Monitoring Bacterial Community Dynamics in Abalone (Haliotis discus hannai) and the Correlations Associated with Aquatic Diseases
Abstract
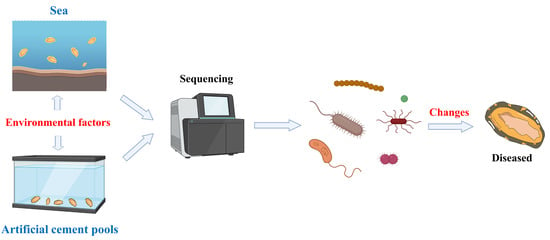
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. DNA Extraction and Purification
2.3. 454 Tag Amplicon Pyrosequencing for Analysis of Bacterial 16S rRNA Gene Sequences
2.4. Data Processing
2.5. Macrogenome Sequencing and Functional Annotation
2.6. Statistical Analyses
3. Results
3.1. Characterization of the Bacterial Community in Seawater Samples and the Intestine of Abalone
3.2. Analysis of the Bacterial Community of Seawater Samples and Abalone Intestine during Different Months
3.3. Comparison of the Number of OTUs in the Intestine of Abalone in Different Months
3.4. Relationship between the Bacterial Community and Environmental Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cook, P.A. Recent trends in worldwide abalone production. J. Shellfish Res. 2016, 35, 581. [Google Scholar] [CrossRef]
- Liang, S.; Luo, X.; You, W.; Ke, C. Hybridization improved bacteria resistance in abalone: Evidence from physiological and molecular responses. Fish Shellfish. Immunol. 2018, 72, 679–689. [Google Scholar] [CrossRef]
- Wang, R.X.; Liu, G.F.; Xu, L.W.; Wang, J.Y. Studies on the stripping off and death of larvaes of Haliotis diversicolor. Reeve Mar. Sci. 2007, 31, 53–57. [Google Scholar]
- Ma, Y.; Wang, Z.Y.; Zhang, Z.X.; Wang, L. Bacterial genetic diversity on settlement substrates during mass mortality of larvae of nona-porous abalone (Haliotis diversicolor supertexta). Aquaculture 2008, 282, 1–6. [Google Scholar] [CrossRef]
- Jo, Y.J.; Jung, K.H.; Lee, M.Y.; Choi, M.J. Effect of high-pressure short-time processing on the physicochemical properties of abalone (Haliotis discus hannai) during refrigerated storage. Inno. Food Sci. Emerg. Technol. 2014, 23, 33–38. [Google Scholar] [CrossRef]
- Wang, R.X.; He, J.; Wang, J.Y. Heterotrophic Bacterial Abundance and Diversity in the Farming Environment and Intestines of the Oyster Crassostrea hongkongensis. J. Shellfish Res. 2016, 35, 343–350. [Google Scholar] [CrossRef]
- Zhang, Q.; Qu, Q.; Lu, T.; Ke, M. The combined toxicity effect of nanoplastics and glyphosate on Microcystis aeruginosa growth. Environ. Pollut. 2018, 243, 1106–1112. [Google Scholar] [CrossRef]
- Lu, T.; Ke, M.J.; Peijnenburg, W.J.G.M.; Zhu, Y.C. Investigation of rhizospheric microbial communities in wheat, barley, and two rice varieties at the seedling stage. J. Agric. Food Chem. 2018, 66, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, L.; Zheng, B.; Zhu, Y. Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar. Pollut. Bull. 2013, 72, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Liu, T. Sediment enzyme activities and microbial community diversity in an oligotrophic drinking water reservoir, eastern China. PLoS ONE 2013, 8, e78571. [Google Scholar] [CrossRef] [PubMed]
- Zhi, E.; Song, Y.; Duan, L.; Yu, H.; Peng, J. Spatial distribution and diversity of microbial community in large-scale constructed wetland of the Liao River Conservation Area. Environ. Earth Sci. 2015, 73, 5085–5094. [Google Scholar] [CrossRef]
- Schreier, H.J.; Mirzoyan, N.; Saito, K. Microbial diversity of biological filters in recirculating aquaculture systems. Curr. Opin. Biotechnol. 2010, 21, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Blancheton, J.P.; Attramadal, K.J.K.; Michaud, L.; D’Orbcastel, E.R. Insight into bacterial population in aquaculture systems and its implication. Aquac. Eng. 2013, 53, 30–39. [Google Scholar] [CrossRef]
- Martins, P.; Cleary, D.F.R.; Pires, A.C.C.; Rodrigues, A.M. Molecular analysis of bacterial communities and detection of potential pathogens in a recirculating aquaculture system for Scophthalmus maximus and Solea senegalensis. PLoS ONE 2013, 8, e80847. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.X.; Wang, J.Y.; Sun, Y.C.; Yang, B.L. Antibiotic resistance monitoring in Vibrio spp. isolated from rearing environment and intestines of abalone Haliotis diversicolor. Mar. Pollut. Bull. 2015, 101, 701–706. [Google Scholar] [CrossRef]
- Wang, R.X.; Wang, A.; Wang, J.Y. Antibiotic resistance monitoring in heterotrophic bacteria from anthropogenic-polluted seawater and the intestines of oyster Crassostrea hongkongensis. Ecotoxicol. Environ. Saf. 2014, 109, 27–31. [Google Scholar] [CrossRef]
- Yashiro, E.; Spear, R.N.; Mcmanus, P.S. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J. Appl. Microbiol. 2011, 110, 1284–1296. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.F.; Ye, L. 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2011, 6, 1137–1147. [Google Scholar] [CrossRef]
- Brown, M.N.; Briones, A.; Diana, J.; Raskin, L. Ammonia-oxidizing archaea and nitrite- oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol. Ecol. 2013, 83, 7–25. [Google Scholar] [CrossRef]
- Xue, S.X.; Xu, W.; Wei, J.L.; Sun, J.S. Impact of environmental bacterial communities on fish health in marine recirculating aquaculture systems. Vet. Microbiol. 2017, 203, 34–39. [Google Scholar] [CrossRef]
- Wu, S.G.; Gao, T.H.; Zheng, Y.Z.; Wang, W.W. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2010, 303, 1–7. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, M.; Kita, Y.; Mizuno, A.; Goto-Yamamoto, N. Evaluation of method bias for determining bacterial populations in bacterial community analyses. J. Biosci. Bioeng. 2017, 124, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Brooker, M.R.; Dowd, S.E.; Camerlengo, T. Target Region Selection Is a Critical Determinant of Community Fingerprints Generated by 16S Pyrosequencing. PLoS ONE 2011, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, G.; Angert, E.R.; Wang, W.; Li, W.; Zou, H. Composition, Diversity, and Origin of the Bacterial Community in Grass Carp Intestine. PLoS ONE 2012, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, B.J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Meth. 2013, 95, 401–414. [Google Scholar]
- Wang, Y.H.; Pyecroft, S.; Barton, M. Seasonal microbial profiling of left kidney and stomach of farmed adult greenlip abalone (Haliotis laevigata). Aquac. Res. 2021, 52, 2085–2096. [Google Scholar] [CrossRef]
- Del’Duca, A.; Cesar, D.E.; Abreu, P.C. Bacterial community of pond’s water, sediment and in the guts of tilapia (Oreochromis niloticus) juveniles characterized by fluorescent in situ hybridization technique. Aquac. Res. 2015, 46, 707–715. [Google Scholar] [CrossRef]
- Giatsis, C.; Sipkema, D.; Smidt, H.; Heilig, H.; Benvenuti, G.; Verreth, J.; Verdegem, M. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci. Rep. 2015, 5, 18206. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Wang, K.; Wu, J.; Qiuqian, L.; Yang, K.; Qian, Y.; Zhang, D. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotechnol. 2015, 99, 6911–6919. [Google Scholar] [CrossRef]
- Ingerslev, H.C.; Strube, M.L.; Jørgensen, L.V.; Dalsgaard, I.; Boye, M.; Madsen, L. Diet type dictates the gut microbiota and the immune response against Yersinia ruckeri in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2014, 40, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hou, J.; Deng, M.; Liu, Q.; Wu, C.; Ji, Y.; He, X. Bacterial abundance and diversity in pond water supplied with different feeds. Sci. Rep. 2016, 6, 35232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAdam, P.R.; Richardson, E.J.; Fitzgerald, J.R. High-throughput sequencing for the study of bacterial pathogen biology. Curr. Opin. Microbiol. 2014, 19, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.J.; Lee, J.J.; Chung, H.Y.; Choi, S.H. Analysis of microbiota on abalone (Haliotis discus hannai) in South Korea for improved product management. Internet J. Food Microbiol. 2016, 234, 45–52. [Google Scholar] [CrossRef]
- Wang, J.H.; Lu, J.; Zhang, Y.X.; Wu, J. High-throughput sequencing analysis of the microbial community in coastal intensive mariculture systems. Aquac. Eng. 2018, 83, 93–102. [Google Scholar] [CrossRef]
- Pang, J.; Wang, Q.; Fei, Y.; Zhu, P.; Qiao, L.; Huang, H.; Dang, C.; Gao, W. A real-time recombinase polymerase amplification assay for the rapid detection of Vibrio harveyi. Mol. Cell Probes 2019, 44, 8–13. [Google Scholar] [CrossRef]
- Wang, R.X.; Yao, T.; Liu, X.J.; Wang, J.Y. Isolation and characterisation of Vibrio harveyi as etiological agent of foot pustule disease in the abalone Haliotis discus hannai Ino 1953. Indian J. Fish. 2018, 65, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, R.; Ootsubo, M.; Sawabe, T.; Ezura, Y. Kenichi TajimaBiodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture- independent techniques. Aquaculture 2004, 241, 453–463. [Google Scholar] [CrossRef]
- Zhao, Z.Y.; Chen, Y.P.; Li, Z.G.; Peng, W.J. Environmental factors have a strong impact on the composition and diversity of the gut bacterial community of Chinese black honeybees. J. Asia Pac. Entomol. 2018, 21, 261–267. [Google Scholar] [CrossRef]
- Wong, S.; Rawls, J.F. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 2012, 21, 3100–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abia, A.; Alisoltani, A.; Keshri, J.; Ubomba-Jaswa, E. Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Sci. Total Environ. 2018, 616–617, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Soriano, E.L.; Ramírez, D.T.; Araujo, D.R.; Gómez-Gil, B. Effect of temperature and dietary lipid proportion on gut microbiota in yellowtail kingfish Seriola lalandi juveniles. Aquaculture 2018, 497, 269–277. [Google Scholar] [CrossRef]
- Gattuso, J.P.; Magnan, A.; Bille, R.; Cheung, W.W.L.; Howes, E.L.; Joos, F.; Allemand, D.; Bopp, L.; Cooley, S.R.; Eakin, C.M.; et al. Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 2015, 349, aac4722. [Google Scholar] [CrossRef]
- Olivaa, R.D.P.; Vasquez-Lavín, F.; Martin, V.A.S.; Hernández, J.I.; Vargas, C.A.; Gonzalez, P.S.; Gelcich, S. Ocean Acidification, Consumers’ Preferences, and Market Adaptation Strategies in the Mussel Aquaculture Industry. Ecol. Econ. 2019, 158, 42–50. [Google Scholar] [CrossRef]
- Wessel, N.; Martin, S.; Badou, A.; Dubois, P.; Huchette, S.; Julia, V.; Nunes, F.; Harney, E.; Paillard, C.; Auzoux-Bordenave, S. Effect of CO2-induced ocean acidification on the early development and shell mineralization of the European abalone (Haliotis tuberculata). J. Exp. Mar. Biol. Ecol. 2018, 508, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Byrne, M.; Ho, M.; Wong, E.; Soars, N.A.; Selvakumaraswamy, P.; Shepard-Brennand, H.; Dworjanyn, S.A.; Davis, A.R. Unshelled abalone and corrupted urchins: Development of marine calcifiers in a changing ocean. Proc. R Soc. B 2011, 278, 2376–2383. [Google Scholar] [CrossRef]
- Guo, X.; Huang, M.; Pu, F.; You, W.; Ke, C. Effects of ocean acidification caused by rising CO2 on the early development of three mollusks. Aquat. Biol. 2015, 23, 147–157. [Google Scholar] [CrossRef]
- Chang, Y.J.; Chang, Y.T.; Hung, C.H.; Lee, J.W.; Liao, H.M.; Chou, H.L. Microbial community analysis of anaerobic bio-corrosion in different ORP profiles. Int. Biodeterior. Biodegrad. 2014, 95, 93–101. [Google Scholar] [CrossRef]
- Vezzulli, L.; Grande, C.; Reid, P.C.; Hélaouët, P.; Edwards, M.; Höfle, M.G.; Brettar, I.; Colwell, R.R.; Pruzzo, C. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. USA 2016, 113, E5062–E5071. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Zhu, H.; Wang, J.; Lin, X.; Wang, J.; Huang, Y.; Li, B.; Mou, H.; Ma, X.; Wang, R. Monitoring Bacterial Community Dynamics in Abalone (Haliotis discus hannai) and the Correlations Associated with Aquatic Diseases. Water 2022, 14, 1769. https://doi.org/10.3390/w14111769
Zhang T, Zhu H, Wang J, Lin X, Wang J, Huang Y, Li B, Mou H, Ma X, Wang R. Monitoring Bacterial Community Dynamics in Abalone (Haliotis discus hannai) and the Correlations Associated with Aquatic Diseases. Water. 2022; 14(11):1769. https://doi.org/10.3390/w14111769
Chicago/Turabian StyleZhang, Ting, Hui Zhu, Juan Wang, Xiaozhi Lin, Jiangyong Wang, Yisheng Huang, Bing Li, Hongli Mou, Xilan Ma, and Ruixuan Wang. 2022. "Monitoring Bacterial Community Dynamics in Abalone (Haliotis discus hannai) and the Correlations Associated with Aquatic Diseases" Water 14, no. 11: 1769. https://doi.org/10.3390/w14111769
APA StyleZhang, T., Zhu, H., Wang, J., Lin, X., Wang, J., Huang, Y., Li, B., Mou, H., Ma, X., & Wang, R. (2022). Monitoring Bacterial Community Dynamics in Abalone (Haliotis discus hannai) and the Correlations Associated with Aquatic Diseases. Water, 14(11), 1769. https://doi.org/10.3390/w14111769