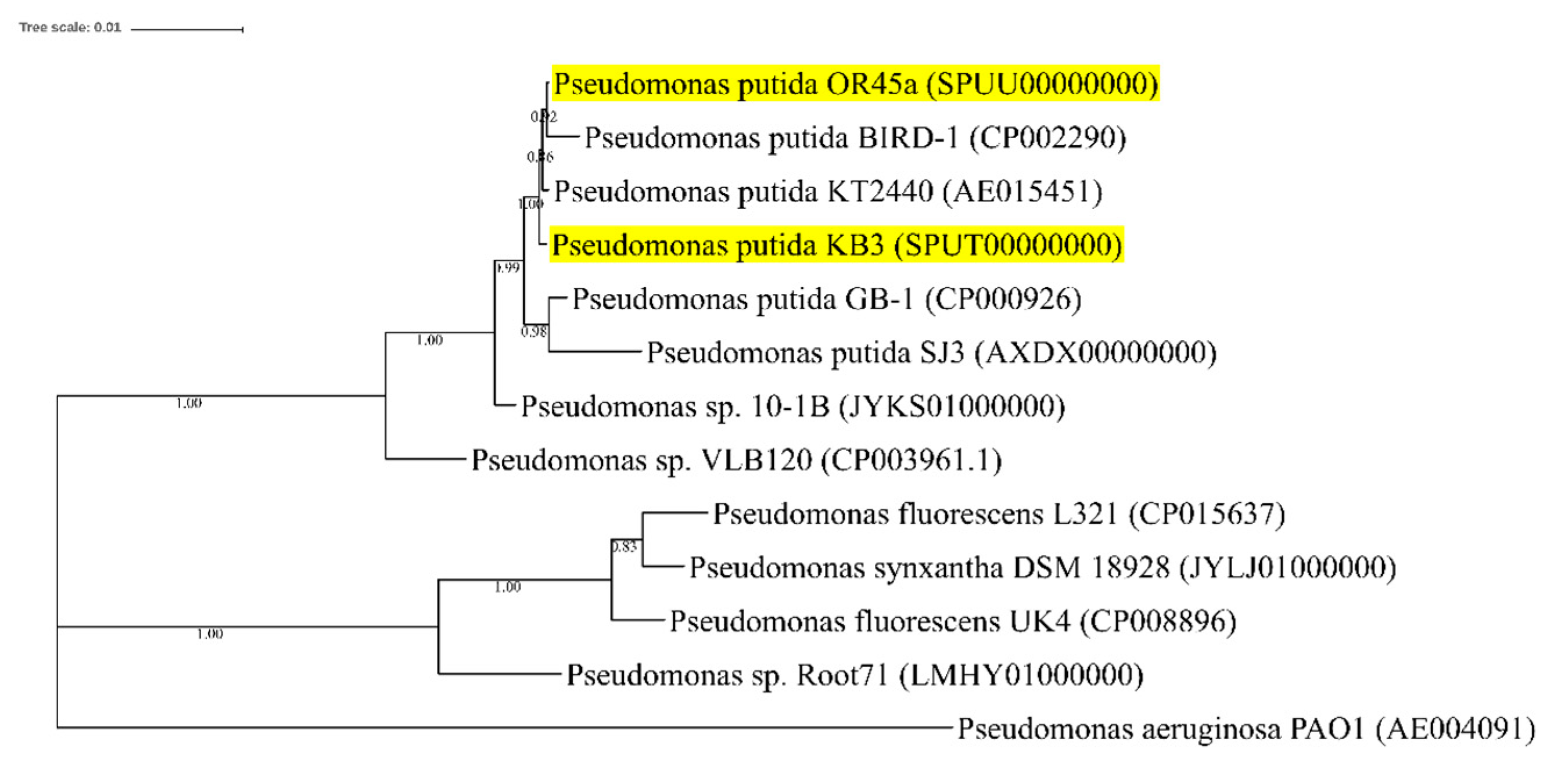
3.8. Genome-Guided Exploration of P. putida OR45a and P. putida KB3
4. Discussion
5. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schiopu, A.M.; Gavrilescu, M. Municipal solid waste landfilling and treatment of resulting liquid effluents. Environ. Eng. Manag. J. 2010, 9, 993–1019. [Google Scholar] [CrossRef]
- Baderna, D.; Caloni, F.; Benfenati, E. Investigating landfill leachate toxicity in vitro: A review of cell models and endpoints. Environ. Int. 2019, 122, 21–30. [Google Scholar] [CrossRef]
- Clifford, E.; Devroedt, C.; Morrison, L.; Healy, M.G. Treatment of landfill leachate in municipal wastewater treatment plants and impacts on effluent ammonium concentrations. J. Environ. Manag. 2017, 188, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, J.M.; Adeloju, S.B.; Ghosh, P.C. Landfill leachate: A promising substrate for microbial fuel cells. Int. J. Hydrog. Energy 2017, 42, 23794–23798. [Google Scholar] [CrossRef] [Green Version]
- Azzouz, L.; Boudiema, N.; Aouichat, F.; Kherat, M.; Mameri, N. Membrane bioreactor performance in treating Algiers’ landfill leachate from using indigenous bacteria and inoculating with activated sludge. Waste Manag. 2018, 75, 384–390. [Google Scholar] [CrossRef]
- Zheng, M.; Li, S.; Dong, Q.; Huang, X.; Liu, Y. Effect of blending landfill leachate with activated sludge on the domestic wastewater treatment process. Environ. Sci. Water Res. Technol. 2019, 5, 268–276. [Google Scholar] [CrossRef]
- Xie, B.; Xiong, S.; Liang, S.; Hu, C.; Zhang, X.; Lu, J. Performance and bacterial compositions of aged refuse reactors treating mature landfill leachate. Bioresour. Technol. 2012, 103, 71–77. [Google Scholar] [CrossRef]
- Öman, B.C.; Junestedt, C. Chemical characterization of landfill leachates–400 parameters and compounds. Waste Manag. 2008, 28, 1876–1891. [Google Scholar] [CrossRef]
- Naveen, B.P.; Mahapatra, D.M.; Sitharam, T.G.; Sivapullaiah, P.V.; Ramachandra, T.V. Physico-chemical and biological characterization of urban municipal landfill leachate. Environ. Pollut. 2017, 220, 1–12. [Google Scholar] [CrossRef]
- Wang, K.; Lusheng, L.; Tan, F.; Wu, D. Treatment of landfill leachate using activated sludge technology: A review. Archaea 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Capodici, M.; Di Trapani, D.; Viviani, G. Co-treatment of landfill leachate in laboratory-scale sequencing batch reactors: Analysis of system performance and biomass activity by means of respirometric techniques. Water Sci. Technol. 2014, 69, 1267–1274. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Q.; Jia, H.; Poveda, M. Study on the effect of landfill leachate on nutrient removal from municipal wastewater. J. Environ. Sci. 2016, 43, 150–158. [Google Scholar] [CrossRef]
- Ren, Y.; Ferraz, F.; Kang, A.J.; Yuan, Q. Treatment of old landfill leachate with high ammonium content using aerobic granular sludge. J. Biol. Eng. 2017, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Çeçen, F.; Tezel, U. Hazardous Pollutants in Biological Treatment Systems: Fundamentals and A Guide to Experimental Research; IWA Publishing: London, UK, 2018. [Google Scholar]
- Tsiligeorgis, J.; Zouboulis, A.; Samaras, P.; Zamoulis, D. Application of a membrane sequencing batch reactor for landfill leachate treatment. Desalination 2008, 221, 483–493. [Google Scholar] [CrossRef]
- Kalka, J. Landfill leachate toxicity removal in combined treatment with municipal wastewater. Sci. World J. 2012, 2012, 1–7. [Google Scholar] [CrossRef] [Green Version]
- De Albuquerque, E.M.; Pozzi, E.; Sakamoto, I.K.; Jurandyr, P. Treatability of landfill leachate combined with sanitary sewage in an activated sludge system. J. Water Process Eng. 2018, 23, 119–128. [Google Scholar] [CrossRef]
- Bolyard, S.C.; Reinhart, D.R.; Lozinski, D.; Motlagh, A.M. Leachate Dissolved Organic Matter Impacts on Wastewater Effluent Quality. In Proceedings of the Global Waste Management Symposium, Palm Springs, CA, USA, 11–14 February 2018. [Google Scholar]
- Michalska, J.; Greń, I.; Żur, J.; Wasilkowski, D.; Mrozik, A. Impact of the biological cotreatment of the Kalina pond leachate on laboratory sequencing batch reactor operation and activated sludge quality. Water 2019, 11, 1539. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Guo, J.; Zhao, L.; Chang, C.; Cui, D. Application of bioaugmentation to improve the activated sludge system into the contact oxidation system treating petrochemical wastewater. Bioresour. Technol. 2009, 100, 597–602. [Google Scholar] [CrossRef]
- Semrany, S.; Faver, L.; Djelal, H.; Taha, S.; Amrane, A. Bioaugmentation: Possible solution in the treatment of bio-refractory organic compounds (Bio-ROCs). Biochem. Eng. J. 2012, 69, 75–86. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, L.; Tao, X.; Hu, K.; Huang, S. Municipal wastewater treatment by the bioaugmentation of Bacillus sp. K5 within a sequencing batch reactor. PLoS ONE 2017, 12, e0178837. [Google Scholar] [CrossRef] [Green Version]
- Quan, X.; Shi, H.; Liu, H.; Wang, J.; Qian, Y. Removal of 2,4-dichlorophenol in a conventional activated sludge system through bioaugmentation. Process Biochem. 2004, 11, 1701–1707. [Google Scholar] [CrossRef]
- Songzhe, F.U.; Hongxia, F.A.N.; Shuangjiang, L.I.U.; Ying, L.I.U.; Zhipei, L.I.U. A bioaugmentation failure caused by phage infection and a weak biofilm formation ability. J. Environ. Sci. 2009, 21, 1153–1161. [Google Scholar] [CrossRef]
- Yu, F.B.; Ali, S.W.; Guan, L.B.; Li, S.P.; Zhou, S. Bioaugmentation of a sequencing batch reactor with Pseudomonas putida ONBA-17 and its impact on reactor bacterial communities. J. Hazard. Mater. 2010, 176, 20–26. [Google Scholar] [CrossRef]
- Fenu, A.; Donckels, B.M.R.; Beffa, T.; Bemfohr, C.; Weemaes, M. Evaluating the application of Microbacterium sp. strain BR1 for the removal of sulfamethoxazole in full-scale membrane bioreactors. Water Sci. Technol. 2015, 72, 1754–1761. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Yang, G.F.; Zhang, L.; Zhang, Z.Z.; Tian, G.M.; Jin, R.C. Bioaugmentation as a useful strategy for performance enhancement in biological wastewater treatment undergoing different stresses: Application and mechanisms. Crit. Rev. Environ. Sci. Technol. 2017, 47, 1877–1899. [Google Scholar] [CrossRef]
- Nguyen, P.Y.; Carvalho, G.; Reis, A.C.; Nunes, O.C.; Reis, M.A.M.; Oehmen, A. Impact of biogenic substrates on sulfamethoxazole biodegradation kinetics by Achromobacter denitrificans strain PR1. Biodegradation 2017, 28, 205–217. [Google Scholar] [CrossRef]
- Yu, Z.; Mohn, W.W. Bioaugmentation with the resin acid-degrading bacterium Zoogloea resiniphila DhA-35 to counteract pH stress in an aerated lagoon treating pulp and paper mill effluent. Water Res. 2002, 36, 2793–2801. [Google Scholar] [CrossRef]
- Monsalvo, V.M.; Tobajas, M.; Mohedano, A.F.; Rodriguez, J.J. Intensification of sequencing batch reactors by cometabolism and bioaugmentation with Pseudomonas putida for the biodegradation of 4-chlorophenol. J. Chem. Technol. Biotechnol. 2012, 87, 1270–1275. [Google Scholar] [CrossRef]
- Cirja, M.; Hommes, G.; Ivashechkin, P.; Prell, J.; Schäffer, A.; Corvini, F.X.; Lenz, M. Impact of bio-augmentation with Sphingomonas sp. strain TTNP3 in membrane bioreactors degrading nonylphenol. Appl. Microbiol. Biotechnol. 2009, 84, 183–189. [Google Scholar] [CrossRef] [Green Version]
- Xenofontos, E.; Tanase, A.M.; Stoica, I.; Vyrides, I. Newly isolated alkalophilic Advenella species bioaugmented in activated sludge for high p-cresol removal. New Biotechnol. 2016, 33, 305–310. [Google Scholar] [CrossRef]
- Ji, J.; Kakade, A.; Zhang, R.; Zhao, S.; Khan, A.; Liu, P.; Li, X. Alcohol ethoxylate degradation of activated sludge is enhanced by bioaugmentation with Pseudomonas sp. LZ-B. Ecotoxicol. Environ. Saf. 2019, 169, 335–343. [Google Scholar] [CrossRef]
- Herrero, M.; Stuckey, D.C. Bioaugmentation and its application in wastewater treatment: A review. Chemosphere 2015, 140, 119–128. [Google Scholar] [CrossRef]
- Singer, A.C.; van der Gast, C.J.; Thompson, I.P. Perspectives and vision for strain selection in bioaugmentation. Trends Biotechnol. 2005, 23, 74–77. [Google Scholar] [CrossRef]
- Guzik, U.; Greń, I.; Wojcieszyńska, D.; Łabużek, S. Isolation and characterization of a novel strain of Stenotrophomonas maltophilia possessing various dioxygenases for monocyclic hydrocarbon degradation. Braz. J. Microbiol. 2009, 40, 285–291. [Google Scholar] [CrossRef]
- Hupert-Kocurek, K.; Guzik, U.; Wojcieszyńska, D. Characterization of catechol 2,3-dioxygenase from Planococcus sp. strain S5 induced by high phenol concentration. Acta Biochim. Pol. 2012, 59, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Gąszczak, A.; Bartelmus, G.; Greń, I.; Janecki, D. Kinetics of vinyl acetate biodegradation by Pseudomonas fluorescens PCM 2123. Ecol. Chem. Eng. 2018, 25, 487–502. [Google Scholar] [CrossRef]
- Bae, H.S.; Lee, J.M.; Kim, Y.B.; Lee, S.T. Biodegradation of the mixtures of 4-chlorophenol and phenol by Camomonas testosteroni CPW301. Biodegradation 1996, 7, 463–469. [Google Scholar] [CrossRef]
- ISO 5667-13:2011. Water Quality—Sampling—Guidance on Sampling of Sludges; International Organization for Standardization: Geneva, Switzerland, 2011. [Google Scholar]
- ISO 19458:2007P. Water Quality—Sampling for Microbiological Analysis; International Organization for Standardization: Geneva, Switzerland, 2007. [Google Scholar]
- Malik, A.; Sakamoto, M.; Hanazaki, S.; Osawa, M.; Suzuki, T.; Tochigi, M.; Kakii, K. Coaggregation among nonflocculating bacteria isolated from activated sludge. Appl. Environ. Microbiol. 2003, 69, 6056–6063. [Google Scholar] [CrossRef] [Green Version]
- Kurane, R.; Hatamochi, K.; Kakuno, T.; Kiyohara, M.; Hirano, M.; Taniguchi, Y. Production of a bioflocculant by Rhodococcus erythropolis S-1 grown on alcohols. Biosci. Biotech. Biochem. 1994, 58, 428–429. [Google Scholar] [CrossRef]
- Zhang, C.L.; Lin, B.; Xia, S.Q.; Wang, X.J.; Yang, A.M. Production and application of a novel bioflocculant by multiple-microorganism consortia using brewery wastewater as carbon source. J. Environ. Sci. 2007, 19, 667–673. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Taghadosi, R.; Shakibbaie, M.R.; Masoumi, S. Biochemical detection of N-Acyl homoserine lactone from biofilm-forming uropathogenic Escherichia coli isolated from urinary tract infection samples. Rep. Biochem. Mol. Biol. 2015, 3, 56–61. [Google Scholar]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [Green Version]
- Déziel, E.; Comeau, Y.; Villemur, R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 2001, 183, 1195–1204. [Google Scholar] [CrossRef] [Green Version]
- Pacwa-Płociniczak, M.; Płociniczak, T.; Iwan, J.; Żarska, M.; Chorążewski, M.; Dzida, M.; Piotrowska-Seget, Z. Isolation of hydrocarbon-degrading and biosurfactant-producing bacteria and assessment their plant growth-promoting traits. J. Environ. Manag. 2016, 168, 175–184. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Poliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ. Sci. Pollut. Res. Int. 2014, 21, 9385–9395. [Google Scholar] [CrossRef] [Green Version]
- Lakshmanan, V.; Shantharaj, D.; Li, G.; Seyfferth, A.L.; Sherrier, J.D.; Bais, H.P. A natural rice rhizospheric bacterium abates arsenic accumulation in rice (Oryza sativa L.). Planta 2015, 4, 1037–1050. [Google Scholar] [CrossRef]
- MicrobesNG. Available online: https://microbesng.uk (accessed on 1 October 2019).
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acid Res. 2019, 47, 309–314. [Google Scholar] [CrossRef] [Green Version]
- BacMet—Antibacterial Biocide and Metal Resistance Genes Database. Available online: http://bacmet.biomedicine.gu.se/index.html (accessed on 1 October 2019).
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acid Res. 2013, 42, 737–743. [Google Scholar] [CrossRef] [Green Version]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Krepesi, C.; Bánky, D.; Grolmusz, V. AmphoraNet: The webserver implementation of the AMPHORA2 metagenomic workflow suite. Gene 2014, 533, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef] [PubMed]
- iTOL: Interactive Tree of Life. Available online: https://itol.embl.de/tree/15515823090118191539864834 (accessed on 1 October 2019).
- Matilla, M.A.; Pizarro-Tobias, P.; Roca, A.; Fernández, M.; Duque, E.; Molina, L.; Wu, X.; van der Lelie, D.; Gómez, M.L.; Segura, A.; et al. Complete genome of the plant growth-promoting rhizobacterium Pseudomonas putida BIRD-1. J. Bacteriol. 2011, 193, 1290. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, V.A.; Heim, S.; Moore, E.R.; Strätz, M.; Timmis, K.N. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004, 6, 1264–1286. [Google Scholar] [CrossRef]
- Kurata, Y.; Ono, Y.; Ono, Y. Occurrence of phenols in leachates from municipal solid waste landfill sites in Japan. J. Mater. Cycles Waste. 2008, 10, 144–152. [Google Scholar] [CrossRef]
- Kotowska, U.; Kapelewska, J.; Sturgulewska, J. Determination of phenols and pharmaceuticals in municipal wastewaters from Polish treatment plants by ultrasound-assisted emulsification-microextraction followed by GC-MS. Environ. Sci. Pollut. Res. 2013, 21, 1642–1652. [Google Scholar] [CrossRef] [Green Version]
- Joshi, H.; Dave, R.; Venugopalan, V.P. Pumping iron to keep fit: Modulation of siderophore secretion helps efficient aromatic utilization in Pseudomonas putida KT2440. Microbiology 2014, 160, 1393–1400. [Google Scholar] [CrossRef]
- Li, C.; Zhu, L.; Pan, D.; Li, S.; Xiao, H.; Zhang, Z.; Shen, X.; Wang, Y.; Long, M. Siderophore-mediated iron acquisition enhances resistance to oxidative and aromatic compound stress in Cupriavidus necator JMP134. Appl. Environ. Microbiol. 2019, 85, e01938-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chen, Y.; Zhang, X.; Wang, L. Aerobic granulation strategy for bioaugmentation of a sequencing batch reactor (SBR) treating high strength pyridine wastewater. J. Hazard. Mater. 2015, 15, 153–160. [Google Scholar] [CrossRef]
- Cheng, Z.; Meng, X.; Wang, H.; Chen, M.; Li, M. Isolation and characterization of broad spectrum coaggregating bacteria from different water systems for potential use in bioaugmentation. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.L.; Tay, J.H.; Maszenian, A.M.; Tay, S.T.L. Enhanced phenol biodegradation and aerobic granulation by two coaggregating bacterial strains. Environ. Sci. Technol. 2006, 40, 6137–6142. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.L.; Lv, Z.M.; Zhu, F.X.; Min, H.; Bian, C.M. Successful bioaugmentation of an activated sludge reactor with Rhodococcus sp. YYL for efficient tetrahydrofuran degradation. J. Hazard. Mater. 2013, 261, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Adav, S.S.; Lee, D.J.; Lai, J.Y. Intergeneric coaggregation of strains isolated from phenol-degrading aerobic granules. Appl. Microbiol. Biotechnol. 2008, 79, 657–661. [Google Scholar] [CrossRef]
- Di Gioia, D.; Fambrini, L.; Coppini, E.; Fava, F.; Barberio, C. Aggregation-based cooperation during bacterial aerobic degradation of polyethoxylated nonylphenols. Res. Microbiol. 2004, 155, 761–769. [Google Scholar] [CrossRef]
- Song, Z.; Ning, T.; Chen, Y.; Cheng, X.; Ren, N. Bioaugmentation of aerobic granular sludge with the addition of a bioflocculant-producing consortium. Adv. Mater. Res. 2013, 726, 2530–2535. [Google Scholar] [CrossRef]
- Guo, J.; Wang, J.; Cui, D.; Wang, L.; Ma, F.; Chang, C.C.; Yang, J. Application of bioaugmentation in the rapid start-up and stable operation of biological processes for municipal wastewater treatment at low temperatures. Bioresour. Technol. 2010, 110, 6622–6629. [Google Scholar] [CrossRef]
- Nie, M.; Yin, X.; Jia, J.; Wang, Y.; Liu, S.; Shen, Q.; Li, P.; Wang, Z. Production of a novel bioflocculant MNXY1 by Klebsiella pneumoniae strain NY1 and application in precipitation of cyanobacteria and municipal wastewater treatment. J. Appl. Microbiol. 2011, 111, 547–558. [Google Scholar] [CrossRef]
- Patil, S.V.; Patil, C.D.; Salunke, B.K.; Salunkhe, R.B. Studies on characterization of bioflocculant exopolysaccharide of Azotobacter indicus and its potential for wastewater treatment. Appl. Biochem. Biotechnol. 2011, 163, 463–472. [Google Scholar] [CrossRef]
- Buthelezi, S.P.; Olaniran, A.O.; Pillay, B. Textile dye removal from wastewater effluents using bioflocculants produced by indigenous bacterial isolates. Molecules 2012, 17, 14260–14274. [Google Scholar] [CrossRef] [Green Version]
- Tawila, Z.M.A.; Ismail, S.; Dadrasnia, A.; Usman, M.M. Production and characterization of a bioflocculant produced by Bacillus salmalaya 139SI-7 and its applications in wastewater treatment. Molecules 2018, 23, 2689. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Jiang, W.; Wang, X.; Ji, X.; Wang, Y.; Zhang, W.; Chen, J. Culture condition effect on bioflocculant production and actual wastewater treatment application by different types of bioflocculants. In Biodegradation and Bioremediation of Polluted Systems-New Advances and Technologies; Chamy, R., Rosenkranz, F., Soler, L., Eds.; IntechOpen: London, UK, 2015. [Google Scholar] [CrossRef] [Green Version]
- Gomaa, E.Z.; El-Meihy, R.M. Bacterial biosurfactant from Citrobacter freundii MG812314.1 as a bioremoval tool of heavy metals from wastewater. Bull. Natl. Res. Cent. 2019, 43, 69. [Google Scholar] [CrossRef]
- Wang, J.; He, H.; Wang, M.; Wang, S.; Zhang, J.; Wei, W.; Xu, H.; Lv, Z.; Shen, D. Bioaugmentation of activated sludge with Acinetobacter sp. TW enhances nicotine degradation in a synthetic tobacco wastewater treatment system. Bioresour. Technol. 2013, 142, 445–453. [Google Scholar] [CrossRef]
- Gnida, A.; Chorvatova, M.; Wanner, J. The role and significance of extracellular polymers in activated sludge. Part I: Literature review. Acta Hydrochim. Hydrobiol. 2006, 34, 411–424. [Google Scholar] [CrossRef]
- Hahn, M.W.; Moore, E.R.B.; Hofle, M.G. Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. MWH1. Microb. Ecol. 2000, 39, 175–185. [Google Scholar] [CrossRef]
- Sheng, G.; Yu, H.; Li, X. Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: A review. Biotechnol. Adv. 2010, 28, 882–894. [Google Scholar] [CrossRef]
- More, T.T.; Yadav, J.S.S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Extracellular polymeric substances of bacteria and their potential environmental applications. J. Environ. Manag. 2014, 144, 1–25. [Google Scholar] [CrossRef]
- Gupta, A.; Thakur, I.S. Study of optimization of wastewater contaminant removal along with extracellular polymeric substances (EPS) production by a thermotolerant Bacillus sp. ISTVK1 isolated from heat shocked sewage sludge. Bioresour. Technol. 2016, 213, 21–30. [Google Scholar] [CrossRef]
- Liu, X.W.; Sheng, G.P.; Yu, H.Q. Physicochemical characteristics of microbial granules. Biotechnol. Adv. 2009, 27, 1061–1070. [Google Scholar] [CrossRef]
- Fernandes, P.; Ferreiea, B.S.; Cabral, J.M.S. Solvent tolerance in bacteria: Role of efflux pumps and cross-resistance with antibiotics. Int. J. Antimicrob. Agents 2003, 22, 211–216. [Google Scholar] [CrossRef]
- Wick, L.Y.; Pasche, N.; Bernasconi, S.M.; Pelz, O.; Harms, H. Characterization of multiple-substrate utilization by anthracene-degrading Mycobacterium frederiksbergense LB501T. Appl. Environ. Microbiol. 2003, 69, 6133–6142. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, C.; da Cruz, A.; Pons, M.; Pinheiro, H.; Cabral, J.; da Fonseca, M.; Ferreira, B.S.; Fernandes, P. Mycobacterium sp., Rhodococcus erythropolis, and Pseudomonas putida behavior in the presence of organic solvents. Microsc. Res. Tech. 2004, 64, 215–222. [Google Scholar] [CrossRef]
- Chen, J.; Zhan, P.; Koopman, B.; Fang, G.; Shi, Y. Bioaugmentation with Gordonia strain JW8 in treatment of pulp and paper wastewater. Clean Technol. Environ. 2012, 14, 899–904. [Google Scholar] [CrossRef]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar] [CrossRef] [Green Version]
- Karmakar, R.; Bhaskar, R.V.S.U.; Jesudasan, R.E.; Tirumkudulu, M.S.; Venkatesh, K.V. Enhancement of swimming speed leads to a more-efficient chemotactic response to repellent. Appl. Environ. Microbiol. 2016, 82, 1205–1214. [Google Scholar] [CrossRef] [Green Version]
- Bouchez, T.; Patureau, D.; Delgenès, J.P.; Moletta, R. Successful bacterial incorporation into activated sludge flocs using alginate. Bioresour. Technol. 2009, 100, 1031–1032. [Google Scholar] [CrossRef]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Wang, L.; Chi, Z.M.; Liu, X.Y. Bacterial alginate role in aerobic granular bio-particles formation and settleability improvement. Sep. Sci. Technol. 2008, 43, 1642–1652. [Google Scholar] [CrossRef]
- Matz, C.; Bergfeld, T.; Rice, S.A.; Kjelleberg, S. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 2004, 6, 218–226. [Google Scholar] [CrossRef]
- Rojas, A.; Duque, E.; Mosqueda, G.; Golden, G.; Hurtado, A.; Ramos, J.L.; Segura, A. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 2001, 183, 3967–3973. [Google Scholar] [CrossRef] [Green Version]
- Stasinakis, A.S.; Mamais, D.; Thomaidis, N.S.; Lekkas, T.D. Effect of chromium (VI) on bacterial kinetics of heterotrophic biomass of activated sludge. Water Res. 2002, 36, 3341–3349. [Google Scholar] [CrossRef]
- Kumar, D.; Alappat, B.J. Evaluating leachate contamination potential of landfill sites using leachate pollution index. Clean Technol. Environ. 2005, 7, 190–197. [Google Scholar] [CrossRef]
- Li, X.Z.; Zhao, Q.L. Inhibition of microbial activity of activated sludge by ammonia in leachate. Environ. Int. 1999, 25, 961–968. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Yang, J.; Teng, F.; Feng, L.; Fang, X.; Ren, H. Bioaugmentation treatment of mature landfill leachate by new isolated ammonia nitrogen and humic acid resistant microorganism. J. Microbiol. Biotechnol. 2014, 24, 987–997. [Google Scholar] [CrossRef]
| Bacterial Strain | Site of Isolation | Selection Factor | Source |
|---|---|---|---|
| Pseudomonas putida OR45a | AS loaded with refinery wastewater, Jedlicze, Poland | Styrene | IBBEP |
| Glutamicibacter soli OR45b | AS loaded with refinery wastewater, Jedlicze, Poland | Styrene | IBBEP |
| Planococcus sp. S5 | AS loaded with municipal wastewater, Miechowice Bytom, Poland | Sodium salicylate | IBBEP |
| Stenotrophomonas maltophilia KB2 | AS loaded with municipal wastewater, Miechowice Bytom, Poland | Phenol | IBBEP |
| Pseudomonas putida N6 | AS loaded with municipal wastewater from a furniture factory, Jasienica Poland | Phenol | IBBEP |
| Pseudomonas putida KB3 | Uncontaminated, agricultural soil, Siewierz, Poland | Phenol + 4-chlorophenol Phenol + 2,4-dichlorophenol Phenol + 4-chlorophenol + 2,4-dichlorophenol | IBBEP |
| Rhodococcus erythropolis KB4 | Uncontaminated, agricultural soil, Siewierz, Poland | Phenol + 4-chlorophenol Phenol + 2,4-dichlorophenol Phenol + 4-chlorophenol + 2,4-dichlorophenol | IBBEP |
| Pseudomonas sp. (VTT E-93486) | AS from a peat biofilter bed | Styrene | VTT |
| Pseudomonas putida mt-2 (VTT E-022150) | nd | nd | VTT |
| Pseudomonas fluorescens (PCM 2123) | nd | nd | PCM |
| Characteristics | Capabilities Supporting Incorporation into the AS | |
|---|---|---|
| Bacterial Strain | N-AHLs | Exopolysaccharides |
| P. putida OR45a | + | + |
| P. putida KB3 | + | + |
| P. putida mt-2 (VTT E-022150) | + | + |
| P. putida N6 | − | − |
| Pseudomonas sp. (VTT E-93486) | + | − |
| P. fluorescens (PCM 2123) | + | + |
| S. maltophilia KB2 | + | + |
| G. soli OR45b | nd | − |
| Planococcus sp. S5 | nd | + |
| R. erythropolis KB4 | nd | − |
| Characteristics | Capabilities Supporting the Degradation of Pollutants in Wastewater | ||
|---|---|---|---|
| Bacterial Strain | Biosurfactant Production | Emulsification Index (E24), % | Siderophore Production |
| P. putida OR45a | + | 57 ± 8 | + |
| P. putida KB3 | + | 49 ± 3 | + |
| P. putida mt-2 (VTT E-022150) | + | 27 ± 5 | − |
| P. putida N6 | − | nd | − |
| Pseudomonas sp. (VTT E-93486) | − | nd | + |
| P. fluorescens (PCM 2123) | + | 16 ± 7 | + |
| S. maltophilia KB2 | + | 21 ± 3 | + |
| G. soli OR45b | − | nd | − |
| Planococcus sp. S5 | − | nd | − |
| R. erythropolis KB4 | − | nd | − |
| NIC, µg/mL | ||
|---|---|---|
| Ions | P. putida OR45a | P. putida KB3 |
| Zn2+ | 80.68 ± 2.50 | 81.15 ± 2.99 |
| Cu2+ | 128.43 ± 19.90 | 104.15 ± 3.56 |
| Cd2+ | 52.13 ± 2.48 | 59.97 ± 13.49 |
| Ba2+ | 172.63 ± 2.94 | 224.65 ± 11.58 |
| Cr6+ | 8.30 ± 0.59 | 11.30 ± 1.38 |
| Pb2+ | 150.00 ±1.29 | 50.00 ± 0.01 |
| NH3-N | 600.00 ± 52.30 | 900.00 ± 86.40 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalska, J.; Piński, A.; Żur, J.; Mrozik, A. Selecting Bacteria Candidates for the Bioaugmentation of Activated Sludge to Improve the Aerobic Treatment of Landfill Leachate. Water 2020, 12, 140. https://doi.org/10.3390/w12010140
Michalska J, Piński A, Żur J, Mrozik A. Selecting Bacteria Candidates for the Bioaugmentation of Activated Sludge to Improve the Aerobic Treatment of Landfill Leachate. Water. 2020; 12(1):140. https://doi.org/10.3390/w12010140
Chicago/Turabian StyleMichalska, Justyna, Artur Piński, Joanna Żur, and Agnieszka Mrozik. 2020. "Selecting Bacteria Candidates for the Bioaugmentation of Activated Sludge to Improve the Aerobic Treatment of Landfill Leachate" Water 12, no. 1: 140. https://doi.org/10.3390/w12010140
APA StyleMichalska, J., Piński, A., Żur, J., & Mrozik, A. (2020). Selecting Bacteria Candidates for the Bioaugmentation of Activated Sludge to Improve the Aerobic Treatment of Landfill Leachate. Water, 12(1), 140. https://doi.org/10.3390/w12010140








