Anaerobic Digestion of the Organic Fraction of Municipal Solid Waste in Plug-Flow Reactors: Focus on Bacterial Community Metabolic Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pilot Scale Plug-Flow Reactor
2.2. Substrate and Inoculum
2.3. Experimental Design
2.4. Analytical Methods
2.5. Process Performance
2.6. Microbial Community Analysis
Data Analysis
3. Results
3.1. Substrate and Inoculum Characterization
3.2. Process Performances
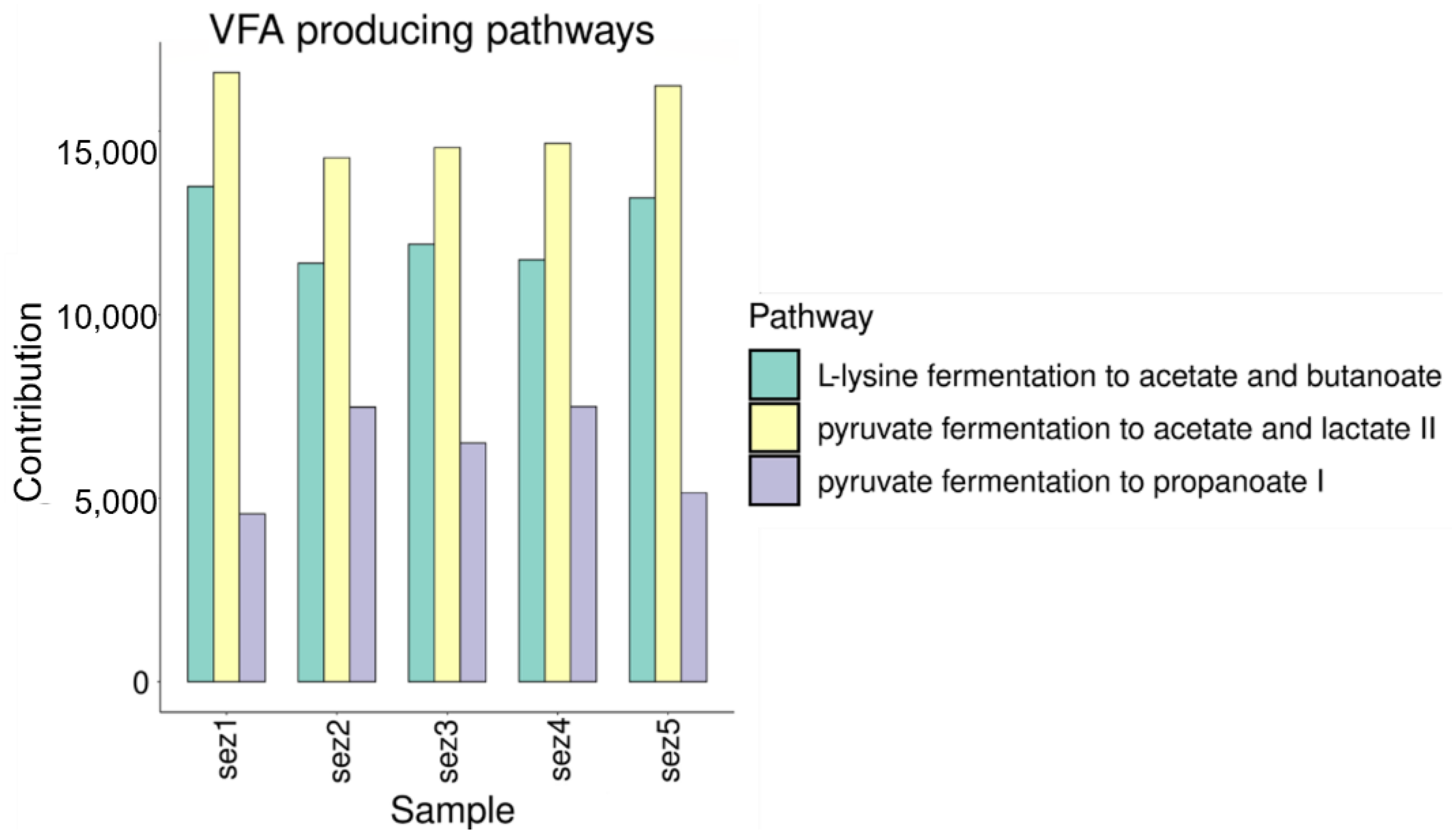
3.2.1. VFA Production
3.2.2. Biogas Production
3.3. Bacterial Community Analysis
Functional Analysis
4. Discussion
4.1. VFA and Biogas Production
4.2. Microbial Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A

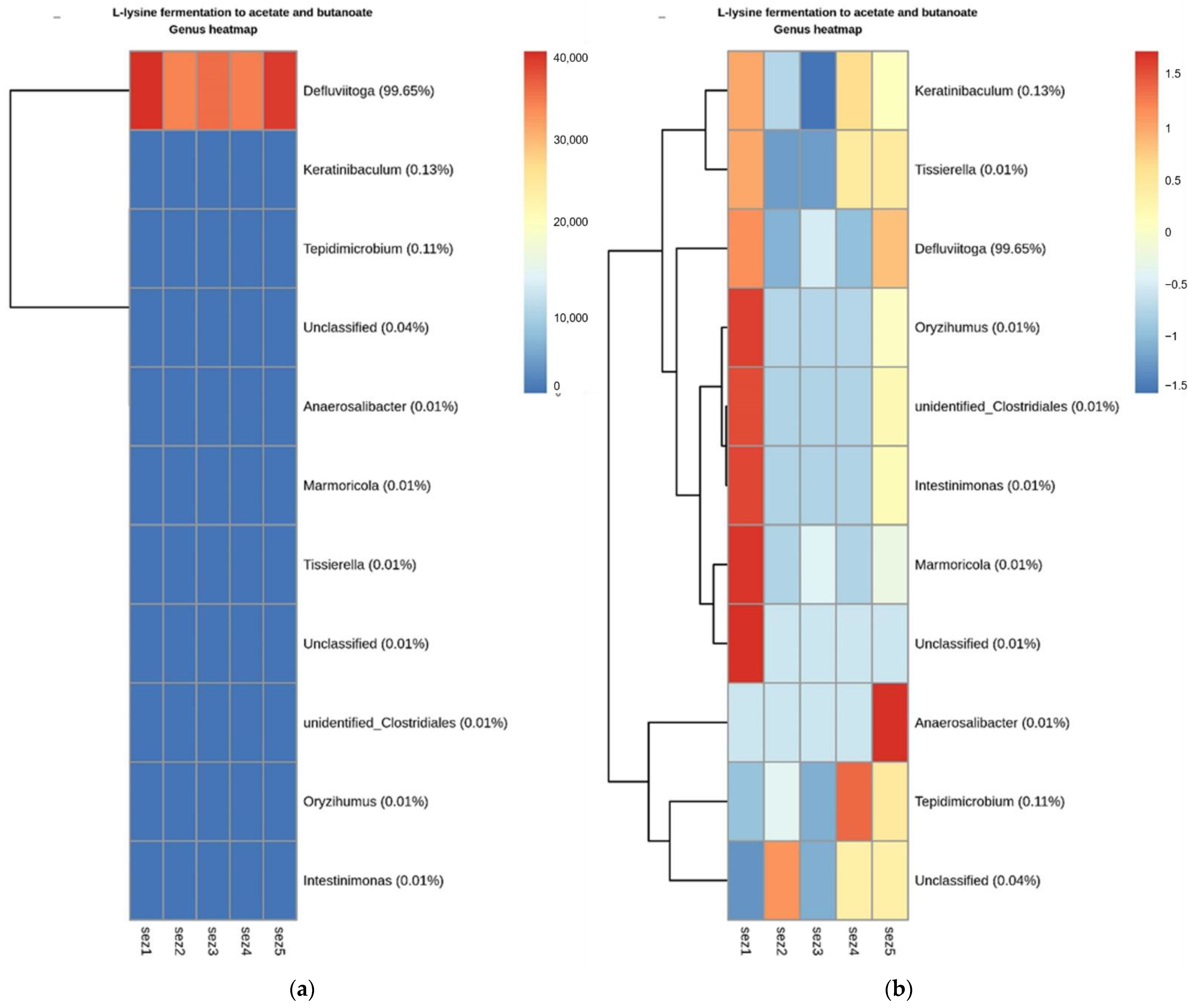








Appendix B
| MetaCyc Code | Pathway |
|---|---|
| 2OXOBUTYRATECAT-PWY | 2-oxobutanoate degradation II |
| PWY-5046 | 2-oxoisovalerate decarboxylation to isobutanoyl-CoA |
| PWY-5535 | acetate formation from acetyl-CoA II |
| PWY-5536 | acetate formation from acetyl-CoA III (succinate) |
| PWY-5676 | acetyl-CoA fermentation to butanoate II |
| PWY0-43 | conversion of succinate to propanoate |
| PROPFERM-PWY | L-alanine fermentation to propanoate and acetate |
| P162-PWY | L-glutamate degradation V (via hydroxyglutarate) |
| PWY-5088 | L-glutamate degradation VIII (to propanoate) |
| PWY-5075 | L-leucine degradation II |
| P163-PWY | L-lysine fermentation to acetate and butanoate |
| PWY-5096 | pyruvate fermentation to acetate and alanine |
| P41-PWY | pyruvate fermentation to acetate and lactate I |
| PWY-5100 | pyruvate fermentation to acetate and lactate II |
| P142-PWY | pyruvate fermentation to acetate I |
| PWY-5482 | pyruvate fermentation to acetate II |
| PWY-5483 | pyruvate fermentation to acetate III |
| PWY-5485 | pyruvate fermentation to acetate IV |
| PWY-5538 | pyruvate fermentation to acetate VI |
| PWY-5600 | pyruvate fermentation to acetate VII |
| PWY-5768 | pyruvate fermentation to acetate VIII |
| P108-PWY | pyruvate fermentation to propanoate I |
| PWY-5494 | pyruvate fermentation to propanoate II (acrylate pathway) |
| PWY-5677 | succinate fermentation to butanoate |
References
- European Commission Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions A new Circular Economy Action Plan for a Cleaner and More Competitive Europe. 2020. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?qid=1583933814386&uri=COM:2020:98:FIN (accessed on 3 September 2021).
- De Schoenmakere, M.; Hoogeveen, Y.; Gillabel, J.; Manshoven, S. European Environment Agency The Circular Economy and the Bioeconomy: Partners in Sustainability. Publications Office. 2018. Available online: https://data.europa.eu/doi/10.2800/00956 (accessed on 25 September 2021).
- Council Directive (EU) 2018/851 of the European Parliament and of the Council of 30 May 2018 Amending Directive 2008/98/EC on Waste (Text with EEA Relevance); 2018; OJ L 150/109. Available online: https://eur-lex.europa.eu/eli/dir/2018/851/oj/eng (accessed on 4 November 2021).
- Directive (EU) 2018/850 of the European Parliament and of the Council of 30 May 2018 Amending Directive 1999/31/EC on the Landfill of Waste (Text with EEA Relevance); 2018; OJ L 150/100. Available online: http://data.europa.eu/eli/dir/2018/850/oj/eng (accessed on 4 November 2021).
- Strazzera, G.; Battista, F.; Garcia, N.H.; Frison, N.; Bolzonella, D. Volatile Fatty Acids Production from Food Wastes for Biorefinery Platforms: A Review. J. Environ. Manag. 2018, 226, 278–288. [Google Scholar] [CrossRef]
- Ubando, A.T.; Felix, C.B.; Chen, W.-H. Biorefineries in Circular Bioeconomy: A Comprehensive Review. Bioresour. Technol. 2020, 299, 122585. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Yang, Y.-H. Microbial Production of Volatile Fatty Acids: Current Status and Future Perspectives. Rev. Environ. Sci. Biotechnol. 2017, 16, 327–345. [Google Scholar] [CrossRef]
- Rocamora, I.; Wagland, S.T.; Villa, R.; Simpson, E.W.; Fernández, O.; Bajón-Fernández, Y. Dry Anaerobic Digestion of Organic Waste: A Review of Operational Parameters and Their Impact on Process Performance. Bioresour. Technol. 2020, 299, 122681. [Google Scholar] [CrossRef]
- Karthikeyan, O.P.; Visvanathan, C. Bio-Energy Recovery from High-Solid Organic Substrates by Dry Anaerobic Bio-Conversion Processes: A Review. Rev. Environ. Sci. Biotechnol. 2013, 12, 257–284. [Google Scholar] [CrossRef]
- Rosenheim, H.; De, I.; Hyvedemm, S. Anaerobic Digestion—Factsheet. 2015. Available online: https://docs.european-bioplastics.org/publications/bp/EUBP_BP_Anaerobic_digestion.pdf (accessed on 11 November 2021).
- Mao, C.; Feng, Y.; Wang, X.; Ren, G. Review on Research Achievements of Biogas from Anaerobic Digestion. Renew. Sustain. Energy Rev. 2015, 45, 540–555. [Google Scholar] [CrossRef]
- Karthikeyan, O.P.; Visvanathan, C. Effect of C/N Ratio and Ammonia-N Accumulation in a Pilot-Scale Thermophilic Dry Anaerobic Digester. Bioresour. Technol. 2012, 113, 294–302. [Google Scholar] [CrossRef]
- Jabeen, M.; Zeshan; Yousaf, S.; Haider, M.R.; Malik, R.N. High-Solids Anaerobic Co-Digestion of Food Waste and Rice Husk at Different Organic Loading Rates. Int. Biodeterior. Biodegrad. 2015, 102, 149–153. [Google Scholar] [CrossRef]
- Patinvoh, R.J.; Kalantar Mehrjerdi, A.; Sárvári Horváth, I.; Taherzadeh, M.J. Dry Fermentation of Manure with Straw in Continuous Plug Flow Reactor: Reactor Development and Process Stability at Different Loading Rates. Bioresour. Technol. 2017, 224, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Aguirre, J.; Aymerich, E.; de Goñi, J.G.M.; Esteban-Gutiérrez, M. Selective VFA Production Potential from Organic Waste Streams: Assessing Temperature and PH Influence. Bioresour. Technol. 2017, 244, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Slezak, R.; Grzelak, J.; Krzystek, L.; Ledakowicz, S. The Effect of Initial Organic Load of the Kitchen Waste on the Production of VFA and H 2 in Dark Fermentation. Waste Manag. 2017, 68, 610–617. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Qiu, Y.; Ren, L.; Jiang, B. Microbial Characteristics in Anaerobic Digestion Process of Food Waste for Methane Production—A Review. Bioresour. Technol. 2018, 248, 29–36. [Google Scholar] [CrossRef]
- Khatami, K.; Atasoy, M.; Ludtke, M.; Baresel, C.; Eyice, Ö.; Cetecioglu, Z. Bioconversion of Food Waste to Volatile Fatty Acids: Impact of Microbial Community, PH and Retention Time. Chemosphere 2021, 275, 129981. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Li, Z.; Feng, J.; Zhao, L.; Yu, J. Effects of Digestate Recirculation Ratios on Biogas Production and Methane Yield of Continuous Dry Anaerobic Digestion. Bioresour. Technol. 2020, 316, 123963. [Google Scholar] [CrossRef]
- Baldi, F.; Pecorini, I.; Iannelli, R. Comparison of Single-Stage and Two-Stage Anaerobic Co-Digestion of Food Waste and Activated Sludge for Hydrogen and Methane Production. Renew. Energy 2019, 143, 1755–1765. [Google Scholar] [CrossRef]
- Angelidaki, I.; Alves, M.; Bolzonella, D.; Borzacconi, L.; Campos, J.L.; Guwy, A.J.; Kalyuzhnyi, S.; Jenicek, P.; van Lier, J.B. Defining the Biomethane Potential (BMP) of Solid Organic Wastes and Energy Crops: A Proposed Protocol for Batch Assays. Water Sci. Technol. 2009, 59, 927–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagbohungbe, M.O.; Dodd, I.C.; Herbert, B.M.J.; Li, H.; Ricketts, L.; Semple, K.T. High Solid Anaerobic Digestion: Operational Challenges and Possibilities. Environ. Technol. Innov. 2015, 4, 268–284. [Google Scholar] [CrossRef]
- Metcalf, N.A.; Eddy, I.; Tchobanoglous, G.; Burton, F.; Stensel, H.D. Wastewater Engineering: Treatment and Reuse; 4° Edizione; McGraw-Hill Education/Asia: Boston, MA, USA, 2002; ISBN 978-0-07-124140-3. [Google Scholar]
- Agenzia Nazionale per Protezione dell’Ambiente, Metodi di Analisi del Compost; ANPA: Roma, Italy, 2001; ISBN 88-448-0258-9.
- Ripley, L.E.; Boyle, W.C.; Converse, J.C. Improved Alkalimetric Monitoring for Anaerobic Digestion of High-Strength Wastes. J. Water Pollut. Control Fed. 1986, 58, 406–411. [Google Scholar]
- Drosg, B. Process Monitoring in Biogas Plants. 40. Available online: https://www.ieabioenergy.com/wp-content/uploads/2013/12/Technical-Brochure-process_montoring.pdf (accessed on 16 October 2021).
- Rajagopal, R.; Massé, D.I.; Singh, G. A Critical Review on Inhibition of Anaerobic Digestion Process by Excess Ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef]
- Baldi, F.; Iannelli, R.; Pecorini, I.; Polettini, A.; Pomi, R.; Rossi, A. Influence of the PH Control Strategy and Reactor Volume on Batch Fermentative Hydrogen Production from the Organic Fraction of Municipal Solid Waste. Waste Manag. Res. 2019, 37, 478–485. [Google Scholar] [CrossRef] [Green Version]
- Kothari, R.; Pandey, A.K.; Kumar, S.; Tyagi, V.V.; Tyagi, S.K. Different Aspects of Dry Anaerobic Digestion for Bio-Energy: An Overview. Renew. Sustain. Energy Rev. 2014, 39, 174–195. [Google Scholar] [CrossRef]
- Microbiome Informatics: OTU vs. ASV. Available online: https://www.zymoresearch.com/blogs/blog/microbiome-informatics-otu-vs-asv (accessed on 6 December 2021).
- Sikora, A.; Detman, A.; Mielecki, D.; Chojnacka, A.; Błaszczyk, M. Searching for Metabolic Pathways of Anaerobic Digestion: A Useful List of the Key Enzymes; IntechOpen: London, UK, 2018; ISBN 978-1-83881-850-0. [Google Scholar]
- Wiechmann, A.; Ciurus, S.; Oswald, F.; Seiler, V.N.; Müller, V. It Does Not Always Take Two to Tango: “Syntrophy” via Hydrogen Cycling in One Bacterial Cell. ISME J. 2020, 14, 1561–1570. [Google Scholar] [CrossRef]
- Lee, W.S.; Chua, A.S.M.; Yeoh, H.K.; Ngoh, G.C. A Review of the Production and Applications of Waste-Derived Volatile Fatty Acids. Chem. Eng. J. 2014, 235, 83–99. [Google Scholar] [CrossRef]
- Zhou, M.; Yan, B.; Wong, J.W.C.; Zhang, Y. Enhanced Volatile Fatty Acids Production from Anaerobic Fermentation of Food Waste: A Mini-Review Focusing on Acidogenic Metabolic Pathways. Bioresour. Technol. 2018, 248, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qin, Y.; Kong, Z.; Wu, J.; Kubota, K.; Li, Y.-Y. Characterization of Microbial Community and Main Functional Groups of Prokaryotes in Thermophilic Anaerobic Co-Digestion of Food Waste and Paper Waste. Sci. Total Environ. 2019, 652, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, C.; Sun, F.; Zhu, W.; Wu, W. A Comparison of Microbial Characteristics between the Thermophilic and Mesophilic Anaerobic Digesters Exposed to Elevated Food Waste Loadings. Bioresour. Technol. 2014, 152, 420–428. [Google Scholar] [CrossRef]
- Dyksma, S.; Jansen, L.; Gallert, C. Syntrophic Acetate Oxidation Replaces Acetoclastic Methanogenesis during Thermophilic Digestion of Biowaste. Microbiome 2020, 8, 105. [Google Scholar] [CrossRef]
- Fernández-Domínguez, D.; Astals, S.; Peces, M.; Frison, N.; Bolzonella, D.; Mata-Alvarez, J.; Dosta, J. Volatile Fatty Acids Production from Biowaste at Mechanical-Biological Treatment Plants: Focusing on Fermentation Temperature. Bioresour. Technol. 2020, 314, 123729. [Google Scholar] [CrossRef]
- Campuzano, R.; González-Martínez, S. Characteristics of the Organic Fraction of Municipal Solid Waste and Methane Production: A Review. Waste Manag. 2016, 54, 3–12. [Google Scholar] [CrossRef]
- Garcia-Aguirre, J.; Esteban-Gutiérrez, M.; Irizar, I.; González-Mtnez de Goñi, J.; Aymerich, E. Continuous Acidogenic Fermentation: Narrowing the Gap between Laboratory Testing and Industrial Application. Bioresour. Technol. 2019, 282, 407–416. [Google Scholar] [CrossRef]
- Papa, G.; Pepè Sciarria, T.; Carrara, A.; Scaglia, B.; D’Imporzano, G.; Adani, F. Implementing Polyhydroxyalkanoates Production to Anaerobic Digestion of Organic Fraction of Municipal Solid Waste to Diversify Products and Increase Total Energy Recovery. Bioresour. Technol. 2020, 318, 124270. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Gutiérrez, M.; Garcia-Aguirre, J.; Irizar, I.; Aymerich, E. From Sewage Sludge and Agri-Food Waste to VFA: Individual Acid Production Potential and up-Scaling. Waste Manag. 2018, 77, 203–212. [Google Scholar] [CrossRef]
- Yoo, C.G.; Meng, X.; Pu, Y.; Ragauskas, A.J. The Critical Role of Lignin in Lignocellulosic Biomass Conversion and Recent Pretreatment Strategies: A Comprehensive Review. Bioresour. Technol. 2020, 301, 122784. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of Anaerobic Digestion Process: A Review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef]
- Liu, T.; Schnürer, A.; Björkmalm, J.; Willquist, K.; Kreuger, E. Diversity and Abundance of Microbial Communities in UASB Reactors during Methane Production from Hydrolyzed Wheat Straw and Lucerne. Microorganisms 2020, 8, 1394. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Y.; Marandola, C.; Krooneman, J.; Euverink, G.J.W. Comparison of the Microbial Communities in Anaerobic Digesters Treating High Alkalinity Synthetic Wastewater at Atmospheric and High-Pressure (11 Bar). Bioresour. Technol. 2020, 318, 124101. [Google Scholar] [CrossRef]
- Svensson, K.; Paruch, L.; Gaby, J.C.; Linjordet, R. Feeding Frequency Influences Process Performance and Microbial Community Composition in Anaerobic Digesters Treating Steam Exploded Food Waste. Bioresour. Technol. 2018, 269, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Ao, T.; Xie, Z.; Zhou, P.; Liu, X.; Wan, L.; Li, D. Comparison of Functional Microbial Profile between Mesophilic and Thermophilic Anaerobic Digestion of Vegetable Waste. 2021. Available online: https://www.researchsquare.com/article/rs-54152/v1 (accessed on 18 October 2021).
- Jang, H.M.; Ha, J.H.; Kim, M.-S.; Kim, J.-O.; Kim, Y.M.; Park, J.M. Effect of Increased Load of High-Strength Food Wastewater in Thermophilic and Mesophilic Anaerobic Co-Digestion of Waste Activated Sludge on Bacterial Community Structure. Water Res. 2016, 99, 140–148. [Google Scholar] [CrossRef]
- Maus, I.; Cibis, K.G.; Bremges, A.; Stolze, Y.; Wibberg, D.; Tomazetto, G.; Blom, J.; Sczyrba, A.; König, H.; Pühler, A.; et al. Genomic Characterization of Defluviitoga tunisiensis L3, a Key Hydrolytic Bacterium in a Thermophilic Biogas Plant and Its Abundance as Determined by Metagenome Fragment Recruitment. J. Biotechnol. 2016, 232, 50–60. [Google Scholar] [CrossRef]
- Sun, L.; Toyonaga, M.; Ohashi, A.; Tourlousse, D.M.; Matsuura, N.; Meng, X.-Y.; Tamaki, H.; Hanada, S.; Cruz, R.; Yamaguchi, T.; et al. Lentimicrobium saccharophilum Gen. Nov., Sp. Nov., a Strictly Anaerobic Bacterium Representing a New Family in the Phylum Bacteroidetes, and Proposal of Lentimicrobiaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 2635–2642. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Dong, X. Proteiniphilum acetatigenes Gen. Nov., Sp. Nov., from a UASB Reactor Treating Brewery Wastewater. Int. J. Syst. Evol. Microbiol. 2005, 55, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomazetto, G.; Hahnke, S.; Wibberg, D.; Pühler, A.; Klocke, M.; Schlüter, A. Proteiniphilum saccharofermentans Str. M3/6T Isolated from a Laboratory Biogas Reactor Is Versatile in Polysaccharide and Oligopeptide Utilization as Deduced from Genome-Based Metabolic Reconstructions. Biotechnol. Rep. 2018, 18, e00254. [Google Scholar] [CrossRef]
- Breure, A.M.; Mooijman, K.A.; van Andel, J.G. Protein Degradation in Anaerobic Digestion: Influence of Volatile Fatty Acids and Carbohydrates on Hydrolysis and Acidogenic Fermentation of Gelatin. Appl. Microbiol. Biotechnol. 1986, 24, 426–431. [Google Scholar] [CrossRef]
- Bouanane-Darenfed, A.; Ben Hania, W.; Cayol, J.-L.; Ollivier, B.; Fardeau, M.-L. Reclassification of Acetomicrobium faecale as Caldicoprobacter faecalis Comb. Nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 3286–3288. [Google Scholar] [CrossRef]
- Hazlewood, G.P.; Gilbert, H.J. Xylan and Cellulose Utilization by the Clostridia. Biotechnol. Read. Mass 1993, 25, 311–341. [Google Scholar]
- Fonknechten, N.; Chaussonnerie, S.; Tricot, S.; Lajus, A.; Andreesen, J.R.; Perchat, N.; Pelletier, E.; Gouyvenoux, M.; Barbe, V.; Salanoubat, M.; et al. Clostridium sticklandii, a Specialist in Amino Acid Degradation:Revisiting Its Metabolism through Its Genome Sequence. BMC Genom. 2010, 11, 555. [Google Scholar] [CrossRef] [Green Version]
- Park, G.W.; Kim, I.; Jung, K.; Seo, C.; Han, J.-I.; Chang, H.N.; Kim, Y.-C. Enhancement of Volatile Fatty Acids Production from Rice Straw via Anaerobic Digestion with Chemical Pretreatment. Bioprocess Biosyst. Eng. 2015, 38, 1623–1627. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Enumeration of Amino Acid Fermenting Bacteria in the Human Large Intestine: Effects of PH and Starch on Peptide Metabolism and Dissimilation of Amino Acids. FEMS Microbiol. Ecol. 1998, 25, 355–368. [Google Scholar] [CrossRef]
- Westerholm, M.; Roos, S.; Schnürer, A. Syntrophaceticus schinkii Gen. Nov., Sp. Nov., an Anaerobic, Syntrophic Acetate-Oxidizing Bacterium Isolated from a Mesophilic Anaerobic Filter. FEMS Microbiol. Lett. 2010, 309, 100–104. [Google Scholar] [CrossRef] [Green Version]
- Wainaina, S.; Lukitawesa; Kumar Awasthi, M.; Taherzadeh, M.J. Bioengineering of Anaerobic Digestion for Volatile Fatty Acids, Hydrogen or Methane Production: A Critical Review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef] [Green Version]
- Amha, Y.M.; Anwar, M.Z.; Brower, A.; Jacobsen, C.S.; Stadler, L.B.; Webster, T.M.; Smith, A.L. Inhibition of Anaerobic Digestion Processes: Applications of Molecular Tools. Bioresour. Technol. 2018, 247, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Einsle, O.; Rees, D.C. Structural Enzymology of Nitrogenase Enzymes. Chem. Rev. 2020, 120, 4969–5004. [Google Scholar] [CrossRef] [PubMed]
- Artz, J.H.; Zadvornyy, O.A.; Mulder, D.W.; Keable, S.M.; Cohen, A.E.; Ratzloff, M.W.; Williams, S.G.; Ginovska, B.; Kumar, N.; Song, J.; et al. Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases. J. Am. Chem. Soc. 2020, 142, 1227–1235. [Google Scholar] [CrossRef]
- Sokolova, T.G.; Henstra, A.-M.; Sipma, J.; Parshina, S.N.; Stams, A.J.M.; Lebedinsky, A.V. Diversity and Ecophysiological Features of Thermophilic Carboxydotrophic Anaerobes: Thermophilic Carboxydotrophic Anaerobes. FEMS Microbiol. Ecol. 2009, 68, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]




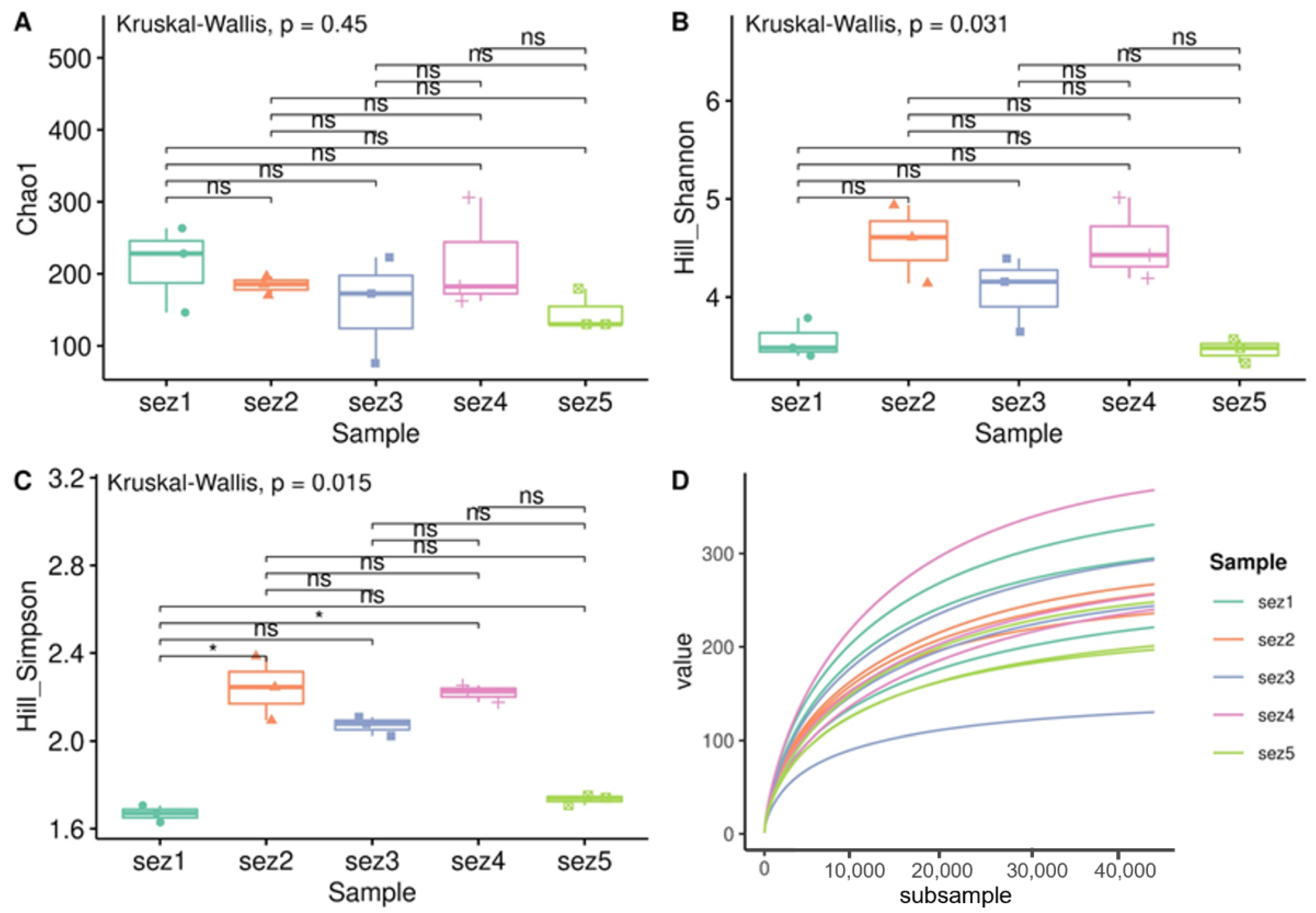




| Parameter ° | T1 | T2 | T3 | T4 | T5 |
|---|---|---|---|---|---|
| C/N [-] | 26.05 ± 2.6 | 18.08 ± 1.8 | 18.08 ± 1.8 | 19.19 ± 1.91 | 32.02 ± 3.2 |
| Carbohydrates [%VS] | 44.63 ± 4.24 | 33.75 ± 3.2 | 33.75 ± 3.2 | 25.96 ± 2.46 | 46.95 ± 4.46 |
| Proteins [%VS] | 13.52 ± 1.55 | 16.98 ± 1.95 | 16.98 ± 1.95 | 10.04 ± 1.15 | 14.41 ± 1.65 |
| Lipid [%VS] [%VS] | 1.51 ± 0.13 | 1.85 ± 0.16 | 1.85 ± 0.16 | 0.84 ± 0.07 | 1.74 ± 0.15 |
| Lignin [%VS] | 24.84 ± 2.73 | 19.81 ± 2.17 | 19.81 ± 2.17 | 16 ± 1.76 | 13.99 ± 1.53 |
| Cellulose [%VS] | 13.1 ± 1.04 | 18.43 ± 1.47 | 18.43 ± 1.47 | 8.83 ± 0.7 | 23.05 ± 1.84 |
| Parameter | T1 | T2 | T3 | T4 | T5 |
|---|---|---|---|---|---|
| pH [-] | 8.37 ± 0.41 | 8.33 ± 0.41 | 8.25 ± 0.41 | 8.24 ± 0.41 | 8.1 ± 0.4 |
| IA/PA [-] | 0.49 ± 0.04 | 0.59 ± 0.05 | 0.59 ± 0.05 | 0.52 ± 0.05 | 0.57 ± 0.05 |
| TAN [mg/L] | 2177.7 ± 108.88 | 3356.1 ± 167.8 | 2877.7 ± 143.88 | 1870.5 ± 93.52 | 1081.1 ± 54.05 |
| FA [mg/L] | 1034.8 ± 51.74 | 1132.3 ± 56.61 | 1092.7 ± 54.63 | 751.3 ± 37.56 | 393.09 ± 19.65 |
| Biogas [NLbiogas/d] | 43.21 ± 2.16 | 75.02 ± 3.75 | 85.08 ± 4.25 | 92.38 ± 4.61 | 145.03 ± 7.25 |
| SGP [NLbiogas/kgVS] | 182.2 ± 9.11 | 339.83 ± 16.99 | 429.99 ± 21.49 | 326.02 ± 16.3 | 364.04 ± 18.2 |
| SMP [NLCH4/kgVS] | 103.87 ± 5.19 | 198.69 ± 9.93 | 247.27 ± 12.36 | 165.08 ± 8.25 | 237.8 ± 11.89 |
| GPR [NLbiogas/(lr d)] | 1.56 ± 0.07 | 2.82 ± 0.14 | 3.12 ± 0.15 | 3.24 ± 0.16 | 5.11 ± 0.25 |
| VSout [%] | 69.78 ± 0.13 | 67.3 ± 0.13 | 63.96 ± 0.12 | 62.06 ± 0.12 | 62.68 ± 0.12 |
| REVS [%] | 52.91 ± 0.21 | 42.33 ± 0.16 | 40.09 ± 0.16 | 45.38 ± 0.18 | 52.1 ± 0.2 |
| CH4 [%] | 56.85 ± 2.53 | 58.47 ± 2.72 | 57.64 ± 2.62 | 50.63 ± 1.84 | 65.32 ± 3.61 |
| CO2 [%] | 43.15 ± 1.16 | 41.53 ± 1.03 | 42.36 ± 1.09 | 49.37 ± 1.72 | 34.68 ± 0.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, E.; Becarelli, S.; Pecorini, I.; Di Gregorio, S.; Iannelli, R. Anaerobic Digestion of the Organic Fraction of Municipal Solid Waste in Plug-Flow Reactors: Focus on Bacterial Community Metabolic Pathways. Water 2022, 14, 195. https://doi.org/10.3390/w14020195
Rossi E, Becarelli S, Pecorini I, Di Gregorio S, Iannelli R. Anaerobic Digestion of the Organic Fraction of Municipal Solid Waste in Plug-Flow Reactors: Focus on Bacterial Community Metabolic Pathways. Water. 2022; 14(2):195. https://doi.org/10.3390/w14020195
Chicago/Turabian StyleRossi, Elena, Simone Becarelli, Isabella Pecorini, Simona Di Gregorio, and Renato Iannelli. 2022. "Anaerobic Digestion of the Organic Fraction of Municipal Solid Waste in Plug-Flow Reactors: Focus on Bacterial Community Metabolic Pathways" Water 14, no. 2: 195. https://doi.org/10.3390/w14020195
APA StyleRossi, E., Becarelli, S., Pecorini, I., Di Gregorio, S., & Iannelli, R. (2022). Anaerobic Digestion of the Organic Fraction of Municipal Solid Waste in Plug-Flow Reactors: Focus on Bacterial Community Metabolic Pathways. Water, 14(2), 195. https://doi.org/10.3390/w14020195








