Nanomaterial-Based Sensors for the Detection of Glyphosate
Abstract
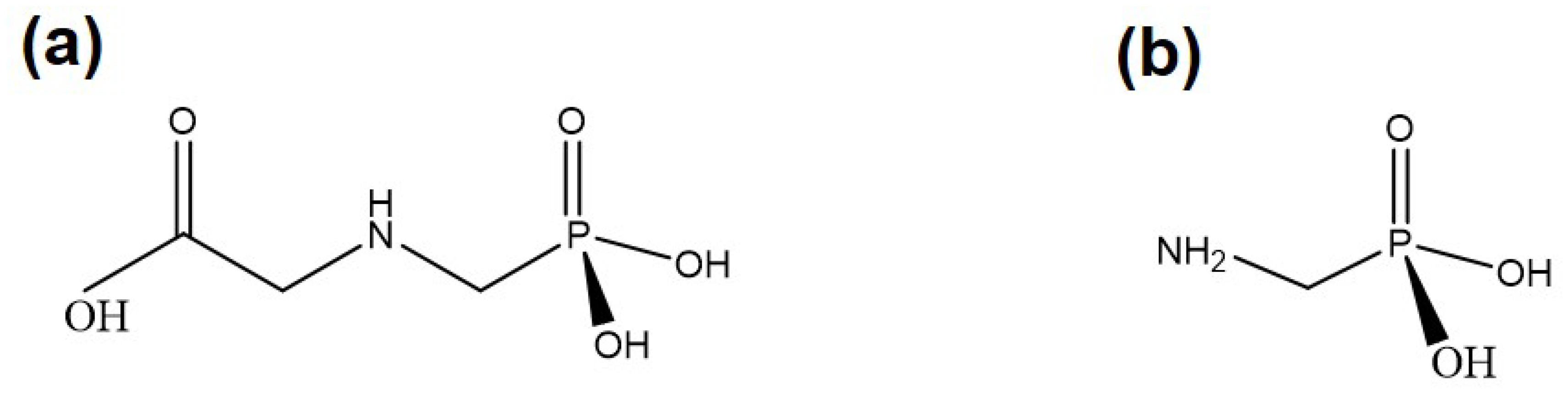
:1. Introduction
2. Glyphosate Detection Techniques
| Matrix | Analytical Technique | LOD | Linear Range | Reference |
|---|---|---|---|---|
| Groundwater | Solid-phase extraction followed by liquid chromatography coupled to tandem mass spectrometry (SPE-LC-MS/MS) | 18.9 pM | 0.3–3 nM | [58] |
| Tap water and irrigation water | Electrochemiluminescence | 0.2 mM | 0.2–16.6 mM | [59] |
| Tap water | High-performance liquid chromatography (HPLC) and ultraviolet spectroscopy | 0.4 µM | 29.6 µM–0.6 mM | [60] |
| Groundwater | Fluorescence spectroscopy | 0.25 µM | 0.59–2.96 µM | [61] |
| Potable, treated wastewater, urban, and groundwater | Spectrophotometric: 5-Phenyldipyrrinate of Nickel (II) | 0.20 µM | 0.59 µM–11 µM | [62] |
| Canal water | Liquid chromatography fluorescence (LC-FLD) + tandem mass spectrometry (MS-MS) | 0.6 nM | 0.6 nM–0.3 µM | [63] |
| Surface water | Chromatography-tandem mass spectrometry (LC-MS/MS) | 0.89 nM | 2.96–2957 nM | [64] |
| Pearl River water | Fluorescence | 47.3 nM | 59.1 nM–47.3 µM | [65] |
| Deionized water | Optical: prism coupling optical waveguide | 1.4 nM | 1.4–5.0 nM | [66] |
| Laser induced fluorescence (LIF) | 0.3 nM | 0.1 nM–5.0 µM | [67] | |
| Enzyme-linked immunosorbent assay (ELISA) | 0.6 nM | 3.2–4.5 nM | [68] | |
| Solid-phase extraction (SPE) Derivatized/Gas chromatography-flame photometric detection (GC-FPD) | 0.6 nM | 59.15–5915 nM | [69] | |
| Sequential-injection reversed-phase chromatography | 30 nM | 0.10–12.8 μM | [70] |
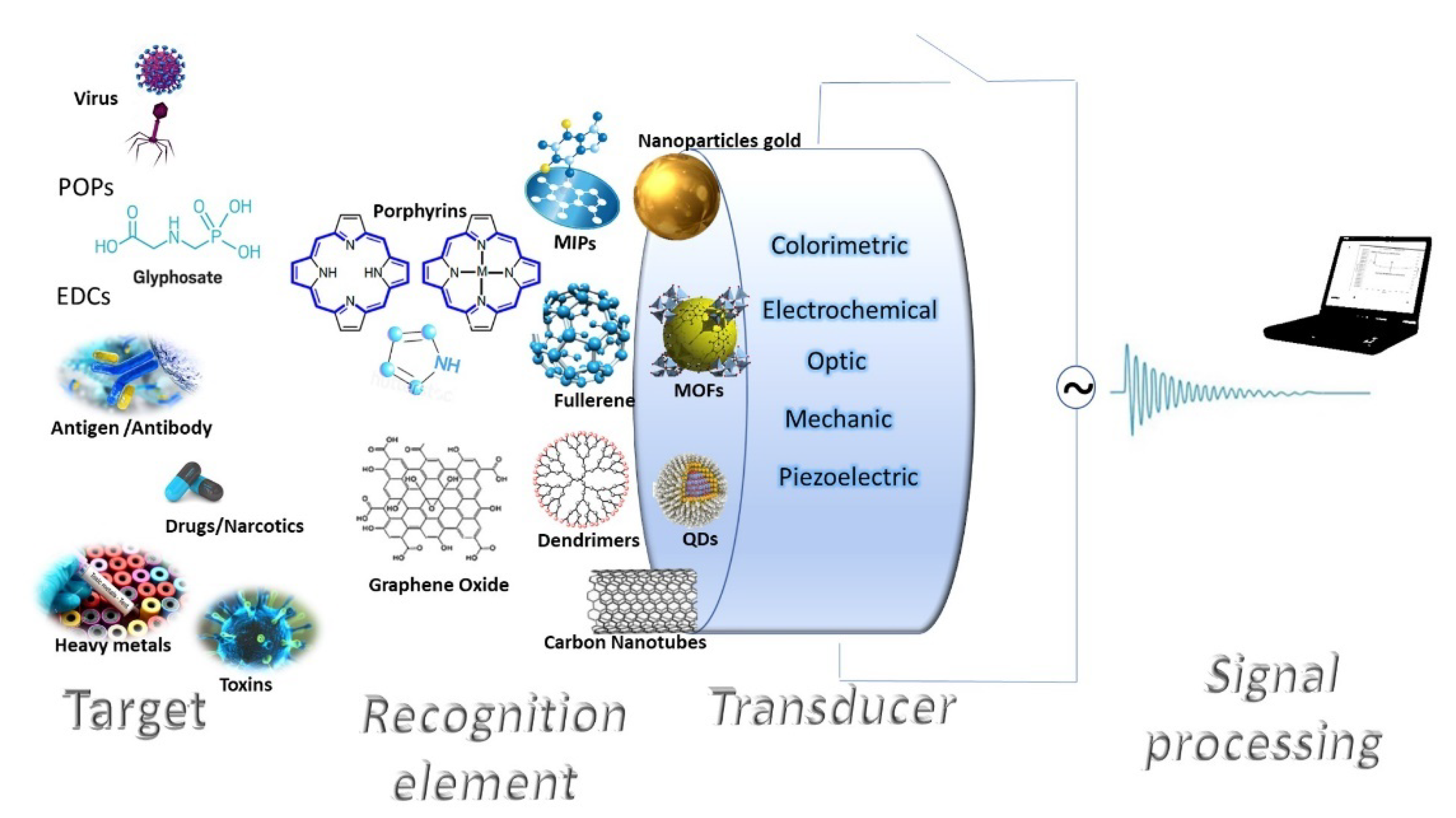
3. Nanomaterial-Based Sensors for Monitoring Pollutants

3.1. 0D Nanomaterials
3.1.1. Gold Nanoparticles (AuNPs)
3.1.2. Other Metallic Nanoparticles
| System | Detection Method (Transducer) | Linear Range | Detection Limit | Reference |
|---|---|---|---|---|
| Lu-AuNPs-Lcys-Cu (II) composites | Electrochemical luminescence | 0.001∼1.0 μM | 0.5 nM. | [102] |
| Organometallic osmium carbonyl clusters (10OsCO-AuNPs) | SERS | NR | 5.9 × 10−7 µM | [105] |
| AgNPs based on inner filter effect | Fluorometric | 0.15–15 µM | 0.07 µM | [106] |
| AgNPs | UV-Vis extinction spectra SERS | 0~30 µM | 6 µM 7.5 µM | [107] |
| GC/rGO-CuNPs | (DPV) Electrochemical | 0.1–1.1 µM | 0.19 µM | [108] |
| Polyethylenimine-capped NaGdF4:Yb,Er upconversion nanoparticles (UCNPs), copper (II), hydrogen peroxide, and 3, 3′, 5, 5′-tetramethylbenzidine | Fluorometric Colorimetric | 0.3–739 µM 30–739 µM | 0.06 µM 5.91µM | [114] |
| Agarose-guar gum entrapped bio-nanoconjugate of urease with AuNPs | Potentiometric | 2.96–296 µM | 2.96 µM | [117] |
3.1.3. Upconversion Nanoparticles (UCNPs)
3.1.4. 0D Biosensors
0D Nanomaterials with Antibodies
0D Nanomaterials with Aptamers
0D Nanomaterials with Enzymes
0D Nanomaterials with Other Biological Elements
3.1.5. Carbon Dots (CDs)
3.1.6. Quantum Dots (QDs)
3.2. 1D Nanomaterials
3.2.1. Carbon Nanotubes (CNTs)
3.2.2. Nanofibers (NFs)
3.2.3. Nanorods (NRs)
3.3. 2D Nanomaterials
3.3.1. Graphene and Graphene Analogs Such as Transition Metal Dichalcogenides and Transition Metal Oxide
| 1D Nanomaterials Nanocomposites | ||||
|---|---|---|---|---|
| Detection Method (Transducer) | Linear Range | Detection Limit | Reference | |
| Cu2+-Cu/GC | Electrochemical | 0.4–10 μM | 0.186 μM | [110] |
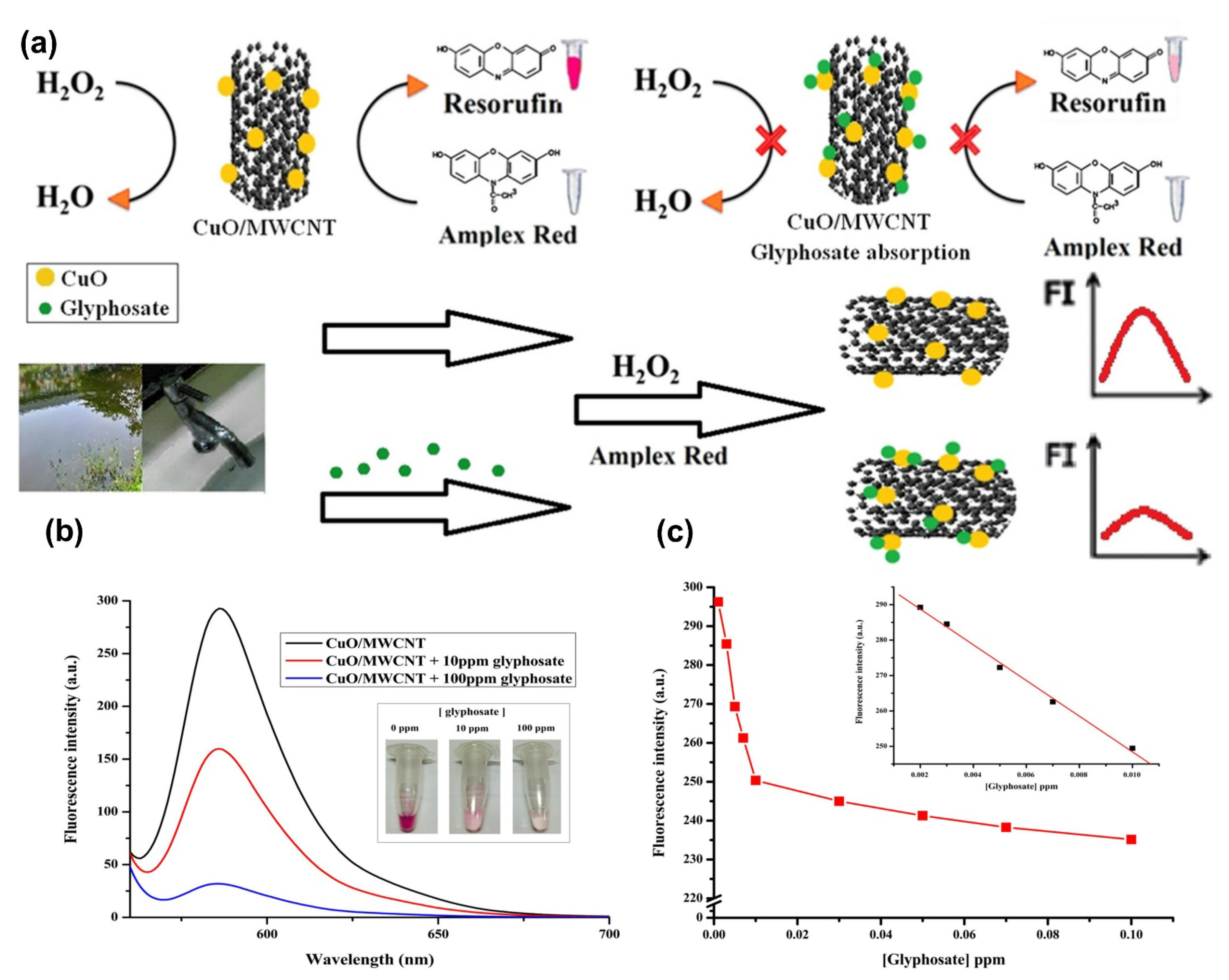
| Inhibiting the catalytic activity of the CuO/MWCNTs | Turn-off fluorescence | 0.012–0.06 µM | 3.96 × 10−6 µM | [112] |
| Carbon dot labeled antibodies (lgG-CDs) magnetic nanoparticles Fe3O4 and GLYP utilized to couple with the excess lgG-CDs | Immune Reaction | 0.06–473 µM | 0.05 µM | [127] |
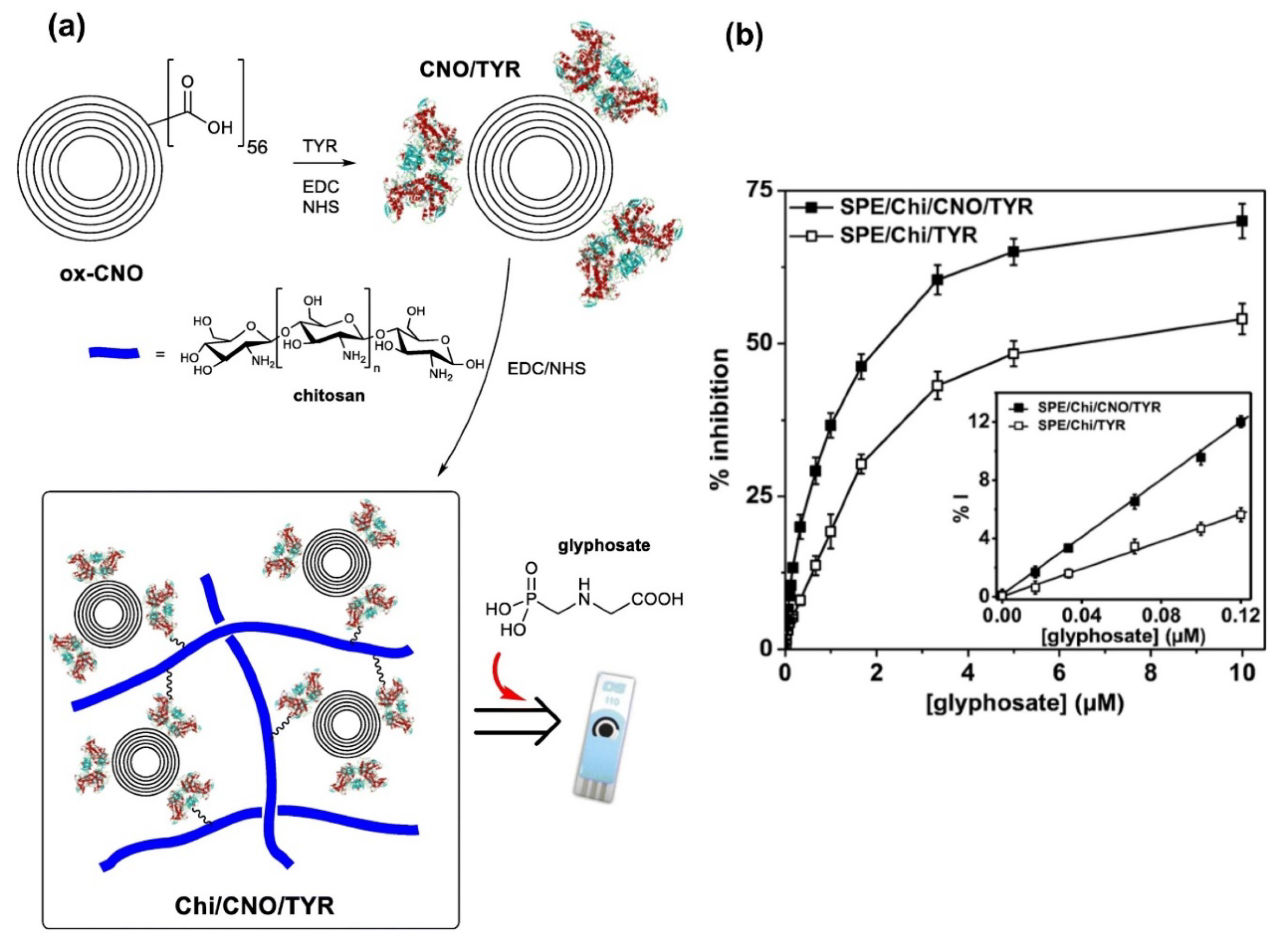
| Carbon nano-onions (CNOs) | Tyrosinase based Amperometric | 0.015–10 μM | 6.5 nM | [150] |
| CdTe-CQD | Photoluminescence (PL) Fluorometric | 0–1000 nM | 2 pM | [157] |
| Pencil graphite electrode modified by hollow fiber pregnant by MWCNTS-ionic liquid composite and CuONPs | Electrochemical | 5 nM–1.1 μM | 1.3 nM | [160] |
| MWCNTs decorated with nano-ZnO. Nano-ZnO | Spectro electrochemical and electrochemical detection | GLYP: 0–100 µM AMPA: 30–100 µM | GLYP:1 μM AMPA:10 µM | [162] |
| GE/MWCNTs-HRP | Electrochemical | 0–4.5 mM | 1.32 pM | [173] |
| Carbon paste electrode (spectroscopic-grade graphite powder) | Electrochemical | 0.044–2.8 µM | 2 × 10−3 µM | [174] |
| Fluorescent CDs | Turn-on Fluorescence | 0.18–59 µM | 0.09 µM | [175] |
| Fluorescent CDs | FRET | 0.02–2.0 µM. | 0.6 µM | [176] |
| CDs | Fluorescent quenching | 1.5 × 10−3–30 µM | Diazinon: 1.5 × 10−3 µM GLYP: 0.012 µM | [177] |
| GQDs-AgNPs system | Fluorometric Luminescence probe | 0.18–11.83 µM | 0.05 µM | [178] |
| CdTe-quantum dots | Fluorometric | 10–118 µM | 3 µM | [179] |
| 2D Nanomaterials Nanocomposites | ||||
| Peroxidase Mimetic Activity of MoS2 Nanosheets | Colorimetric | 2.4–12 µM | 0.51 µM | [171] |
| Ultrathin two-dimensional metal-organic framework nanosheets, decorated with tetra-pyridyl calix[4]arene (MOF-Calix) | Fluorescence | 2.5–45 μM | 2.25 μM | [180] |
| GLYP/Ru(bpy)32+system on gold electrodes modified with SAM. | ECL | 0–100 µM | 0.01 µM | [181] |
| Calixarene-functionalized luminescent silica nanoparticles [Ru(bpy)3] 2+ complex. | FRET | 0–2 µM | 0.8 µM | [182] |
| 3D Nanomaterials Nanocomposites | ||||
| Nanoporous Copper film (microelectrode) | Electrochemical | 0.030–0.065 μM | 4 × 10−3 µM | [111] |
| Nanobody (CP4-EPSPS protein) and Mesoporous Carbon | Electrochemical immunosensor | 6 × 10−6–0.6 µM | 4.3 × 10−6 µM | [123] |
| ZnS-QDs on ordered mesoporous carbons substrate | HRP and ECL | 0.1 nM–10 mM | NR | [183] |
3.3.2. Meso-/Nanoporous
Metal-Organic Frameworks (MOFs)
3.4. 3D Nanomaterials Such as Nanocomposites in Sensors for GLYP Detection
Nanocomposites
| Metal-Organic Framework (MOFs) System Nanomaterial | ||||
|---|---|---|---|---|
| Detection Method (Transducer) | Linear Range | Detection Limit | Reference | |
| Hierarchically porous Cu-BTC MOF platform | Electrochemical | 1.0 × 10−6–0.01 µM and 0.01~1000 µM | 1.4 × 10−7 µM | [73] |
| (CuOx@mC/GCE) mesoporous with MOF | Electrochemical | 1.0 × 10−9–100 µM | 7.69 × 10−10 µM | [198] |
| Molecularly Imprinted Polymer (MIP) Nanomaterial System | ||||
| PAP-MIP-MOF films- Gly as template molecule/ (AuNPS) / Gold electrode | Electrochemical | 6x10−9–6 × 10−3 µM | 5 fM | [100] |
| Coumarin-based ligand (CL) quenched by Cu2+ due to the process of photoinduced electron transfer. | Fluorescence | 0.12–8.87 µM | 0.11 µM | [113] |
| antibody-modified magnetic particles, using TMB as an enzymatic substrate. | Electrochemical immunoassay (competitive) | 0–0.06 µM | 3 × 10−5 µM | [129] |
| Fe3O4/molecular-imprinted nanocomposite | Electrochemical | NR | 10 µM | [201] |
| Ppy matrix/molecules GLYP templates | Electrochemical | NR | 1 × 10−7 µM | [208] |
| Molecularly imprinted mesoporous organosilica (MIMO)/QD-encapsulated GLYP imprinted mesoporous organosilica | Fluorescence | 0 nM–800 µM | 0.1 nM | [209] |
| Ppy-MIP | Gravimetric a and electrochemical b | 1 pM–1 nM | 1 pM | [51] |
| Ppy-MIP | Electrochemical | 0.03–4.73 µM | 1.6 µM | [213] |
| Composite AuNPs-Ppy -MIP on the surface of the ITO electrode | Voltammetric | 2.4–7.1 µM | 0.5 µM | [214] |
| Disubstituted polyacetylenes (TZP and PBP) with Cu ions | Fluorescence | NR | 0.08 µM | [215] |
| Poly(2,5dimethoxyaniline) (PDMA) doped with poly (4-styrenesulfonic acid) (PSS)/ HRP | Amperometric | NR | 0.09 µM | [216] |
| Molecularly imprinted polymer (MIPs) made of chitosan (CS) | EIS | 2 × 10−6–0.3 µM | 6 × 10−9 µM | [217] |
4. Nanosensors Performance Using Real Samples
| Sample Matrix | Interferences | Accuracy (Range/AVERAGE Recovery) | Precision | Reference |
|---|---|---|---|---|
| Tap water samples | AMPA | 98.7–102.6% | 3.33–4.54% | [100] |
| Water samples, including local river water and lake water. | Deltamethrin, acetamiprid, chlorpyrifos, carbendazim | 98.9–105 %. | 3.59–6.52% | [102] |
| Tap water | Pyrethroid cypermethrin and deltamethrin, diazinon and malathion and mevinphos | 96–104% | NR | [108] |
| River water samples | Simazine, propazine, and atrazine, AMPA, Na+, K+, Ni2+, Ca2+, Mg2+, | 98.7–105.2% | 1.58–4.76% | [111] |
| Real water samples from the Taitung Flowing Lake in the Taitung Forest Park and the tap water of school | Chlorothalonil,Cyanofenphos, Propanil, Chlorpyrifos, Carbendazim, Acetamiprid, Fenvalerate, Carbaryl, Dimethoate | 96–107% | 1.6–4.1% | [112] |
| Real samples river and lake water | Dimethoate, malathion, fenitrothion, carbendazim, fluazinam, chlorpyrifos, triadimefon, trichlorfon, and methamidophos. | 91.31–105.28% | 1.10–3.48% | [113] |
| Tap water and rice | K+, Na+, Ca2+, Zn2+, Mg2+, Fe3+, Pb2+, Ni2+, Co2+, Cr2+, Ag+, Hg2+, and Cd2+, trichlorfon, profenofos, malathion, fenitrothion, chlorpyrifos, glyphosate. ascorbic acid and glutathione | Cu2+ = 92.41–108.38% GLYP = 89.87–109.39% | NR | [116] |
| Tap water | Dichlorvos, dimethoate, 2–4D, paraquat dichloride, hexaconazole | 86% | 6% | [117] |
| Pearl River water, tea, and soil | Na+, K+, NH4+, NO3−, PO43−, F−, Mg2+, Zn2+, Ca2+, and Fe3+ pmida, glyphosine, omethoate, phosmet | 87.4–103.7%. | 4.67% | [127] |
| River water soil | Zn2+, Cd2+, Ca2+, Mg2+, Na+, NH4+, Br−, NO3−, SO42–,PO43−, Glufosinate. Bialaphos, Tridemorph, Chlorpyrifos, Cypermethrin (Aminomethyl) phosphonic acid | 92.19–103.25 % | 4.3–5.10% | [160] |
| Environmental water samples | Na+, K+, Mg2+, Ca2+, NO−3, CO32−, SO42−, dimethoate, isocarbophos, phosalone carbaryl, bendiocarb | 99–108% | NR | [171] |
| Water samples collected from Qing Lake, Guanlan Lake and Yan Lake (Changchun, China). | Na+,K+,Mg2+,Ca2+,Ba2+,Zn2+,Ag+,Cd2+, Fe3+,Hg2+,Mn2+ Pb2+, Hg2+, Cd2+,Fe3+, Ag+, Ametryn, Metsulfuron-methyl, Metaflumizone, Dinotefuran, Chlorfenapyr,Carbendazim, Pymetrozine, Imidacloprid, Chlorothalonil,Chlorpyrifos, Glufosinate | 93.3–106.7% | 2.1% | [175] |
| Environmental water samples | NaCl, KCl, CaCl2, NH4Cl, MgCl2, CdCl2, ZnSO4, ascorbic acid, pyridoxine, glycine, lysine, aspartic acid, arginine glucose, sucrose, maltose, quercetin, puerarin, trifluralin, dicamba, acetochlor, atrazine, and AMPA | 93.7–102.6% | 1.7–3.3%, | [176] |
| Environmental simples water and cereal samples (amaranth, barley, oat, and quinoa). | K+, Na+, Cl−, NO3- Mg2+, PO43−, CO32−, SO42− Ni2+ Carbaryl, carbendazim, fipronil, imidacloprid, malathion, nitenpyram, o-phenylphenol, pyraclostrobin, thiabendazole, thiacloprid, thiamethoxam | 92–108% | 3.7% | [179] |
| Ground water and rice | Mancozeb, Thiamethoxam, Cartap Hydrochloride, Emamectin Benzoate, Alphamethrin, Fenpropathrin, Triazophos, Imidacloprid, and Chlorpyrifos | 94–98% | NR | [182] |
| Water-quality control and on-site applications | Phenol, caffeine, uric acid, ascorbic acid, hydroquinon, profenofos, chlorpyrifos, and carbofuran | 100.00–108.00% | 0.10–3.79% | [212] |
| Cucumber and tap water | AMPA, chlorpyrifos, aldicarb | 72.70–98.96% | 1.07–4.48% | [213] |
| Potato, Tap water | Malathion, Fenamiphos, Parathion Zn2+, Ni2+, Hg2+, Fe3+, Al3+, Cd2+, Co2+, Cu2+ Na+ | 81.3–101% | 0.49–4.61% | [218] |
| Groundwater samples, soybean extracts, and lettuce extracts | Glufosinate, carbaryl, bentazon, monocrotophos, thiram, and carbofuran, AMPA) and sarcosine, Mn4+,Fe2+,Fe3+, K+,Cl−,Zn2+,SO42−, BO33−, Na+, NO3−, PO4 3−, and Ca2+ | 97.9–102.1%. | 0.2–2.3% | [219] |
| Drinking water | Mn+ = Al3+, Fe3+, Cr3+, Hg2+, Ni2+, Cu2+, Cd2+, Mn2+, Zn2+, Co2+, Ca2+, Ba2+,Mg2+,K+, and Na+ fenthion, parathion-ethyl, parathion-methyl, chlorpyrifos, dichlorvos, profe- nofos, azinphosethyl, azinphosmethyl, fenitrothion, malathion, diazinon | NR | NR | [220] |
5. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Storck, V.; Karpouzas, D.G.; Martin-laurent, F. Towards a better pesticide policy for the European Union. Sci. Total Environ. 2017, 575, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kabir, E.; Ara, S. Exposure to pesticides and the associated human health effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef]
- Landrigan, P.J.; Fuller, R.; Acosta, N.J.R.; Adeyi, O.; Arnold, R.; Basu, N.N.; Baldé, A.B.; Bertollini, R.; Bose-O’Reilly, S.; Boufford, J.I.; et al. The Lancet Commission on pollution and health. Lancet 2018, 391, 462–512. [Google Scholar] [CrossRef] [Green Version]
- Steingrímsdóttir, M.M.; Petersen, A.; Fantke, P. A screening framework for pesticide substitution in agriculture. J. Clean. Prod. 2018, 192, 306–315. [Google Scholar] [CrossRef]
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M. Recent perspective of herbicide: Review of demand and adoption in world agriculture. J. Bangladesh Agric. Univ. 2016, 13, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An international database for pesticide risk assessments and management. Hum. Ecol. Risk Assess. 2016, 22, 1050–1064. [Google Scholar] [CrossRef] [Green Version]
- Myers, J.P.; Antoniou, M.N.; Blumberg, B.; Carroll, L.; Colborn, T.; Everett, L.G.; Hansen, M.; Landrigan, P.J.; Lanphear, B.P.; Mesnage, R.; et al. Concerns over use of glyphosate-based herbicides and risks associated with exposures: A consensus statement. Environ. Health A Glob. Access Sci. Source 2016, 15, 19. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.K. Toxicity of Herbicides. In Veterinary Toxicology: Basic and Clinical Principles, 3rd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 553–567. ISBN 9780128114100. [Google Scholar]
- Leyva-Soto, L.A.; Balderrama-Carmona, A.P.; Moran-Palacio, E.F.; Diaz-Tenorio, L.M.; Gortares-Moroyoqui, P. Glyphosate and aminomethylphosphonic acid in population of agricultural fields: Health risk assessment overview. Appl. Ecol. Environ. Res. 2018, 16, 5127–5140. [Google Scholar] [CrossRef]
- Richmond, M.E. Glyphosate: A review of its global use, environmental impact, and potential health effects on humans and other species. J. Environ. Stud. Sci. 2018, 8, 416–434. [Google Scholar] [CrossRef]
- Heap, I.; Duke, S.O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 2018, 74, 1040–1049. [Google Scholar] [CrossRef]
- Torretta, V.; Katsoyiannis, I.A.; Viotti, P.; Rada, E.C. Critical review of the effects of glyphosate exposure to the environment and humans through the food supply chain. Sustainability 2018, 10, 950. [Google Scholar] [CrossRef] [Green Version]
- Gillezeau, C.; Van Gerwen, M.; Shaffer, R.M.; Rana, I.; Zhang, L.; Sheppard, L.; Taioli, E. The evidence of human exposure to glyphosate: A review. Environ. Health A Glob. Access Sci. Source 2019, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Peruzzo, P.J.; Porta, A.A.; Ronco, A.E. Levels of glyphosate in surface waters, sediments and soils associated with direct sowing soybean cultivation in north pampasic region of Argentina. Environ. Pollut. 2008, 156, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Vereecken, H. Mobility and leaching of glyphosate: A review. Pest Manag. Sci. Former. Pestic. Sci. 2005, 61, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Annett, R.; Habibi, H.R.; Hontela, A. Impact of glyphosate and glyphosate-based herbicides on the freshwater environment. J. Appl. Toxicol. 2014, 34, 458–479. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Montanarella, L.; Jones, A.; Fernández-Ugalde, O.; Mol, H.G.J.; Ritsema, C.J.; Geissen, V. Distribution of glyphosate and aminomethylphosphonic acid (AMPA) in agricultural topsoils of the European Union. Sci. Total Environ. 2018, 621, 1352–1359. [Google Scholar] [CrossRef]
- Gill, J.P.K.; Sethi, N.; Mohan, A. Analysis of the glyphosate herbicide in water, soil and food using derivatising agents. Environ. Chem. Lett. Rev. 2017, 15, 85–100. [Google Scholar] [CrossRef]
- Valavanidis, A. Glyphosate, the Most Widely Used Herbicide. Health and safety issues. Why scientist differ in their evaluation of its adverse health effects. Sci. Rev. 2018, 1, 1–36. [Google Scholar]
- Mariane, R.; Souza, D.; Seibert, D.; Beatriz, H.; De Jesus, F.; Fagundes-klen, M.R.; Bergamasco, R. Occurrence, impacts and general aspects of pesticides in surface water: A review. Process. Saf. Environ. Prot. 2020, 135, 22–37. [Google Scholar] [CrossRef]
- Reynoso, E.C.; Torres, E.; Bettazzi, F.; Palchetti, I. Trends and perspectives in immunosensors for determination of currently-used pesticides: The case of glyphosate, organophosphates, and neonicotinoids. Biosensors 2019, 9, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA. Conclusion on the peer review of the pesticide risk assessment of the active substance glyphosate. EFSA J. 2017, 13, 4302. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Evaluation of Five Organophosphate Insecticides and Herbicides; World Health Organization: Lyon, France, 2015. [Google Scholar]
- Guyton, K.Z.; Loomis, D.; Grosse, Y.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Scoccianti, C.; Mattock, H.; Straif, K.; Blair, A.; et al. Carcinogenicity of tetrachlorvinphos, parathion, malathion, diazinon, and glyphosate. Lancet Oncol. 2015, 16, 490–491. [Google Scholar] [CrossRef]
- Mesnage, R.; Defarge, N.; Spiroux de Vendômois, J.; Séralini, G.E. Potential toxic effects of glyphosate and its commercial formulations below regulatory limits. Food Chem. Toxicol. 2015, 84, 133–153. [Google Scholar] [CrossRef] [Green Version]
- Banaee, M.; Akhlaghi, M.; Soltanian, S.; Sureda, A.; Gholamhosseini, A.; Rakhshaninejad, M. Combined effects of exposure to sub-lethal concentration of the insecticide chlorpyrifos and the herbicide glyphosate on the biochemical changes in the freshwater crayfish Pontastacus leptodactylus. Ecotoxicology 2020, 29, 1500–1515. [Google Scholar] [CrossRef] [PubMed]
- Ingaramo, P.; Alarcón, R.; Muñoz-de-Toro, M.; Luque, E.H. Are glyphosate and glyphosate-based herbicides endocrine disruptors that alter female fertility? Mol. Cell. Endocrinol. 2020, 518, 110934. [Google Scholar] [CrossRef] [PubMed]
- Meftaul, I.M.; Venkateswarlu, K.; Dharmarajan, R.; Annamalai, P.; Asaduzzaman, M.; Parven, A.; Megharaj, M. Controversies over human health and ecological impacts of glyphosate: Is it to be banned in modern agriculture? Environ. Pollut. 2020, 263, 114372. [Google Scholar] [CrossRef]
- Peillex, C.; Pelletier, M. The impact and toxicity of glyphosate and glyphosate-based herbicides on health and immunity. J. Immunotoxicol. 2020, 17, 163–174. [Google Scholar] [CrossRef]
- Lu, T.; Xu, N.; Zhang, Q.; Zhang, Z.; Debognies, A.; Zhou, Z.; Sun, L.; Qian, H. Understanding the influence of glyphosate on the structure and function of freshwater microbial community in a microcosm. Environ. Pollut. 2020, 260, 114012. [Google Scholar] [CrossRef]
- Valle, A.L.; Mello, F.C.C.; Alves-Balvedi, R.P.; Rodrigues, L.P.; Goulart, L.R. Glyphosate detection: Methods, needs and challenges. Environ. Chem. Lett. 2019, 17, 291–317. [Google Scholar] [CrossRef]
- European Commission. Council Directive 98/83/EC on the Quality of Water Intended for Human Consumption. Friends of Earth Europe 2013, Friends of Earth Europe. The Environmental Effects of Glyphosate (2013). Available online: https://www.foeeurope.org/sites/default/files/press_releases/foee_5_environmental_impacts_glyphosate.pdf (accessed on 1 April 2022).
- Environmental Protection Agency. National Primary Drinking Water Regulations EPA 816 F 09 0004. In Epa; 2009. Available online: https://www.epa.gov/sites/default/files/2016-06/documents/npwdr_complete_table.pdf (accessed on 1 April 2022).
- US EPA Drinking Water Contaminants: National Primary Drinking Water Regulations. US EPA. 2015. Available online: http//water.epa.gov/drink/contaminants/index.cfm (accessed on 1 April 2022).
- Health Canada. Guidelines for Canadian Drinking Water Quality—Summary Table. Water and Air Quality Bureau, Healthy Environments and Consumer Safety Branch; Health Canada: Ottawa, ON, Canada, 2020; Available online: https://www.canada.ca/content/dam/hc-sc/migration/hc-sc/ewh-semt/alt_formats/pdf/pubs/water-eau/sum_guide-res_recom/summary-table-EN-2020-02-11.pdf (accessed on 1 April 2022).
- Secretaría de Salud, S. NOM-201-SSA1-2015, Productos y Servicios. Agua y hielo para Consumo Humano, Envasados y a Granel. Especificaciones Sanitarias. D. Of. Fed. Available online: https://dof.gob.mx/nota_detalle.php?codigo=5420977&fecha=22/12/2015#gsc.t (accessed on 1 April 2022).
- Hamilton, D.J.; Ambrus, Á.; Dieterle, R.M.; Felsot, A.S.; Harris, C.A.; Holland, P.T.; Katayama, A.; Kuriharas, N.; Linders, J.; Unsworth, J.; et al. Regulatory limits for pesticide residues in water (IUPAC technical report). Pure Appl. Chem. 2003, 75, 1123–1155. [Google Scholar] [CrossRef]
- Li, Z.; Jennings, A. Worldwide regulations of standard values of pesticides for human health risk control: A review. Int. J. Environ. Res. Public Health 2017, 14, 826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarazona, J.V.; Court-Marques, D.; Tiramani, M.; Reich, H.; Pfeil, R.; Istace, F.; Crivellente, F. Glyphosate toxicity and carcinogenicity: A review of the scientific basis of the European Union assessment and its differences with IARC. Arch. Toxicol. 2017, 91, 2723–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winfield, T.W.; Bashe, W.J.; Baker, T.V. Method 547 Determination of Glyphosate in Drinking Water By Direct-Aqueous- Injection Hplc, Post-Column Derivatization, and Fluorescence Detection. Technol. Appl. 1990, 1, 1–16. [Google Scholar]
- Mitra, S.; Patnaik, P.; Kebbekus, B.B. Environmental Chemical Analysis; CRC Press: New York, NY, USA, 2019; ISBN 9780849338380. [Google Scholar]
- Okada, E.; Coggan, T.; Anumol, T.; Clarke, B.; Allinson, G. A simple and rapid direct injection method for the determination of glyphosate and AMPA in environmental water samples. Anal. Bioanal. Chem. 2019, 411, 715–724. [Google Scholar] [CrossRef]
- Begum, S.S.; Sushmaa, B.S.; Vijayaraja, S. Bioanalytical Techniques–An Overview. PharmaTutor 2015, 3, 14–24. [Google Scholar]
- Peng, J.; Tang, F.; Zhou, R.; Xie, X.; Li, S.; Xie, F.; Yu, P.; Mu, L. New techniques of on-line biological sample processing and their application in the field of biopharmaceutical analysis. Acta Pharm. Sin. B 2016, 6, 540–551. [Google Scholar] [CrossRef] [Green Version]
- Koskinen, W.C.; Marek, L.J.; Hall, K.E. Analysis of glyphosate and aminomethylphosphonic acid in water, plant materials and soil. Pest Manag. Sci. 2016, 72, 423–432. [Google Scholar] [CrossRef]
- Zouaoui, K.; Dulaurent, S.; Gaulier, J.M.; Moesch, C.; Lacha, G. Determination of glyphosate and AMPA in blood and urine from humans: About 13 cases of acute intoxication. Forensic Sci. Int. J. 2013, 226, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Oulkar, D.P.; Hingmire, S.; Goon, A.; Jadhav, M.; Ugare, B.; Thekkumpurath, A.S.; Banerjee, K. Optimization and validation of a residue analysis method for glyphosate, glufosinate, and their metabolites in plant matrixes by liquid chromatography with tandem mass spectrometry. J. AOAC Int. 2017, 100, 631–639. [Google Scholar] [CrossRef]
- Fontàs, C.; Sanchez, J.M. Evaluation and optimization of the derivatization reaction conditions of glyphosate and aminomethylphosphonic acid with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate using reversed-phase liquid chromatography. J. Sep. Sci. 2020, 43, 3931–3939. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.L.; Passos, C.J.S.; Morgado, M.G.A.; Mello, D.C.; Infante, C.M.C.; Caldas, E.D. Determination of glyphosate, AMPA and glufosinate by high performance liquid chromatography with fluorescence detection in waters of the Santarém Plateau, Brazilian Amazon. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2020, 55, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Mazouz, Z.; Rahali, S.; Fourati, N.; Zerrouki, C.; Aloui, N.; Seydou, M.; Yaakoubi, N.; Chehimi, M.M.; Othmane, A.; Kalfat, R. Highly selective polypyrrole MIP-based gravimetric and electrochemical sensors for picomolar detection of glyphosate. Sensors 2017, 17, 2586. [Google Scholar] [CrossRef] [Green Version]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. International union of pure and applied chemistry physical chemistry division, steering committee on biophysical chemistry analytical chemistry division, commission V.5 (electroanalytical chemistry) Electrochemical biosensors: Proposed definitions and cla. Sens. Actuators B Chem. 1996, 30, 81. [Google Scholar] [CrossRef]
- Saxena, U.; Das, A.B. Nanomaterials towards fabrication of cholesterol biosensors: Key roles and design approaches. Biosens. Bioelectron. 2016, 75, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Rawtani, D.; Khatri, N.; Tyagi, S.; Pandey, G. Nanotechnology-based recent approaches for sensing and remediation of pesticides. J. Environ. Manag. 2018, 206, 749–762. [Google Scholar] [CrossRef]
- Bǎnicǎ, F.G. Chemical Sensors and Biosensors: Fundamentals and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2012; ISBN 9780470710661. [Google Scholar]
- Korotkaya, E.V. Biosensors: Design, classification, and applications in the food industry. Foods Raw Mater. 2014, 2, 161–171. [Google Scholar] [CrossRef]
- Arduini, F.; Cinti, S.; Scognamiglio, V.; Moscone, D.; Palleschi, G. How cutting-edge technologies impact the design of electrochemical (bio)sensors for environmental analysis. A review. Anal. Chim. Acta 2017, 959, 15–42. [Google Scholar] [CrossRef]
- Sanchís, J.; Kantiani, L.; Llorca, M.; Rubio, F.; Ginebreda, A.; Fraile, J.; Garrido, T.; Farré, M. Determination of glyphosate in groundwater samples using an ultrasensitive immunoassay and confirmation by on-line solid-phase extraction followed by liquid chromatography coupled to tandem mass spectrometry. Anal. Bioanal. Chem. 2012, 402, 2335–2345. [Google Scholar] [CrossRef]
- Chuang, H.Y.; Hong, T.P.; Whang, C.W. A simple and rapid screening method for glyphosate in water using flow-injection with electrochemiluminescence detection. Anal. Methods 2013, 5, 6186–6191. [Google Scholar] [CrossRef]
- Fang, F.; Wei, R.; Liu, X. Novel pre-column derivatisation reagent for glyphosate by high-performance liquid chromatography and ultraviolet detection. Int. J. Environ. Anal. Chem. 2014, 94, 661–667. [Google Scholar] [CrossRef]
- Pérez, A.L.; Tibaldo, G.; Sánchez, G.H.; Siano, G.G.; Marsili, N.R.; Schenone, A.V. A novel fluorimetric method for glyphosate and AMPA determination with NBD-Cl and MCR-ALS. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 214, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Romero-Natale, A.; Palchetti, I.; Avelar, M.; González-Vergara, E.; Garate-Morales, J.L.; Torres, E. Spectrophotometric detection of glyphosate in water by complex formation between bis 5-phenyldipyrrinate of nickel (II) and glyphosate. Water 2019, 11, 719. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, C.E.; Bellmund, S.; Gardinali, P.R. A simple method for routine monitoring of glyphosate and its main metabolite in surface waters using lyophilization and LC-FLD + MS/MS. Case study: Canals with influence on Biscayne National Park. Sci. Total Environ. 2014, 496, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Gao, Y.; Guo, D.; Liu, W.; Wang, J.; Zheng, J.; Zhong, J.; Zhao, Q. Sensitive, rapid and non-derivatized determination of glyphosate, glufosinate, bialaphos and metabolites in surface water by LC–MS/MS. SN Appl. Sci. 2019, 1, 305. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Liu, B.; Yuan, D.; Ma, J. A simple method for the determination of glyphosate and aminomethylphosphonic acid in seawater matrix with high performance liquid chromatography and fluorescence detection. Talanta 2016, 161, 700–706. [Google Scholar] [CrossRef]
- Dai, H.; Sang, M.; Wang, Y.; Du, R.; Yuan, W.; Jia, Z.; Cao, Z.; Chen, X. Determination of trace glyphosate in water with a prism coupling optical waveguide configuration. Sens. Actuators A Phys. 2014, 218, 88–93. [Google Scholar] [CrossRef]
- Wei, X.; Gao, X.; Zhao, L.; Peng, X.; Zhou, L.; Wang, J.; Pu, Q. Fast and interference-free determination of glyphosate and glufosinate residues through electrophoresis in disposable microfluidic chips. J. Chromatogr. A 2013, 1281, 148–154. [Google Scholar] [CrossRef]
- Mörtl, M.; Németh, G.; Juracsek, J.; Darvas, B.; Kamp, L.; Rubio, F.; Székács, A. Determination of glyphosate residues in Hungarian water samples by immunoassay. Microchem. J. 2013, 107, 143–151. [Google Scholar] [CrossRef]
- Zhang, W.; Feng, Y.; Ma, L.; An, J.; Zhang, H.; Cao, M. A method for determining glyphosate and its metabolite aminomethyl phosphonic acid by gas chromatography-flame photometric detection. J. Chromatogr. A 2019, 1589, 116–121. [Google Scholar] [CrossRef]
- Oliveira Pereira, E.A.; Freitas Melo, V.; Abate, G.; Masini, J.C. Determination of glyphosate and aminomethylphosphonic acid by sequential-injection reversed-phase chromatography: Method improvements and application in adsorption studies. Anal. Bioanal. Chem. 2019, 411, 2317–2326. [Google Scholar] [CrossRef]
- Sharma, P.; Pandey, V.; Sharma, M.M.M.; Patra, A.; Singh, B.; Mehta, S.; Husen, A. A Review on Biosensors and Nanosensors Application in Agroecosystems. Nanoscale Res. Lett. 2021, 16, 136. [Google Scholar] [CrossRef] [PubMed]
- Vikesland, P.J. Nanosensors for water quality monitoring. Nat. Nanotechnol. 2018, 13, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, L.; Shen, C.; Wang, C.; Hu, X.; Wang, G. An electrochemical sensor on the hierarchically porous Cu-BTC MOF platform for glyphosate determination. Sens. Actuators B Chem. 2019, 283, 487–494. [Google Scholar] [CrossRef]
- Gupta, A.; Eral, H.B.; Hatton, T.A.; Doyle, P.S. Nanoemulsions: Formation, properties and applications. Soft Matter 2016, 12, 2826–2841. [Google Scholar] [CrossRef] [Green Version]
- Kurbanoglu, S.; Ozkan, S.A.; Merkoçi, A. Nanomaterials-based enzyme electrochemical biosensors operating through inhibition for biosensing applications. Biosens. Bioelectron. 2017, 89, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Jampílek, J.; Kráľová, K.; Campos, E.V.R.; Fraceto, L.F. Bio-Based Nanoemulsion Formulations Applicable in Agriculture, Medicine, and Food Industry. In Nanobiotechnology in Bioformulations; Springer: Cham, Switzerland, 2019; pp. 33–84. [Google Scholar]
- Dasgupta, N.; Ranjan, S.; Chakraborty, A.R.; Ramalingam, C.; Shanker, R.; Kumar, A. Nanoagriculture and Water Quality Managenent. In Nanoscience in Food and Agriculture 1; Springer: Cham, Switzerland, 2016; pp. 1–43. ISBN 9783319393056. [Google Scholar]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef]
- Reynoso, E.C.; Romero-Guido, C.; Rebollar-Pérez, G.; Torres, E. Chapter 16—Enzymatic biosensors for the detection of water pollutants. In Enzymatic Biosensors for the Detection of Water Pollutants; Elsevier: Amsterdam, The Netherlands, 2022; pp. 463–511. [Google Scholar]
- Pokropivny, V.V.; Skorokhod, V.V. Classification of nanostructures by dimensionality and concept of surface forms engineering in nanomaterial science. Mater. Sci. Eng. C 2007, 27, 990–993. [Google Scholar] [CrossRef]
- Saleh, T.A. Nanomaterials: Classification, properties, and environmental toxicities. Environ. Technol. Innov. 2020, 20, 101067. [Google Scholar] [CrossRef]
- Wang, Z.; Hu, T.; Liang, R.; Wei, M. Application of Zero-Dimensional Nanomaterials in Biosensing. Front. Chem. 2020, 8, 320. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, L.; Zeng, G.; Zhang, C.; Zhang, Y.; Xie, X. Sensors and Actuators B: Chemical Current progress in biosensors for heavy metal ions based on DNAzymes / DNA molecules functionalized nanostructures: A review. Sens. Actuators B. Chem. 2016, 223, 280–294. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Dev, A.; Karmakar, S. Nanosensors and nanobiosensors in food and agriculture. Environ. Chem. Lett. 2018, 16, 161–182. [Google Scholar] [CrossRef]
- Campbell, F.W.; Compton, R.G. The use of nanoparticles in electroanalysis: An updated review. Anal. Bioanal. Chem. 2010, 396, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Jian, M.; Geleta, G.S.; Wang, Z. Two-Dimensional Layered Nanomaterial-Based Electrochemical Biosensors for Detecting Microbial Toxins. Toxins 2019, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Su, S.; Sun, Q.; Gu, X.; Xu, Y.; Shen, J.; Zhu, D.; Chao, J.; Fan, C.; Wang, L. Two-dimensional nanomaterials for biosensing applications. TrAC Trends Anal. Chem. 2019, 119, 115610. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhu, Z.; Du, D.; Lin, Y. Nanomaterial-based electrochemical biosensors for food safety. J. Electroanal. Chem. 2016, 781, 147–154. [Google Scholar] [CrossRef]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef]
- Banwaskar, M.R.; Dachawar, S.N. Dachawar Nanotechnology: A New Perspective for Management of Environment. Int. Journal Res. Anal. Rev. 2019, 6, 78–82. [Google Scholar]
- Messina, G.A.; Regiart, M.; Pereira, S.V.; Bertolino, F.A.; Aranda, P.R.; Raba, J.; Fernández-Baldo, M.A. Nanomaterials in the Development of Biosensor and Application in the Determination of Pollutants in Water. In Advanced Research in Nanosciences for Water Technology; Springer: Cham, Switzerland, 2019; pp. 195–215. [Google Scholar]
- Chen, H.; Zhang, L.; Hu, Y.; Zhou, C.; Lan, W.; Fu, H.; She, Y. Nanomaterials as optical sensors for application in rapid detection of food contaminants, quality and authenticity. Sens. Actuators B. Chem. 2021, 329, 129135. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, S.; Wu, W. Shape control of inorganic nanoparticles from solution. Nanoscale 2016, 8, 1237–1259. [Google Scholar] [CrossRef]
- Liu, G.; Lu, M.; Huang, X.; Li, T.; Xu, D. Application of gold-nanoparticle colorimetric sensing to rapid food safety screening. Sensors 2018, 18, 4166. [Google Scholar] [CrossRef] [Green Version]
- Priyadarshini, E.; Pradhan, N. Gold nanoparticles as efficient sensors in colorimetric detection of toxic metal ions: A review. Sens. Actuators B Chem. 2017, 238, 888–902. [Google Scholar] [CrossRef]
- Gaviña, P.; Parra, M.; Gil, S.; Costero, A.M. Red or Blue? Gold Nanoparticles in Colorimetric Sensing; IntechOpen: London, UK, 2018; pp. 3–15. [Google Scholar] [CrossRef]
- Csáki, A.; Stranik, O.; Fritzsche, W. Localized surface plasmon resonance based biosensing. Expert Rev. Mol. Diagn. 2018, 18, 279–296. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, H.; Qu, J.; Zhu, Q.; Chen, X. Visual detection of glyphosate in environmental water samples using cysteamine-stabilized gold nanoparticles as colorimetric probe. Anal. Methods 2013, 5, 917–924. [Google Scholar] [CrossRef]
- Do, M.H.; Florea, A.; Farre, C.; Bonhomme, A.; Bessueille, F.; Vocanson, F.; Tran-Thi, N.T.; Jaffrezic-Renault, N. Molecularly imprinted polymer-based electrochemical sensor for the sensitive detection of glyphosate herbicide. Int. J. Environ. Anal. Chem. 2015, 95, 1489–1501. [Google Scholar] [CrossRef]
- Nafisah, S.; Morsin, M.; Jumadi, N.A.; Nayan, N.; Mohd Shah, N.S.; Razali, N.L.; An’nisa, N.Z. Improved Sensitivity and Selectivity of Direct Localized Surface Plasmon Resonance Sensor Using Gold Nanobipyramids for Glyphosate Detection. IEEE Sens. J. 2020, 20, 2378–2389. [Google Scholar] [CrossRef]
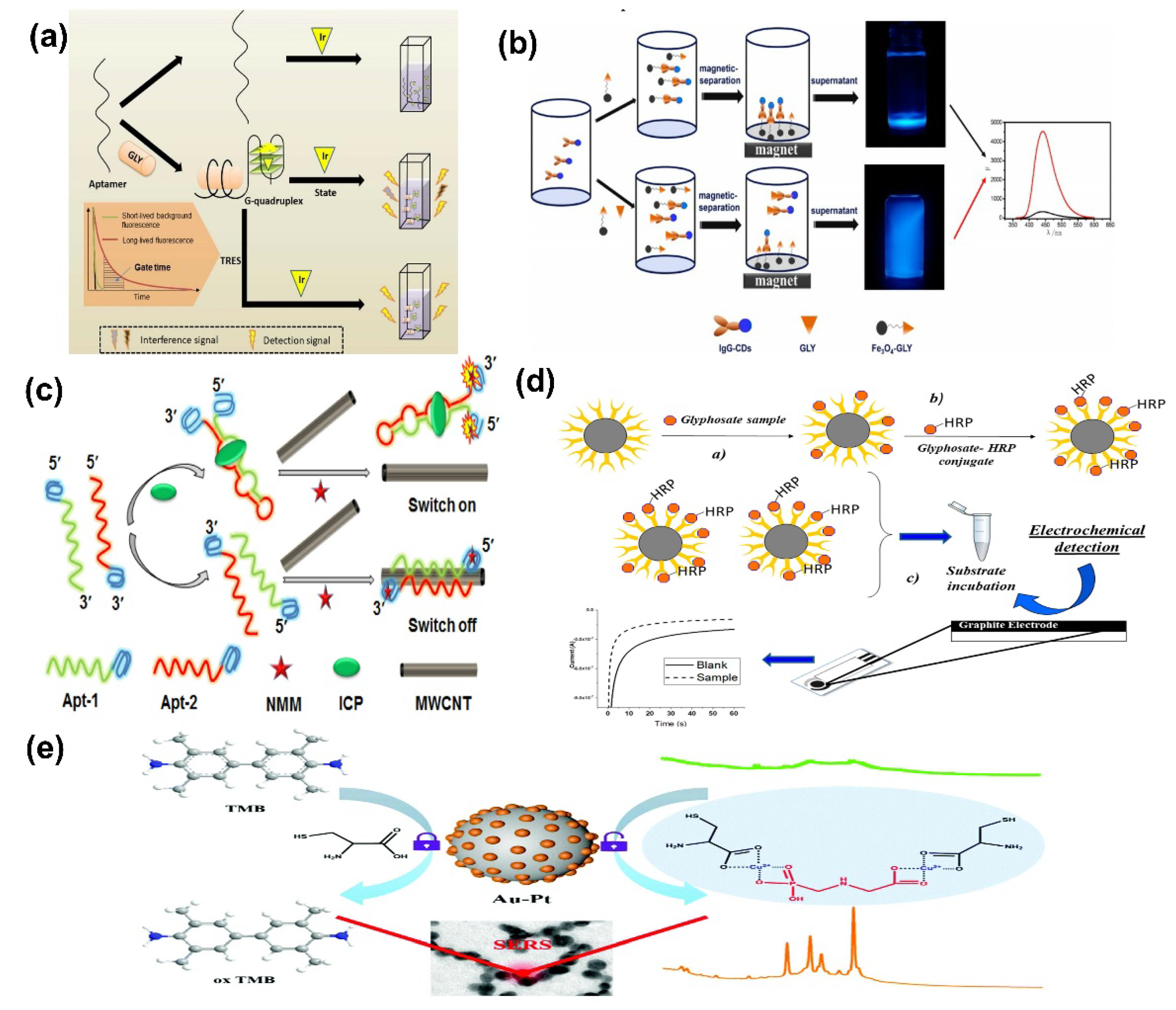
- Liu, H.; Chen, P.; Liu, Z.; Liu, J.; Yi, J.; Xia, F.; Zhou, C. Electrochemical luminescence sensor based on double suppression for highly sensitive detection of glyphosate. Sens. Actuators B Chem. 2020, 304, 127364. [Google Scholar] [CrossRef]
- Song, D.; Wang, Y.; Lu, X.; Gao, Y.; Li, Y.; Gao, F. Ag nanoparticles-decorated nitrogen-fluorine co-doped monolayer MoS2 nanosheet for highly sensitive electrochemical sensing of organophosphorus pesticides. Sens. Actuators B Chem. 2018, 267, 5–13. [Google Scholar] [CrossRef]
- Xie, Y.; Yu, Y.; Lu, L.; Ma, X.; Gong, L.; Huang, X.; Liu, G.; Yu, Y. CuO nanoparticles decorated 3D graphene nanocomposite as non-enzymatic electrochemical sensing platform for malathion detection. J. Electroanal. Chem. 2018, 812, 82–89. [Google Scholar] [CrossRef]
- Tan, M.J.; Hong, Z.Y.; Chang, M.H.; Liu, C.C.; Cheng, H.F.; Loh, X.J.; Chen, C.H.; Liao, C.D.; Kong, K.V. Metal carbonyl-gold nanoparticle conjugates for highly sensitive SERS detection of organophosphorus pesticides. Biosens. Bioelectron. 2017, 96, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bi, Y.; Hou, J.; Li, H.; Xu, Y.; Wang, B.; Ding, H.; Ding, L. Facile, green and clean one-step synthesis of carbon dots from wool: Application as a sensor for glyphosate detection based on the inner filter effect. Talanta 2016, 160, 268–275. [Google Scholar] [CrossRef] [PubMed]
- De Goes, R.E.; Possetti, G.R.C.; Muller, M.; Fabris, J.L. Tuning of Citrate-Stabilized Laser Ablated Silver Nanoparticles for Glyphosate Detection. IEEE Sens. J. 2020, 20, 1843–1850. [Google Scholar] [CrossRef]
- Setznagl, S.; Cesarino, I. Copper nanoparticles and reduced graphene oxide modified a glassy carbon electrode for the determination of glyphosate in water samples. Int. J. Environ. Anal. Chem. 2022, 102, 293–305. [Google Scholar] [CrossRef]
- Bataller, R.; Campos, I.; Laguarda-Miro, N.; Alcañiz, M.; Soto, J.; Martínez-Máñez, R.; Gil, L.; García-Breijo, E.; Ibáñez-Civera, J. Glyphosate detection by means of a voltammetric electronic tongue and discrimination of potential interferents. Sensors 2012, 12, 17553–17568. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Aguirre, M.; Urreta, S.E.; Gomez, C.G. A Cu2+-Cu/glassy carbon system for glyphosate determination. Sens. Actuators B Chem. 2019, 284, 675–683. [Google Scholar] [CrossRef]
- Regiart, M.; Kumar, A.; Gonçalves, J.M.; Junior, G.J.S.; Masini, J.C.; Angnes, L.; Bertottii, M. An Electrochemically Synthesized Nanoporous Copper Microsensor for Highly Sensitive and Selective Determination of Glyphosate. ChemElectroChem 2020, 7, 1558–1566. [Google Scholar] [CrossRef]
- Chang, Y.C.; Lin, Y.S.; Xiao, G.T.; Chiu, T.C.; Hu, C.C. A highly selective and sensitive nanosensor for the detection of glyphosate. Talanta 2016, 161, 94–98. [Google Scholar] [CrossRef]
- Wang, X.; Sakinati, M.; Yang, Y.; Ma, Y.; Yang, M.; Luo, H.; Hou, C.; Huo, D. The construction of a CND/Cu2+ fluorescence sensing system for the ultrasensitive detection of glyphosate. Anal. Methods 2020, 12, 520–527. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, L.; Sharma, A.S.; Chen, M.; Chen, Q. A system composed of polyethylenimine-capped upconversion nanoparticles, copper(II), hydrogen peroxide and 3,3′,5,5′-tetramethylbenzidine for colorimetric and fluorometric determination of glyphosate. Microchim. Acta 2019, 186, 835. [Google Scholar] [CrossRef]
- Jiang, R.; Pang, Y.H.; Yang, Q.Y.; Wan, C.Q.; Shen, X.F. Copper porphyrin metal-organic framework modified carbon paper for electrochemical sensing of glyphosate. Sens. Actuators B Chem. 2022, 358, 131492. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; Lin, L.; Wang, X.; Li, J.; Liu, H.; Liu, X.; Huo, D.; Hou, C. A dual-signal sensing strategy based on ratiometric fluorescence and colorimetry for determination of Cu2+ and glyphosate. Anal. Bioanal. Chem. 2022, 414, 2619–2628. [Google Scholar] [CrossRef] [PubMed]
- Vaghela, C.; Kulkarni, M.; Haram, S.; Aiyer, R.; Karve, M. A novel inhibition based biosensor using urease nanoconjugate entrapped biocomposite membrane for potentiometric glyphosate detection. Int. J. Biol. Macromol. 2018, 108, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zeng, K.; Liu, J. Immunochemical detection of emerging organic contaminants in environmental waters. TrAC Trends Anal. Chem. 2017, 87, 49–57. [Google Scholar] [CrossRef]
- Stavra, E.; Petrou, P.S.; Koukouvinos, G.; Economou, A.; Goustouridis, D.; Misiakos, K.; Raptis, I.; Kakabakos, S.E. Fast, sensitive and selective determination of herbicide glyphosate in water samples with a White Light Reflectance Spectroscopy immunosensor. Talanta 2020, 214, 120854. [Google Scholar] [CrossRef]
- Sharma, R.; Ragavan, K.V.; Thakur, M.S.; Raghavarao, K.S.M.S. Recent advances in nanoparticle based aptasensors for food contaminants. Biosens. Bioelectron. 2015, 74, 612–627. [Google Scholar] [CrossRef]
- Viirlaid, E.; Ilisson, M.; Kopanchuk, S.; Mäeorg, U.; Rinken, A.; Rinken, T. Immunoassay for rapid on-site detection of glyphosate herbicide. Environ. Monit. Assess. 2019, 191, 507. [Google Scholar] [CrossRef]
- Chen, F.; Li, G.; Liu, H.; Leung, C.; Ma, D. G-quadruplex-based detection of glyphosate in complex biological systems by a time-resolved luminescent assay. Sens. Actuators B Chem. 2020, 320, 128393. [Google Scholar] [CrossRef]
- Zhang, M.; Li, G.; Zhou, Q.; Pan, D.; Zhu, M.; Xiao, R.; Zhang, Y.; Wu, G.; Wan, Y.; Shen, Y. Boosted Electrochemical Immunosensing of Genetically Modified Crop Markers Using Nanobody and Mesoporous Carbon. ACS Sens. 2018, 3, 684–691. [Google Scholar] [CrossRef]
- Wang, X.; Qin, L.; Zhou, M.; Lou, Z.; Wei, H. Nanozyme Sensor Arrays for Detecting Versatile Analytes from Small Molecules to Proteins and Cells. Anal. Chem. 2018, 90, 11696–11702. [Google Scholar] [CrossRef]
- Qiu, L.; Lv, P.; Zhao, C.; Feng, X.; Fang, G.; Liu, J.; Wang, S. Electrochemical detection of organophosphorus pesticides based on amino acids conjugated nanoenzyme modified electrodes. Sens. Actuators B Chem. 2019, 286, 386–393. [Google Scholar] [CrossRef]
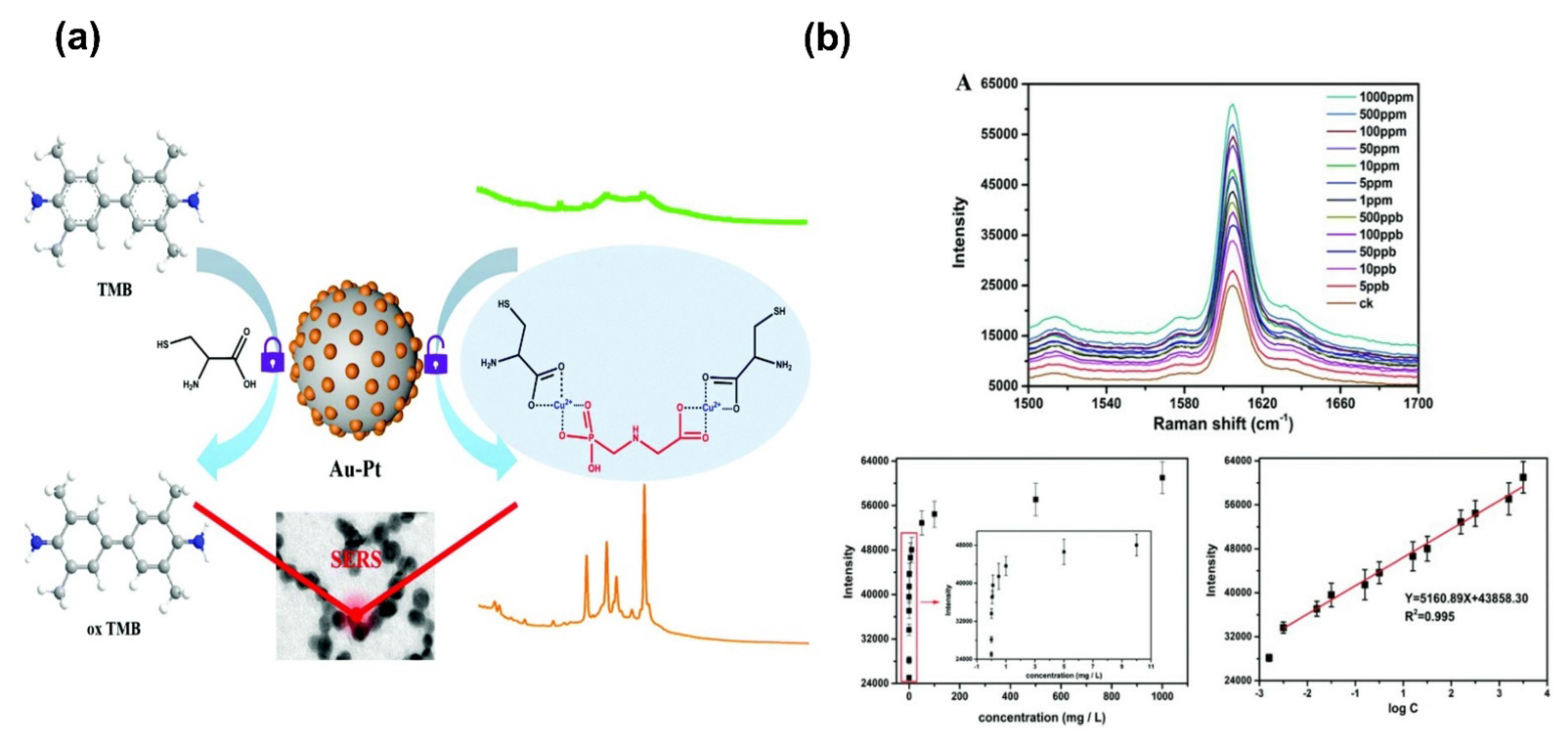
- Ma, J.; Feng, G.; Ying, Y.; Shao, Y.; She, Y.; Zheng, L.; Ei-aty, A.M.A.; Wang, J. Sensitive SERS assay for glyphosate based on the prevention of L -cysteine inhibition of a Au–Pt nanozyme. Analyst 2021, 146, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lin, B.; Cao, Y.; Guo, M.; Yu, Y. A Highly Selective and Sensitive Fluorescence Detection Method of Glyphosate Based on an Immune Reaction Strategy of Carbon Dot Labeled Antibody and Antigen Magnetic Beads. J. Agric. Food Chem. 2016, 64, 6042–6050. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, X.; Chen, X.; Qu, B.; Lu, L. Label-free and enzyme-free fluorescent isocarbophos aptasensor based on MWCNTs and G-quadruplex. Talanta 2018, 188, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Bettazzi, F.; Natale, A.R.; Torres, E.; Palchetti, I. Glyphosate determination by coupling an immuno-magnetic assay with electrochemical sensors. Sensors 2018, 18, 2965. [Google Scholar] [CrossRef] [Green Version]
- Guan, N.; Li, Y.; Yang, H.; Hu, P.; Lu, S.; Ren, H.; Liu, Z.; Soo Park, K.; Zhou, Y. Dual-functionalized gold nanoparticles probe based bio-barcode immuno-PCR for the detection of glyphosate. Food Chem. 2021, 338, 128133. [Google Scholar] [CrossRef]
- Bala, R.; Kumar, M.; Bansal, K.; Sharma, R.K.; Wangoo, N. Ultrasensitive aptamer biosensor for malathion detection based on cationic polymer and gold nanoparticles. Biosens. Bioelectron. 2016, 85, 445–449. [Google Scholar] [CrossRef]
- Bala, R.; Sharma, R.K.; Wangoo, N. Development of gold nanoparticles-based aptasensor for the colorimetric detection of organophosphorus pesticide phorate. Anal. Bioanal. Chem. 2016, 408, 333–338. [Google Scholar] [CrossRef]
- Bala, R.; Dhingra, S.; Kumar, M.; Bansal, K.; Mittal, S.; Sharma, R.K.; Wangoo, N. Detection of organophosphorus pesticide–Malathion in environmental samples using peptide and aptamer based nanoprobes. Chem. Eng. J. 2017, 311, 111–116. [Google Scholar] [CrossRef]
- Bala, R.; Mittal, S.; Sharma, R.K.; Wangoo, N. A supersensitive silver nanoprobe based aptasensor for low cost detection of malathion residues in water and food samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 196, 268–273. [Google Scholar] [CrossRef]
- Bala, R.; Swami, A.; Tabujew, I.; Peneva, K.; Wangoo, N.; Sharma, R.K. Ultra-sensitive detection of malathion using quantum dots-polymer based fluorescence aptasensor. Biosens. Bioelectron. 2018, 104, 45–49. [Google Scholar] [CrossRef]
- Madianos, L.; Skotadis, E.; Tsekenis, G.; Patsiouras, L.; Tsigkourakos, M.; Tsoukalas, D. Ιmpedimetric Nanoparticle Aptasensor for Selective and Label Free Pesticide Detection. Microelectron. Eng. 2018, 189, 39–45. [Google Scholar] [CrossRef]
- Madianos, L.; Tsekenis, G.; Skotadis, E.; Patsiouras, L.; Tsoukalas, D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018, 101, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; An, X.; Yao, Y.; Guo, Y.; Sun, X. Electrochemical aptasensor based on one step co-electrodeposition of aptamer and GO-CuNPs nanocomposite for organophosphorus pesticide detection. Sens. Actuators B Chem. 2019, 287, 503–509. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, S.; Shi, X.; Zhang, H.; Zhao, Q.; Dong, H.; Guo, Y.; Sun, X. Electrochemiluminescence Aptasensor for Profenofos Detection Based on Silver Nanoparticles Enhanced Luminol Luminescence System. J. Electrochem. Soc. 2019, 166, B1562–B1566. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, C.; He, J.; Zhang, H.; Xu, Z. Fluorescence assay for three organophosphorus pesticides in agricultural products based on Magnetic-Assisted fluorescence labeling aptamer probe. Food Chem. 2020, 307, 125534. [Google Scholar] [CrossRef] [PubMed]
- Bucur, B.; Munteanu, F.D.; Marty, J.L.; Vasilescu, A. Advances in enzyme-based biosensors for pesticide detection. Biosensors 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Wei, J.; Zou, J.; Cheng, Z.; Huang, Z.; Gu, L.; Zhong, Z.; Li, S.; Wang, Y.; Li, P. Electrochemical detection of methyl-paraoxon based on bifunctional cerium oxide nanozyme with catalytic activity and signal amplification effect. J. Pharm. Anal. 2021, 11, 653–660. [Google Scholar] [CrossRef]
- Boruah, P.K.; Darabdhara, G.; Das, M.R. Polydopamine functionalized graphene sheets decorated with magnetic metal oxide nanoparticles as efficient nanozyme for the detection and degradation of harmful triazine pesticides. Chemosphere 2021, 268, 129328. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, J.; Qu, X. Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chem. Rev. 2019, 119, 4357–4412. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yang, L.; Luo, M.; Wang, Y.; Li, P. Nanozyme-assisted technique for dual mode detection of organophosphorus pesticide. Ecotoxicol. Environ. Saf. 2019, 179, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Huang, X.; Liu, B.; Zou, W.; Wu, Y. Facile Colorimetric Nanozyme Sheet for the Rapid Detection of Glyphosate in Agricultural Products Based on Inhibiting Peroxidase-Like Catalytic Activity of Porous Co3O4 Nanoplates. J. Agric. Food Chem. 2021, 69, 3537–3547. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ghalandari, B.; Lin, L.; Sang, X.; Su, W.; Divsalar, A.; Ding, X. A turn-on fluorescence sensor based on Cu2+ modulated DNA-templated silver nanoclusters for glyphosate detection and mechanism investigation. Food Chem. 2022, 367, 130617. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, H.; Li, M.; Lu, Q.; Zhang, Y.; Yao, S. Template protection of gold nanoclusters for the detection of organophosphorus pesticides. New J. Chem. 2019, 43, 5423–5428. [Google Scholar] [CrossRef]
- Sok, V.; Fragoso, A. Amperometric biosensor for glyphosate based on the inhibition of tyrosinase conjugated to carbon nano-onions in a chitosan matrix on a screen-printed electrode. Microchim. Acta 2019, 186, 569. [Google Scholar] [CrossRef]
- Prasad, R. Advanced Research in Nanosciences for Water Technology; Springer: Cham, Switzerland, 2019; ISBN 978-3-030-02380-5. [Google Scholar]
- Faridbod, F.; Sanati, A.L. Graphene Quantum Dots in Electrochemical Sensors/Biosensors. Curr. Anal. Chem. 2018, 15, 103–123. [Google Scholar] [CrossRef]
- Masteri-Farahani, M.; Mahdavi, S.; Khanmohammadi, H. Chemically functionalized ZnS quantum dots as new optical nanosensor of herbicides. Mater. Res. Express 2018, 5, 035055. [Google Scholar] [CrossRef]
- Lesiak, A.; Drzozga, K.; Cabaj, J.; Ba’nski, M.; Malecha, K.; Podhorodecki, A. Optical Sensors Based on II-VI Quantum Dots. Nanomaterials 2019, 9, 192. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, D.; Mandal, A.; Mitra, T.; Chakraborty, K.; Bardhan, M.; Dasgupta, A.K. Nanosensing of Pesticides by Zinc Oxide Quantum Dot: An Optical and Electrochemical Approach for the Detection of Pesticides in Water. J. Agric. Food Chem. 2018, 66, 414–423. [Google Scholar] [CrossRef]
- Bonilla, J.C.; Bozkurt, F.; Ansari, S.; Sozer, N.; Kokini, J.L. Applications of quantum dots in Food Science and biology. Trends Food Sci. Technol. 2016, 53, 75–89. [Google Scholar] [CrossRef]
- Bera, M.K.; Mohapatra, S. Ultrasensitive detection of glyphosate through effective photoelectron transfer between CdTe and chitosan derived carbon dot. Colloids Surf. A Physicochem. Eng. Asp. 2020, 596, 124710. [Google Scholar] [CrossRef]
- Xing, Y.; Dittrich, P.S. One-dimensional nanostructures: Microfluidic-based synthesis, alignment and integration towards functional sensing devices. Sensors 2018, 18, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, B.; Lu, M.; Xu, M. Amperometric sensing of organophosphorus pesticides based on covalently attached multilayer assemblies of diazo-resin, Prussian blue single-walled carbon nanotubes, and acetylcholinesterase. Rev. Roum. Chim. 2019, 64, 763–774. [Google Scholar] [CrossRef]
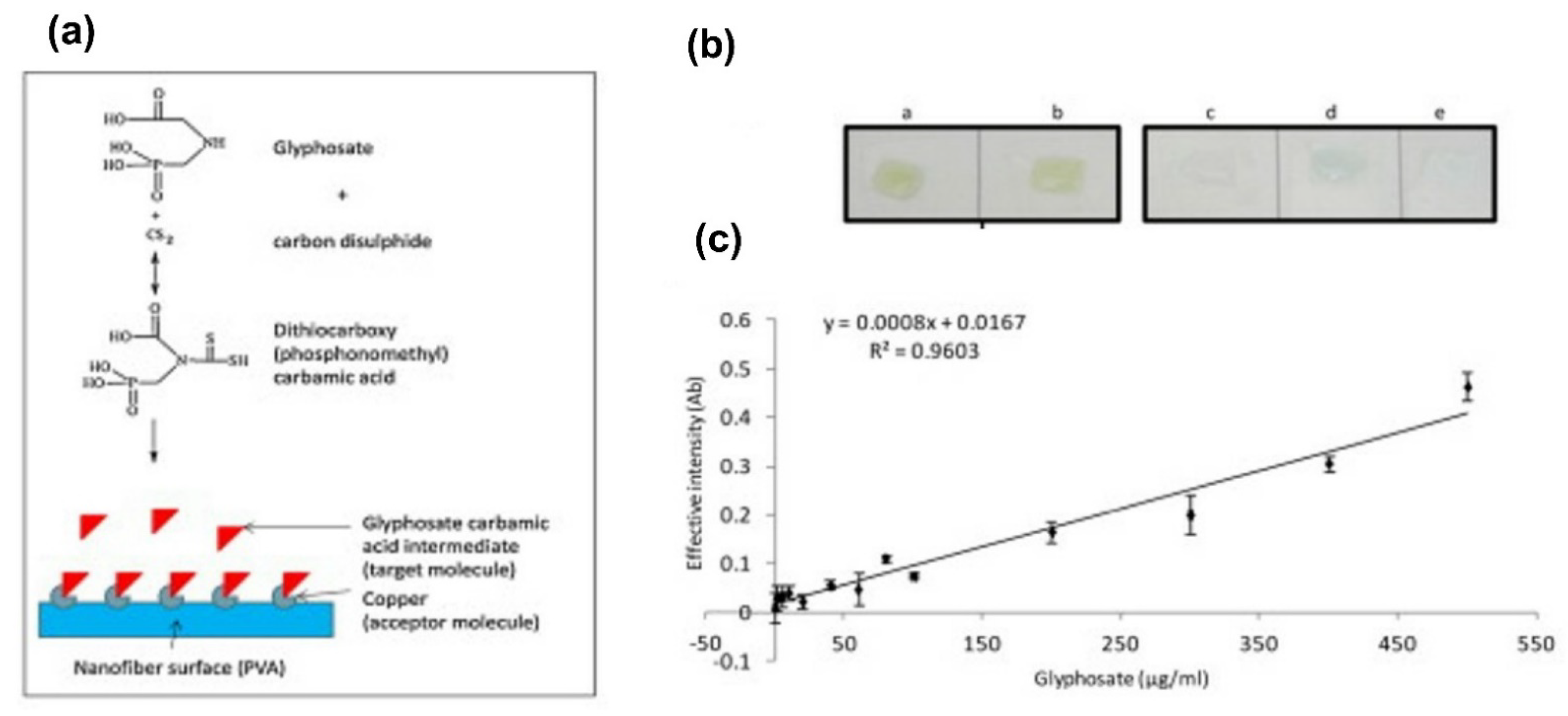
- Gholivand, M.B.; Akbari, A.; Norouzi, L. Development of a novel hollow fiber- pencil graphite modified electrochemical sensor for the ultra-trace analysis of glyphosate. Sens. Actuators B Chem. 2018, 272, 415–424. [Google Scholar] [CrossRef]
- Renedo, O.D.; Alonso-Lomillo, M.A.; Martínez, M.A. Recent developments in the field of screen-printed electrodes and their related applications. Talanta 2007, 73, 202–219. [Google Scholar] [CrossRef]
- Habekost, A. Rapid and sensitive spectroelectrochemical and electrochemical detection of glyphosate and AMPA with screen-printed electrodes. Talanta 2017, 162, 583–588. [Google Scholar] [CrossRef]
- Fang, J.; Wang, X.; Lin, T. Functional Applications of Electrospun Nano. Nanofibers Prod. Prop. Funct. Appls. 2011, 14, 287–302. [Google Scholar]
- Srivastava, A.K.; Bajpai, P.; Awasthi, P.; Kumar, A.; Prasad, N.E. Nanofiber Based Sensors for Water Pollution Monitoring. In Sensors in Water Pollutants Monitoring: Role of Material; Springer: Singapore, 2020. [Google Scholar]
- De Almeida, L.K.S.; Chigome, S.; Torto, N.; Frost, C.L.; Pletschke, B.I. A novel colorimetric sensor strip for the detection of glyphosate in water. Sens. Actuators B Chem. 2015, 206, 357–363. [Google Scholar] [CrossRef]
- Cui, H.F.; Zhang, T.T.; Lv, Q.Y.; Song, X.; Zhai, X.J.; Wang, G.G. An acetylcholinesterase biosensor based on doping Au nanorod@SiO2 nanoparticles into TiO2-chitosan hydrogel for detection of organophosphate pesticides. Biosens. Bioelectron. 2019, 141, 111452. [Google Scholar] [CrossRef]
- Torul, H.; Boyaci, I.H.; Tamer, U. Attomole detection of glyphosate by surface-enhanced raman spectroscopy using gold nanorods. Fabad J. Pharm. Sci. 2010, 35, 179–184. [Google Scholar]
- Yola, M.L. Electrochemical activity enhancement of monodisperse boron nitride quantum dots on graphene oxide: Its application for simultaneous detection of organophosphate pesticides in real samples. J. Mol. Liq. 2019, 277, 50–57. [Google Scholar] [CrossRef]
- Zeng, M.; Chen, M.; Huang, D.; Lei, S.; Zhang, X.; Wang, L.; Cheng, Z. Engineered two-dimensional nanomaterials: An emerging paradigm for water purification and monitoring. Mater. Horizons 2021, 8, 758–802. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yan, Y.; Hao, N.; Liu, Q.; Qian, J.; Chen, S.; Wang, K. Dual signal amplification coupling dual inhibition effect for fabricating photoelectrochemical chlorpyrifos biosensor. Sens. Actuators B Chem. 2017, 238, 239–248. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, H.; Li, Z.; Pang, J.; Lin, T.; Guo, L.; Fu, F.F. Colorimetric sensing of glyphosate in environmental water based on peroxidase mimetic activity of MoS2 nanosheets. J. Nanosci. Nanotechnol. 2017, 17, 5730–5734. [Google Scholar] [CrossRef]
- Wu, Z.; Hu, Y.; Pan, X.; Tang, Y.; Dai, Y.; Wu, Y. A liquid colorimetric chemosensor for ultrasensitive detection of glyphosate residues in vegetables using a metal oxide with intrinsic peroxidase catalytic activity. Food Addit. Contam. Part A 2022, 39, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Cahuantzi-Muñoz, S.L.; González-Fuentes, M.A.; Ortiz-Frade, L.A.; Torres, E.; Ţălu, Ş.; Trejo, G.; Méndez-Albores, A. Electrochemical Biosensor for Sensitive Quantification of Glyphosate in Maize Kernels. Electroanalysis 2019, 31, 927–935. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, P.C.; Maximiano, E.M.; Oliveira, P.A.; Camargo, J.S.; Fiorucci, A.R.; Arruda, G.J. Direct electrochemical detection of glyphosate at carbon paste electrode and its determination in samples of milk, orange juice, and agricultural formulation. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2018, 53, 817–823. [Google Scholar] [CrossRef]
- Wang, L.; Bi, Y.; Gao, J.; Li, Y.; Ding, H.; Ding, L. Carbon dots based turn-on fluorescent probes for the sensitive determination of glyphosate in environmental water samples. RSC Adv. 2016, 6, 85820–85828. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, J.; Liu, S.; Yang, J.; Zhang, H.; Yan, J.; Hu, X. Fluorescent carbon dots for glyphosate determination based on fluorescence resonance energy transfer and logic gate operation. Sens. Actuators B Chem. 2017, 242, 545–553. [Google Scholar] [CrossRef]
- Ashrafi Tafreshi, F.; Fatahi, Z.; Ghasemi, S.F.; Taherian, A.; Esfandiari, N. Ultrasensitive fluorescent detection of pesticides in real sample by using green carbon dots. PLoS ONE 2020, 15, e0230646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-López, J.; Llorent-Martínez, E.J.; Ortega-Barrales, P.; Ruiz-Medina, A. Graphene quantum dots-silver nanoparticles as a novel sensitive and selective luminescence probe for the detection of glyphosate in food samples. Talanta 2020, 207, 120344. [Google Scholar] [CrossRef]
- Jiménez-López, J.; Llorent-Martínez, E.J.; Ortega-Barrales, P.; Ruiz-Medina, A. Multicommutated Flow System for the Determination of Glyphosate Based on Its Quenching Effect on CdTe-Quantum Dots Fluorescence. Food Anal. Methods 2018, 11, 1840–1848. [Google Scholar] [CrossRef]
- Yu, C.X.; Hu, F.L.; Song, J.G.; Zhang, J.L.; Liu, S.S.; Wang, B.X.; Meng, H.; Liu, L.L.; Ma, L.F. Ultrathin two-dimensional metal-organic framework nanosheets decorated with tetra-pyridyl calix[4]arene: Design, synthesis and application in pesticide detection. Sens. Actuators B Chem. 2020, 310, 127819. [Google Scholar] [CrossRef]
- Marzari, G.; Cappellari, M.V.; Morales, G.M.; Fungo, F. Electrochemiluminescent detection of glyphosate using electrodes modified with self-assembled monolayers. Anal. Methods 2017, 9, 2452–2457. [Google Scholar] [CrossRef] [Green Version]
- Ashwin, B.C.M.A.; Saravanan, C.; Stalin, T.; Muthu Mareeswaran, P.; Rajagopal, S. FRET-based Solid-state Luminescent Glyphosate Sensor Using Calixarene-grafted Ruthenium(II)bipyridine Doped Silica Nanoparticles. ChemPhysChem 2018, 19, 2768–2775. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, G.; Gong, L.; Dai, H.; Zhang, S.; Li, Y.; Lin, Y. An enzyme-assisted electrochemiluminescent biosensor developed on order mesoporous carbons substrate for ultrasensitive glyphosate sensing. Electrochim. Acta 2015, 186, 624–630. [Google Scholar] [CrossRef]
- Duan, L.; Wang, C.; Zhang, W.; Ma, B.; Deng, Y.; Li, W.; Zhao, D. Interfacial Assembly and Applications of Functional Mesoporous Materials. Chem. Rev. 2021, 121, 14349–14429. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, R.; Yu, B.; Liang, A.; Jiang, Z. A highly sensitive gold nanosol SERS aptamer assay for glyphosate with a new COF nanocatalytic reaction of glycol-Au(III). Sens. Actuators B Chem. 2021, 344, 130288. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Tang, L.; Long, B.; Zeng, G. Nanoporous Materials Based Sensors for Pollutant Detection. In Nanohybrid and Nanoporous Materials for Aquatic Pollution Control; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 265–291. ISBN 9780128141557. [Google Scholar]
- Song, D.; Jiang, X.; Li, Y.; Lu, X.; Luan, S.; Wang, Y.; Li, Y.; Gao, F. Metal−organic frameworks-derived MnO2/Mn3O4 microcuboids with hierarchically ordered nanosheets and Ti3C2 MXene / Au NPs composites for electrochemical pesticide detection. J. Hazard. Mater. 2019, 373, 367–376. [Google Scholar] [CrossRef]
- He, K.; Li, Z.; Wang, L.; Fu, Y.; Quan, H.; Li, Y.; Wang, X.; Gunasekaran, S.; Xu, X. A Water-Stable Luminescent Metal − Organic Framework for Rapid and Visible Sensing of Organophosphorus Pesticides. Appl. Mater. Interfaces 2019, 11, 26250–26260. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Chen, X.; Li, X.; Jin, M. Nonderivatization method for determination of glyphosate, glufosinate, bialaphos, and their main metabolites in environmental waters based on magnetic metal-organic framework pretreatment. J. Sep. Sci. 2019, 42, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, E.; Fakhri, H.; Hajian, A.; Afkhami, A.; Bagheri, H. High-performance electrochemical enzyme sensor for organophosphate pesticide detection using modified metal-organic framework sensing platforms. Bioelectrochemistry 2019, 130, 107348. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Xu, W.; Cui, X.; Wang, Y. Recent Progress in Metal–Organic Frameworks and Their Derived Nanostructures for Energy and Environmental Applications. ChemSusChem 2017, 10, 1645–1663. [Google Scholar] [CrossRef] [PubMed]
- Vikrant, K.; Tsang, D.C.W.; Raza, N.; Giri, B.S.; Kukkar, D.; Kim, K.H. Potential Utility of Metal-Organic Framework-Based Platform for Sensing Pesticides. ACS Appl. Mater. Interfaces 2018, 10, 8797–8817. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, J.; Chen, X.; Yang, W.; Pei, H.; Hu, N.; Li, Z.; Suo, Y.; Li, T.; Wang, J. The simultaneous detection and removal of organophosphorus pesticides by a novel Zr-MOF based smart adsorbent. J. Mater. Chem. A 2018, 6, 2184–2192. [Google Scholar] [CrossRef]
- Marin, P.; Bergamasco, R.; Módenes, A.N.; Paraiso, P.R.; Hamoudi, S. Synthesis and characterization of graphene oxide functionalized with MnFe2O4 and supported on activated carbon for glyphosate adsorption in fixed bed column. Process Saf. Environ. Prot. 2019, 123, 59–71. [Google Scholar] [CrossRef]
- Santos, T.R.T.; Andrade, M.B.; Silva, M.F.; Bergamasco, R.; Hamoudi, S. Development of α- and γ-Fe2O3 decorated graphene oxides for glyphosate removal from water. Environ. Technol. 2019, 40, 1118–1137. [Google Scholar] [CrossRef]
- Park, H.; May, A.; Portilla, L.; Dietrich, H.; Münch, F.; Rejek, T.; Sarcletti, M.; Banspach, L.; Zahn, D.; Halik, M. Magnetite nanoparticles as efficient materials for removal of glyphosate from water. Nat. Sustain. 2020, 3, 129–135. [Google Scholar] [CrossRef]
- Li, S.; Liu, X.; Chai, H.; Huang, Y. Recent advances in the construction and analytical applications of metal-organic frameworks-based nanozymes. TrAC Trends Anal. Chem. 2018, 105, 391–403. [Google Scholar] [CrossRef]
- Gu, C.; Wang, Q.; Zhang, L.; Yang, P.; Xie, Y.; Fei, J. Ultrasensitive non-enzymatic pesticide electrochemical sensor based on HKUST-1-derived copper oxide @ mesoporous carbon composite. Sens. Actuators B Chem. 2020, 305, 127478. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, T.; Wang, X.; Huo, Q.; Liu, Y. Facile Fabricating Hierarchically Porous Metal−Organic Frameworks via a Template-Free Strategy. Cryst. Growth Des. 2016, 16, 504–510. [Google Scholar] [CrossRef]
- Bao, J.; Huang, T.; Wang, Z.; Yang, H.; Geng, X.; Xu, G.; Samalo, M.; Sakinati, M.; Huo, D.; Hou, C. 3D graphene/copper oxide nano-flowers based acetylcholinesterase biosensor for sensitive detection of organophosphate pesticides. Sens. Actuators B Chem. 2019, 279, 95–101. [Google Scholar] [CrossRef]
- Duan, G.W.; Zhang, J.; Li, Y.; Xu, Y.M.; Yin, F.; Fu, Y.Z. The preparation of Fe3O4/molecular-imprinted nanocomposite and the application on the recognition and separation of glyphosate. Inorg. Nano-Metal Chem. 2017, 47, 481–487. [Google Scholar] [CrossRef]
- Wu, S.; Huang, F.; Lan, X.; Wang, X.; Wang, J.; Meng, C. Electrochemically reduced graphene oxide and Nafion nanocomposite for ultralow potential detection of organophosphate pesticide. Sens. Actuators B Chem. 2013, 177, 724–729. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.; Zhang, Y.; Ma, Z.; Zhu, M.; Gao, E. A lanthanide metal–organic framework as ratio fluorescence probe to detect pesticides in water. Inorganica Chim. Acta 2021, 528, 120632. [Google Scholar] [CrossRef]
- Pan, J.; Chen, W.; Ma, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587. [Google Scholar] [CrossRef]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular imprinting of macromolecules for sensor applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef]
- Prasad, B.B.; Jauhari, D.; Tiwari, M.P. Doubly imprinted polymer nanofilm-modified electrochemical sensor for ultra-trace simultaneous analysis of glyphosate and glufosinate. Biosens. Bioelectron. 2014, 59, 81–88. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef]
- Mazouz, Z.; Touchente, Z.A.; Laradi, H.; Fourati, N.; Yaakoubi, N.; Touzani, R.; Chehimi, M.M.; Kalfat, R.; Othmane, A.; Zerrouki, C. Design of Novel Electrochemical Sensors for the Selective Detection of Glyphosate. Proceedings 2017, 1, 483. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Lee, J.; Shin, I.S. Advanced method for fabrication of molecularly imprinted mesoporous organosilica with highly sensitive and selective recognition of glyphosate. Sci. Rep. 2019, 9, 10293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawetwong, P.; Chairam, S.; Jarujamrus, P.; Amatatongchai, M. Enhanced selectivity and sensitivity for colorimetric determination of glyphosate using Mn–ZnS quantum dot embedded molecularly imprinted polymers combined with a 3D-microfluidic paper-based analytical device. Talanta 2021, 225, 122077. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Lyu, Z.; Li, S.; Ruan, X.; Fei, M.; Zhou, Y.; Niu, X.; Zhu, W.; Du, D.; Lin, Y. Molecularly imprinted polypyrrole nanotubes based electrochemical sensor for glyphosate detection. Biosens. Bioelectron. 2021, 191, 113434. [Google Scholar] [CrossRef]
- Thimoonnee, S.; Somnet, K.; Ngaosri, P.; Chairam, S.; Karuwan, C.; Kamsong, W.; Tuantranont, A.; Amatatongchai, M. Fast, sensitive and selective simultaneous determination of paraquat and glyphosate herbicides in water samples using a compact electrochemical sensor. Anal. Methods 2022, 14, 820–833. [Google Scholar] [CrossRef]
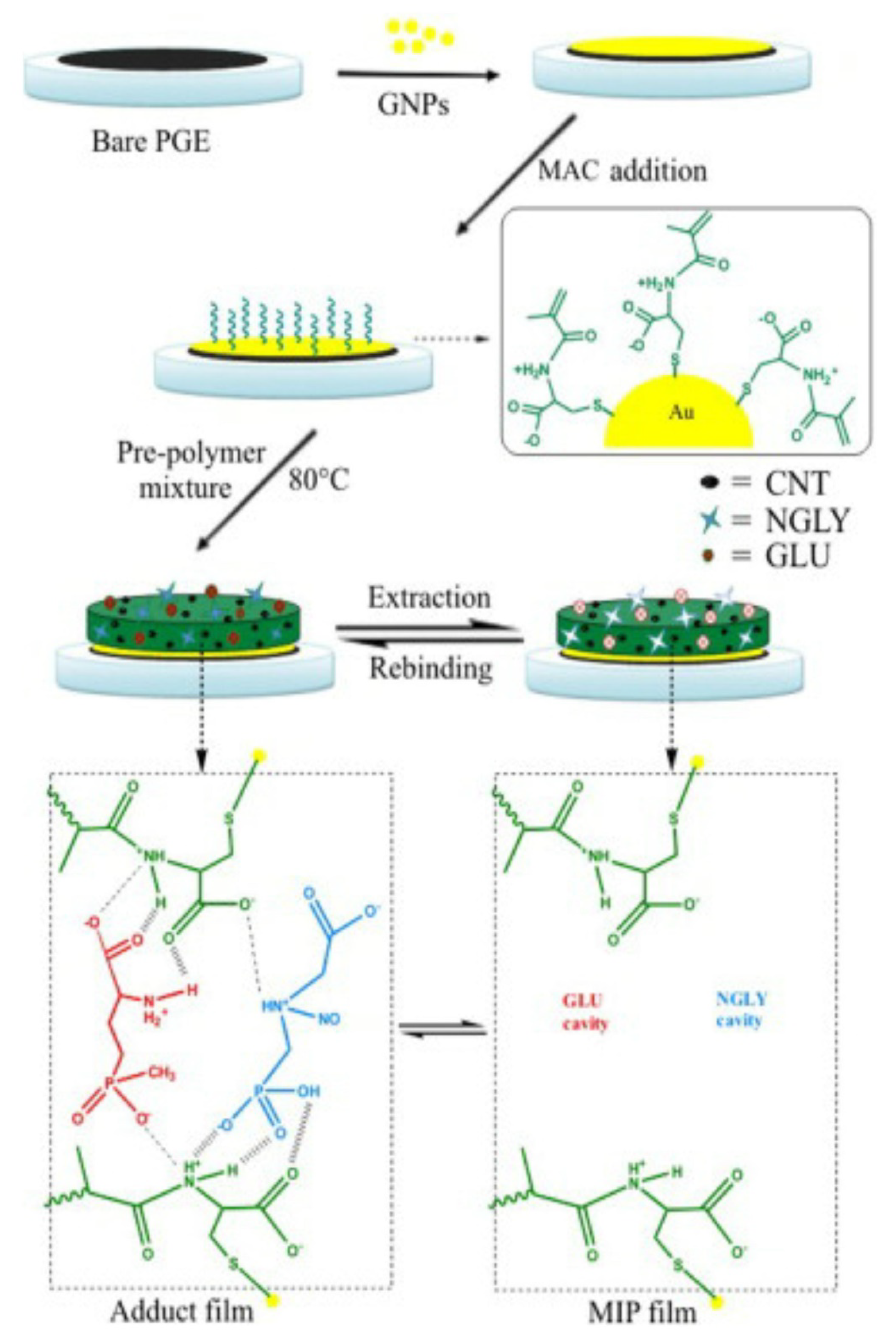
- Zhang, C.; She, Y.; Li, T.; Zhao, F.; Jin, M.; Guo, Y.; Zheng, L.; Wang, S.; Jin, F.; Shao, H.; et al. A highly selective electrochemical sensor based on molecularly imprinted polypyrrole-modified gold electrode for the determination of glyphosate in cucumber and tap water. Anal. Bioanal. Chem. 2017, 409, 7133–7144. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Wu, K.; Zhang, L.; Ge, S.; Yu, J. A molecularly imprinted polypyrrole for ultrasensitive voltammetric determination of glyphosate. Microchim. Acta 2017, 184, 1959–1967. [Google Scholar] [CrossRef]
- Gui, M.; Jiang, J.; Wang, X.; Yan, Y.; Li, S.; Xiao, X.; Liu, T.; Liu, T.; Feng, Y. Copper ion-mediated glyphosate detection with N-heterocycle based polyacetylene as a sensing platform. Sens. Actuators B Chem. 2017, 243, 696–703. [Google Scholar] [CrossRef]
- Songa, E.A.; Waryo, T.; Jahed, N.; Baker, P.G.L.; Kgarebe, B.V.; Iwuoha, E.I. Electrochemical nanobiosensor for glyphosate herbicide and Its metabolite. Electroanalysis 2009, 21, 671–674. [Google Scholar] [CrossRef]
- Zouaoui, F.; Bourouina-Bacha, S.; Bourouina, M.; Abroa-Nemeir, I.; Halima, H.B.; Gallardo-Gonzalez, J.; El Alami El Hassani, N.; Alcacer, A.; Bausells, J.; Jaffrezic-Renault, N.; et al. Electrochemical impedance spectroscopy determination of glyphosate using a molecularly imprinted chitosan. Sens. Actuators B Chem. 2020, 309, 127753. [Google Scholar] [CrossRef]
- Aydin, Z.; Keleş, M. A reaction-based system for the colorimetric detection of glyphosate in real samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120501. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.S.; Pontes, M.S.; Santiago, E.F.; Fiorucci, A.R.; Arruda, G.J. An efficient and simple method using a graphite oxide electrochemical sensor for the determination of glyphosate in environmental samples. Sci. Total Environ. 2020, 749, 142385. [Google Scholar] [CrossRef]
- Wiwasuku, T.; Boonmak, J.; Burakham, R.; Hadsadee, S.; Jungsuttiwong, S.; Bureekaew, S.; Promarak, V.; Youngme, S. Turn-on fluorescent probe towards glyphosate and Cr3+based on Cd(ii)-metal organic framework with Lewis basic sites. Inorg. Chem. Front. 2021, 8, 977–988. [Google Scholar] [CrossRef]
- Hu, T.; Xu, J.; Ye, Y.; Han, Y.; Li, X.; Wang, Z.; Sun, D.; Zhou, Y.; Ni, Z. Visual detection of mixed organophosphorous pesticide using QD-AChE aerogel based microfluidic arrays sensor. Biosens. Bioelectron. 2019, 136, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Y.; Wang, C.; He, M.; Lin, Q. Selective detection of water pollutants using a differential aptamer-based graphene biosensor. Biosens. Bioelectron. 2019, 126, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bonizzoni, M. A supramolecular sensing array for qualitative and quantitative analysis of organophosphates in water. J. Am. Chem. Soc. 2014, 136, 14223–14229. [Google Scholar] [CrossRef] [PubMed]
- Banna, M.; Bera, K.; Sochol, R.; Lin, L.; Najjaran, H.; Sadiq, R.; Hoorfar, M. 3D printing-based integrated water quality sensing system. Sensors 2017, 17, 1336. [Google Scholar] [CrossRef] [PubMed]
- Saftics, A.; Türk, B.; Sulyok, A.; Nagy, N.; Gerecsei, T.; Szekacs, I.; Kurunczi, S.; Horvath, R. Biomimetic Dextran-Based Hydrogel Layers for Cell Micropatterning over Large Areas Using the FluidFM BOT Technology. Langmuir 2019, 35, 2412–2421. [Google Scholar] [CrossRef]
- Fan, Y.; Dong, D.; Li, Q.; Si, H.; Pei, H.; Li, L.; Tang, B. Fluorescent analysis of bioactive molecules in single cells based on microfluidic chips. Lab Chip 2018, 18, 1151–1173. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, J.; Zhang, P.; Zhang, Y.; Miao, Y.; Gao, S.; Deng, Y.; Geng, L. Recent advances in microfluidic chip integrated electronic biosensors for multiplexed detection. Biosens. Bioelectron. 2018, 121, 272–280. [Google Scholar] [CrossRef]
- Da Silva Freire, C.; Da Silva Moreira, C.; De Souza Filho, C.A.; Moreno Santa Cruz, R.; Falqueto, A.; Valle, A.L.; Goulart Filho, L.R.; Souto De Medeiros, E.; Nascimento Ferreira, K. Do Application of a Smartphone-based SPR platform for Glyphosate detection. In Proceedings of the 2019 IEEE Sensors Applications Symposium (SAS), Sophia Antipolis, France, 11–13 March 2019; pp. 1–6. [Google Scholar] [CrossRef]



















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zúñiga, K.; Rebollar, G.; Avelar, M.; Campos-Terán, J.; Torres, E. Nanomaterial-Based Sensors for the Detection of Glyphosate. Water 2022, 14, 2436. https://doi.org/10.3390/w14152436
Zúñiga K, Rebollar G, Avelar M, Campos-Terán J, Torres E. Nanomaterial-Based Sensors for the Detection of Glyphosate. Water. 2022; 14(15):2436. https://doi.org/10.3390/w14152436
Chicago/Turabian StyleZúñiga, Karem, Georgette Rebollar, Mayra Avelar, José Campos-Terán, and Eduardo Torres. 2022. "Nanomaterial-Based Sensors for the Detection of Glyphosate" Water 14, no. 15: 2436. https://doi.org/10.3390/w14152436
APA StyleZúñiga, K., Rebollar, G., Avelar, M., Campos-Terán, J., & Torres, E. (2022). Nanomaterial-Based Sensors for the Detection of Glyphosate. Water, 14(15), 2436. https://doi.org/10.3390/w14152436






