Ni2+ and Cu2+ Biosorption by EPS-Producing Serratia plymuthica Strains and Potential Bio-Catalysis of the Organo–Metal Complexes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture
2.2. Biomass Production
2.3. EPS Extraction and Quantification
2.4. EPS Nuclear Magnetic Resonance and Fourier-Transform Infrared Spectroscopy
2.5. Biosorption Experiments
2.6. Analytical Methods
2.7. Statistical Analysis
2.8. Adsorption Isotherms
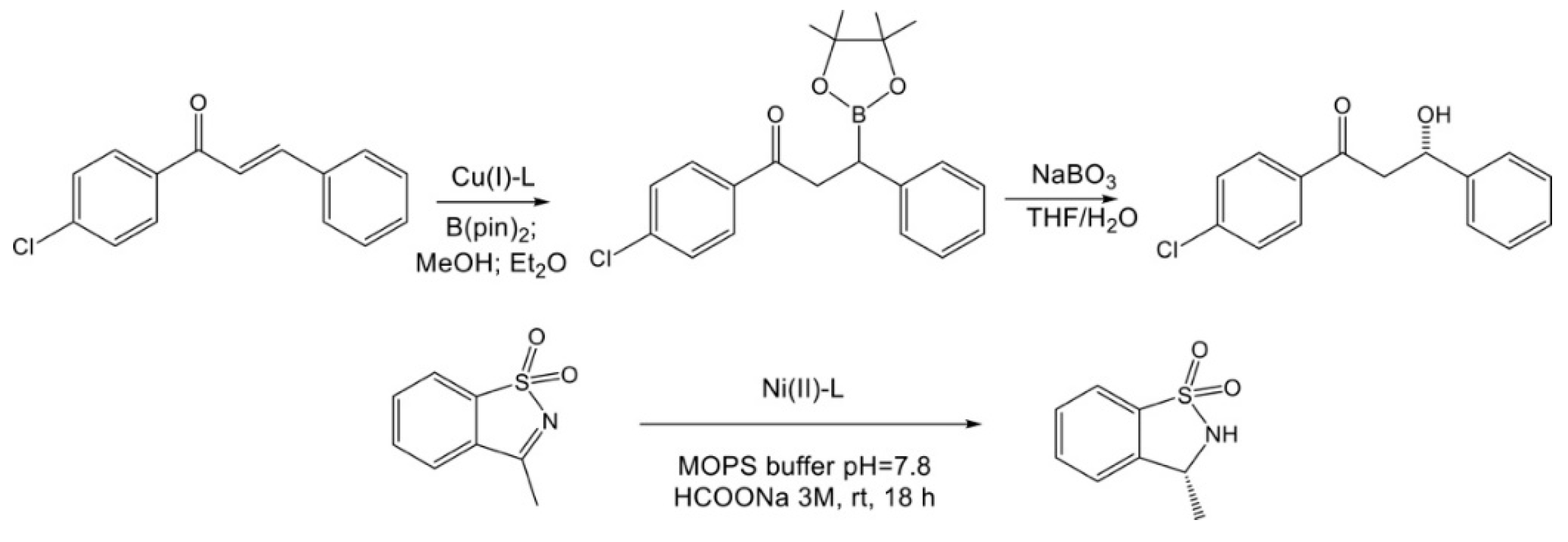
2.9. General Procedure for Catalyzed Addition of Bis(Pinacolate)Diboron on (E)-3-(4-Chlorophenyl)-1-phenylprop-2-en-1-one
2.10. General Procedure for Transfer Hydrogenation on 3-Methylbenzo[d]isothiazole 1,1-dioxide
3. Results and Discussion
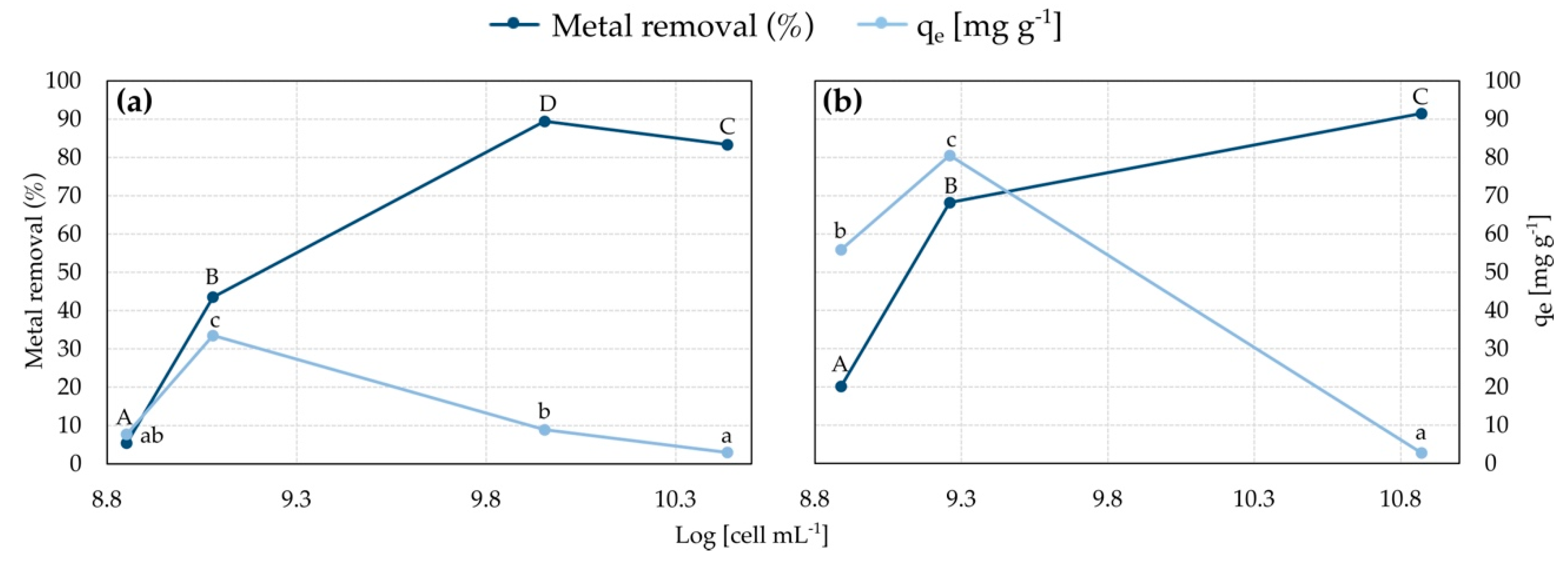
3.1. Metal Adsorption in the Biofilm-Based System
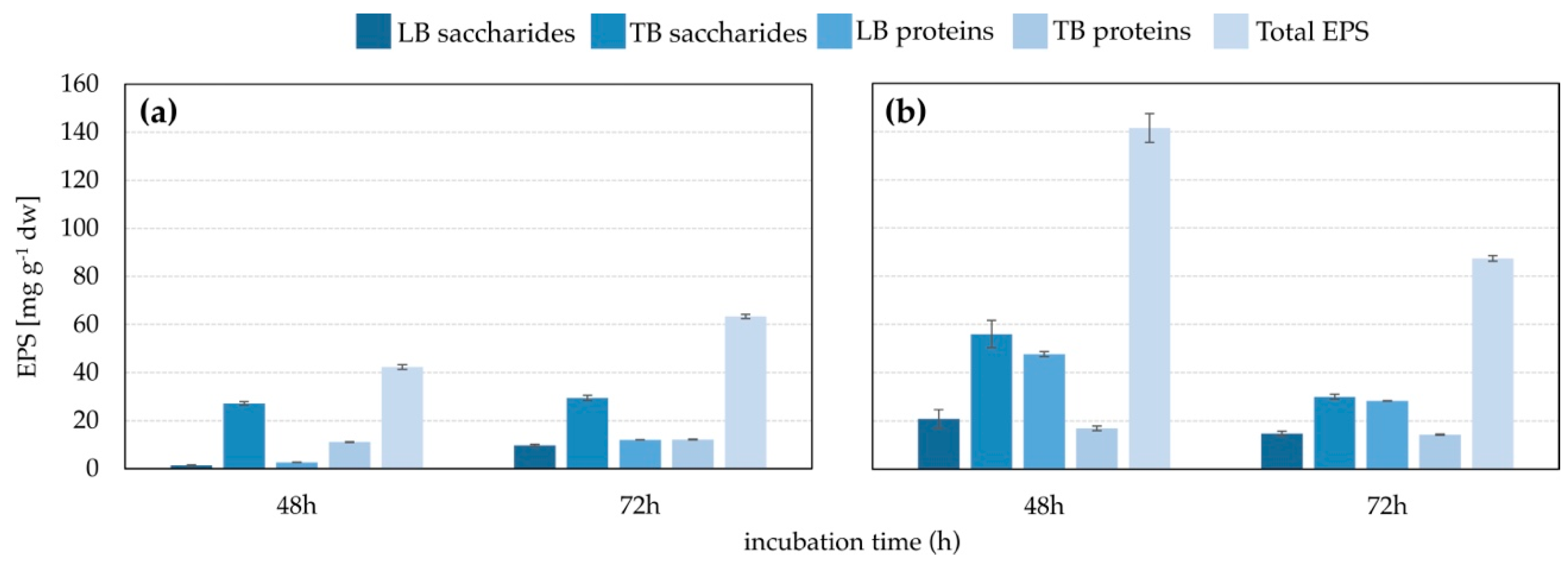
3.2. EPS Production
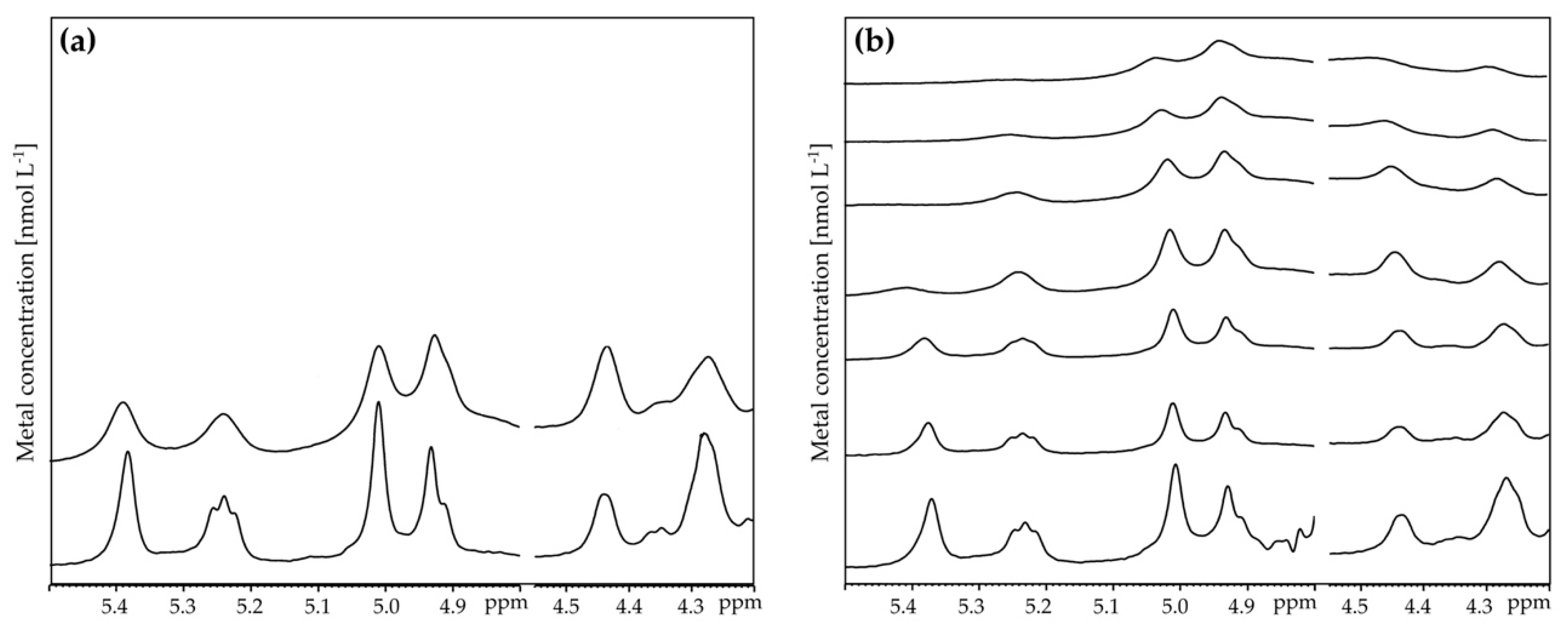
3.3. Analysis of the Interaction of EPS with Metals by NMR
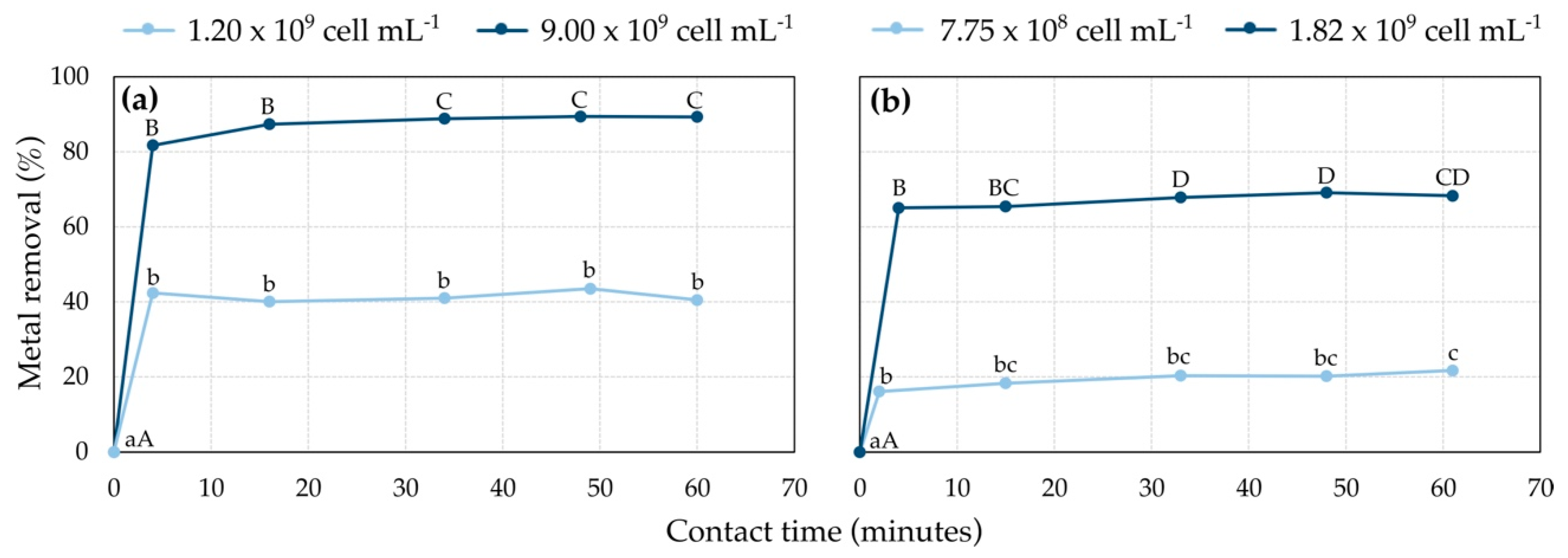
3.4. Cell Metal Adsorption: Effect of Contact Time
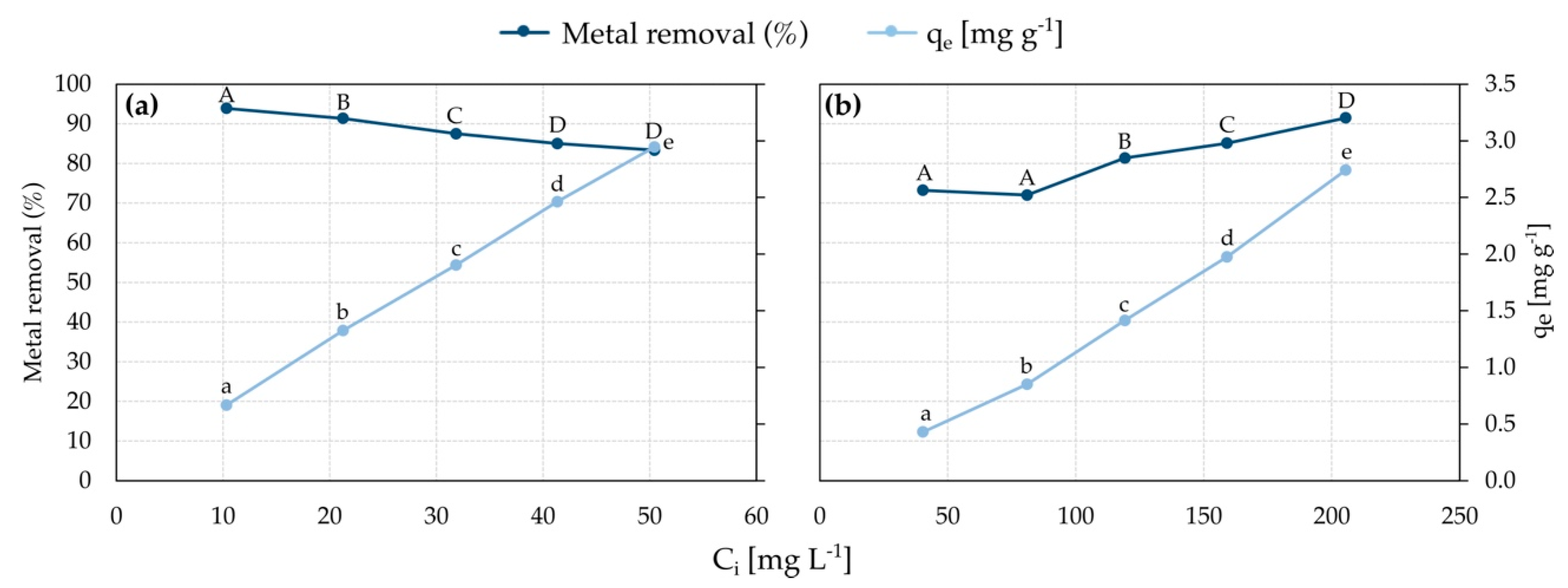
3.5. Cell Metal Adsorption: Effect of Initial Metal Concentration
3.6. Cell Metal Adsorption: Effect of Biomass Dose
3.7. Adsorption Isotherms
3.8. Catalytic Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rana, A.; Sindhu, M.; Kumar, A.; Dhaka, R.K.; Chahar, M.; Singh, S.; Nain, L. Restoration of heavy metal-contaminated soil and water through biosorbents: A review of current understanding and future challenges. Physiol. Plant. 2021, 173, 394–417. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, A.B.; Yanar, A.; Alkan, E.N. Review of heavy metal accumulation on aquatic environment in Northern East Mediterrenean Sea part I: Some essential metals. Rev. Environ. Health 2017, 32, 119–163. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [PubMed]
- Mudd, G.M. Global trends and environmental issues in nickel mining: Sulfides versus laterites. Ore Geol. Rev. 2010, 38, 9–26. [Google Scholar] [CrossRef]
- Park, I.; Tabelin, C.B.; Jeon, S.; Li, X.; Seno, K.; Ito, M.; Hiroyoshi, N. A review of recent strategies for acid mine drainage prevention and mine tailings recycling. Chemosphere 2019, 219, 588–606. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Kamika, I.; Tekere, M. Evaluation of Heavy Metal Removal from Wastewater in a Modified Packed Bed Biofilm Reactor. PLoS ONE 2016, 11, e0155462. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, C.P. Application of Aspergillus oryze and Rhizopus oryzae for Cu2+ removal. Water Res. 1996, 30, 1985–1990. [Google Scholar] [CrossRef]
- Ayangbenro, A.S.; Babalola, O.O. A New Strategy for Heavy Metal Polluted Environments: A Review of Microbial Biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef]
- Oyewole, O.A.; Zobeashia, S.S.L.T.; Oladoja, E.O.; Raji, R.O.; Odiniya, E.E.; Musa, A.M. Biosorption of heavy metal polluted soil using bacteria and fungi isolated from soil. SN Appl. Sci. 2017, 1, 857. [Google Scholar] [CrossRef]
- Gadd, G.M. Biosorption: Critical review of scientific rationale, environmental importance and significance for pollution treatment. J. Chem. Technol. Biotechnol. 2009, 84, 13–28. [Google Scholar] [CrossRef]
- Monge-Amaya, O.; Certucha-Barragán, M.T.; Almendariz-Tapia, F.J.; Figueroa-Torres, G.M. Removal of Heavy Metals from Aqueous Solutions by Aerobic and Anaerobic Biomass. In Biomass Production and Uses; IntechOpen: London, UK, 2015. [Google Scholar]
- Rumo, C.; Stein, A.; Klehr, J.; Tachibana, R.; Prescimone, A.; Häussinger, D.; Ward, T.R. An Artificial Metalloenzyme Based on a Copper Heteroscorpionate Enables sp3 C–H Functionalization via Intramolecular Carbene Insertion. J. Am. Chem. Soc. 2022, 144, 11676–11684. [Google Scholar] [CrossRef] [PubMed]
- Facchetti, G.; Rimoldi, I. 8-Amino-5,6,7,8-tetrahydroquinoline in iridium(iii) biotinylated Cp* complex as artificial imine reductase. New J. Chem. 2018, 42, 18773–18776. [Google Scholar] [CrossRef]
- Pellegrino, S.; Facchetti, G.; Contini, A.; Gelmi, M.L.; Erba, E.; Gandolfi, R.; Rimoldi, I. Ctr-1 Mets7 motif inspiring new peptide ligands for Cu(i)-catalyzed asymmetric Henry reactions under green conditions. RSC Adv. 2016, 6, 71529–71533. [Google Scholar] [CrossRef]
- Vermaak, V.; Vosloo, H.C.M.; Swarts, A.J. Fast and Efficient Nickel(II)-catalysed Transfer Hydrogenation of Quinolines with Ammonia Borane. Adv. Synth. Catal. 2020, 362, 5788–5793. [Google Scholar] [CrossRef]
- Li, B.; Chen, J.; Zhang, Z.; Gridnev, I.D.; Zhang, W. Nickel-Catalyzed Asymmetric Hydrogenation of N-Sulfonyl Imines. Angew. Chem. Int. Ed. 2019, 58, 7329–7334. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Hu, T.; Zhai, Y.; Lu, N.; Aliyeva, J. The improved methods of heavy metals removal by biosorbents: A review. Environ. Pollut. 2020, 258, 113777. [Google Scholar] [CrossRef]
- Sutherland, C.; Venkobachar, C. A diffusion-chemisorption kinetic model for simulating biosorption using forest macrofungus, Fomes fasciatus. Int. Res. J. Plant Sci. 2010, 1, 107–117. [Google Scholar]
- Teng, Z.; Shao, W.; Zhang, K.; Huo, Y.; Zhu, J.; Li, M. Pb biosorption by Leclercia adecarboxylata: Protective and immobilized mechanisms of extracellular polymeric substances. Chem. Eng. J. 2019, 375, 122113. [Google Scholar] [CrossRef]
- Sag, Y.; Kutsal, T. Recent trends in the biosorption of heavy metals: A review. Biotechnol. Bioprocess. Eng. 2001, 6, 376. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C. Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 2009, 27, 195–226. [Google Scholar] [CrossRef]
- Wang, L.L.; Wang, L.F.; Ren, X.M.; Ye, X.D.; Li, W.W.; Yuan, S.J.; Sun, M.; Sheng, G.P.; Yu, H.Q.; Wang, X.K. pH Dependence of Structure and Surface Properties of Microbial EPS. Environ. Sci. Technol. 2012, 46, 737–744. [Google Scholar] [CrossRef] [PubMed]
- De Philippis, R.; Micheletti, E. Heavy metal removal with exopolysaccharide-producing cyanobacteria. Heavy Met. Environ. 2009, 29, 89–122. [Google Scholar]
- Vishan, I.; Kalamdhad, A.S. Heavy metal removal through bacterial biomass isolated from various contaminated sites. Int. J. Environ. Sci. 2016, 7, 1–18. [Google Scholar]
- Sadouk, A.; Mergeay, M. Chromosome mapping in Alkaligens eutrophus CH34. Mol. Gen. Genet. 1993, 240, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.W.; Forster, C.F.; Evison, L. A comparative study of the nature of biopolymers extracted from anaerobic and activated sludge. Water Res. 1990, 24, 743–750. [Google Scholar] [CrossRef]
- Xia, L.; Zheng, X.L.; Shao, H.B.; Xin, J.; Sun, Z.Y.; Wang, L.Y. Effects of bacterial cells and two types of extracellular polymers on bioclogging of sand columns. J. Hydrol. 2016, 535, 293–300. [Google Scholar] [CrossRef]
- Dreywood, R. Qualitative test for carbohydrate material. Ind. Eng. Chem. 1946, 18, 499. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Crini, G.; Peindy, H.N.; Gimbert, F.; Robert, C. Removal of C.I. Basic Green 4 (Malachite Green) from aqueous solutions by adsorption using cyclodextrin-based adsorbent: Kinetic and equilibrium studies. Sep. Purif. Technol. 2007, 53, 97–110. [Google Scholar] [CrossRef]
- Lacerda, E.C.M.; dos Passos Galluzzi Baltazar, M.; dos Reis, T.A.; do Nascimento, C.A.O.; Côrrea, B.; Jandelli Gimenes, L. Copper biosorption from an aqueous solution by the dead biomass of Penicillium ochrochloron. Env. Monit. Assess. 2019, 191, 247. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, W.; Liu, D.; Liu, T.; Wang, Z. Biosorption isotherm study of Cd2+, Pb2+ and Zn2+ biosorption onto marine bacterium Pseudoalteromonas sp. SCSE709-6 in multiple systems. J. Mol. Liq. 2017, 247, 230–237. [Google Scholar] [CrossRef]
- Gandolfi, R.; Facchetti, G.; Cavalca, L.; Mazzini, S.; Colombo, M.; Coffetti, G.; Borgonovo, G.; Scaglioni, L.; Zecchin, S.; Rimoldi, I. Hybrid Catalysts from Copper Biosorbing Bacterial Strains and Their Recycling for Catalytic Application in the Asymmetric Addition Reaction of B2(pin)2 on α,β-Unsaturated Chalcones. Catalysts 2022, 12, 433. [Google Scholar] [CrossRef]
- Facchetti, G.; Bucci, R.; Fusè, M.; Rimoldi, I. Asymmetric Hydrogenation vs. Transfer Hydrogenation in the Reduction of Cyclic Imines. ChemistrySelect 2018, 3, 8797–8800. [Google Scholar] [CrossRef]
- Hong, T.; Yin, J.Y.; Nie, S.P.; Xie, M.Y. Applications of infrared spectroscopy in polysaccharide structural analysis: Progress, challenge and perspective. Food Chem. X 2021, 12, 100168. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Yang, S.; Huang, Q.; Xue, A.; Cai, P. Biosorption mechanisms of Cu(II) by extracellular polymeric substances from Bacillus subtilis. Chem. Geol. 2014, 386, 143–151. [Google Scholar] [CrossRef]
- Rajeswari Kulkarni, M.; Vidya Shetty, K.; Srinikethan, G. Kinetic and equilibrium modeling of biosorption of nickel (II) and cadmium (II) on brewery sludge. Water Sci. Technol. 2019, 79, 888–894. [Google Scholar] [CrossRef]
- Nagarajan, N.; Gunasekaran, P.; Rajendran, P. Genetic characterization, nickel tolerance, biosorption, kinetics, and uptake mechanism of a bacterium isolated from electroplating industrial effluent. Can. J. Microbiol. 2015, 61, 297–306. [Google Scholar] [CrossRef]
- Bhainsa, K.C.; D’Souza, S.F. Removal of copper ions by the filamentous fungus, Rhizopus oryzae from aqueous solution. Bioresour. Technol. 2008, 99, 3829–3835. [Google Scholar] [CrossRef]
- Verma, A.; Shalu Singh, A.; Bishnoi, N.R.; Gupta, A. Biosorption of Cu (II) using free and immobilized biomass of Penicillium citrinum. Ecol. Eng. 2013, 61, 486–490. [Google Scholar] [CrossRef]
- Sethuraman, P.; Dharmendira Kumar, M. Biosorption Kinetics of Cu (II) Ions Removal from Aqueous Solution using Bacteria. Pak. J. Biol. Sci. 2011, 14, 327–335. [Google Scholar] [CrossRef]
- Kashyap, S.; Chandra, R.; Kumar, B.; Verma, P. Biosorption efficiency of nickel by various endophytic bacterial strains for removal of nickel from electroplating industry effluents: An operational study. Ecotoxicology 2021, 31, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Dursun, A.Y. A comparative study on determination of the equilibrium, kinetic and thermodynamic parameters of biosorption of copper(II) and lead(II) ions onto pretreated Aspergillus niger. Biochem. Eng. J. 2006, 28, 187–195. [Google Scholar] [CrossRef]
- Puyen, Z.M.; Villagrasa, E.; Maldonado, J.; Diestra, E.; Esteve, I.; Solé, A. Biosorption of lead and copper by heavy-metal tolerant Micrococcus luteus DE2008. Bioresour. Technol. 2012, 126, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Akhter, K.; Ghous, T.; Andleeb, S.; Ejaz, S.; Khan, B.A.; Ahmed, M.N. Bioaccumulation of heavy metals by metal-resistant bacteria isolated from Tagetes minuta rhizosphere, growing in soil adjoining automobile workshops. Pak. J. Zool. 2017, 49, 1841–1846. [Google Scholar] [CrossRef]
- Naskar, A.; Bera, D. Mechani.istic exploration of Ni (II) removal by immobilized bacterial biomass and interactive influence of coexisting surfactants. Environ. Prog. Sustain. Energy 2018, 37, 342–354. [Google Scholar] [CrossRef]
- Gheethi, A.A.; Efaq, A.N.; Mohamed, R.M.; Abdel-Monem, M.O.; Abdullah, A.H.; Amir Hashim, M.K. Bio-removal of Nickel ions by Sporosarcina pasteurii and Bacillus megaterium, a Comparative Study. IOP Conf. Ser. Mater. Sci. Eng. 2017, 226, 012044. [Google Scholar] [CrossRef]
- Gadd, G.M. Heavy metal accumulation by bacteria and other microorganisms. Experientia 1990, 46, 834–840. [Google Scholar] [CrossRef]
- Díaz, A.; Marrero, J.; Cabrera, G.; Coto, O.; Gómez, J.M. Biosorption of nickel, cobalt, zinc and copper ions by Serratia marcescens strain 16 in mono and multimetallic systems. Biodegradation 2022, 33, 33–43. [Google Scholar] [CrossRef]
- Cristani, M.; Naccari, C.; Nostro, A.; Pizzimenti, A.; Trombetta, D.; Pizzimenti, F. Possible use of Serratia marcescens in toxic metal biosorption (removal). Env. Sci. Pollut. Res. 2012, 19, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Q.; Wu, C.; Luo, X.; Yu, X.; Chen, M. The inappropriate application of the regression Langmuir Qm for adsorption capacity comparison. Sci. Total Environ. 2020, 699, 134222. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, X. Adsorption isotherm models: Classification, physical meaning, application and solving method. Chemosphere 2020, 258, 127279. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.N.; You, S.-J.; Hosseini-Bandegharaei, A.; Chao, H.-P. Mistakes and inconsistencies regarding adsorption of contaminants from aqueous solutions: A critical review. Water Res. 2017, 120, 88–116. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.C.; Wei, X.; Cai, P.; Huang, Q.Y.; Chen, H.; Liang, W.; Rong, X.M. Role of extracellular polymeric substances in Cu(II) adsorption on Bacillus subtilis and Pseudomonas putida. Bioresour. Technol. 2011, 102, 1137–1141. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.K.; Guo, Y.; Byrne, J.M.; Zeitvogel, F.; Schmid, G.; Ingino, P.; Li, J.L.; Neu, T.R.; Swanner, E.D.; Kappler, A.; et al. Binding of heavy metal ions in aggregates of microbial cells, EPS and biogenic iron minerals measured in-situ using metal- and glycoconjugates-specific fluorophores. Geochim. Cosmochim. Acta 2016, 180, 66–96. [Google Scholar] [CrossRef]



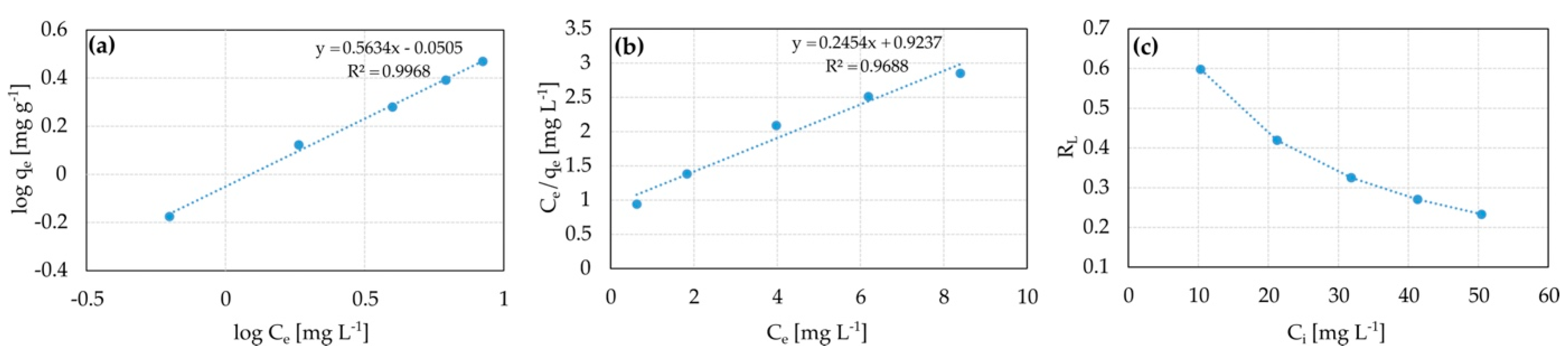
| Sorption System. | Model Isotherm | Parameters | R2 |
|---|---|---|---|
| Ni2+—S. plymuthica SC3I(2) | Freundlich | Kf = 0.89(L g−1), ß = 0.50, nF = 1.98 | 0.9968 |
| Langmuir | Qm = 4.1 (mg g−1), Kl = 0.27(L g−1), al = 0.07(L mg−1), RL = 0.23 − 0.60 | 0.9688 | |
| Cu2+—S. plymuthica As3-5a(5) | Freundlich | nd 1 | 0.3789 |
| Langmuir | nd | 0.0528 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanetti, R.; Zecchin, S.; Colombo, M.; Borgonovo, G.; Mazzini, S.; Scaglioni, L.; Facchetti, G.; Gandolfi, R.; Rimoldi, I.; Cavalca, L. Ni2+ and Cu2+ Biosorption by EPS-Producing Serratia plymuthica Strains and Potential Bio-Catalysis of the Organo–Metal Complexes. Water 2022, 14, 3410. https://doi.org/10.3390/w14213410
Zanetti R, Zecchin S, Colombo M, Borgonovo G, Mazzini S, Scaglioni L, Facchetti G, Gandolfi R, Rimoldi I, Cavalca L. Ni2+ and Cu2+ Biosorption by EPS-Producing Serratia plymuthica Strains and Potential Bio-Catalysis of the Organo–Metal Complexes. Water. 2022; 14(21):3410. https://doi.org/10.3390/w14213410
Chicago/Turabian StyleZanetti, Rocco, Sarah Zecchin, Milena Colombo, Gigliola Borgonovo, Stefania Mazzini, Leonardo Scaglioni, Giorgio Facchetti, Raffaella Gandolfi, Isabella Rimoldi, and Lucia Cavalca. 2022. "Ni2+ and Cu2+ Biosorption by EPS-Producing Serratia plymuthica Strains and Potential Bio-Catalysis of the Organo–Metal Complexes" Water 14, no. 21: 3410. https://doi.org/10.3390/w14213410
APA StyleZanetti, R., Zecchin, S., Colombo, M., Borgonovo, G., Mazzini, S., Scaglioni, L., Facchetti, G., Gandolfi, R., Rimoldi, I., & Cavalca, L. (2022). Ni2+ and Cu2+ Biosorption by EPS-Producing Serratia plymuthica Strains and Potential Bio-Catalysis of the Organo–Metal Complexes. Water, 14(21), 3410. https://doi.org/10.3390/w14213410








