Antimicrobial Resistance of Heterotrophic Bacteria in Drinking Water-Associated Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enumeration of HPC Bacteria and ARB in Drinking Water and Drinking Water-Associated Biofilms
2.2. Study Area and Drinking Water Sampling
2.3. Biofilm Formation in Static Conditions
2.4. Biofilm Analysis
2.5. Drinking Water Analysis
2.6. Antibiotic Susceptibility Testing of Isolated Bacteria
2.7. Assessment of Biofilm Formation Ability of Bacteria Isolated from Drinking Water
2.8. Statistical Analysis of the Data
3. Results
3.1. Total Number of HPC and ARB in Drinking Water and Water-Associated Biofilms
3.2. Phenotype of AMR of Bacterial Isolates from Water and Biofilm
3.3. Single-Species Biofilm Formation of Bacterial Isolates
3.4. Biofilm Formation and MDR
4. Discussion
4.1. Prevalence of ARB in Drinking Water-Associated Biofilms
4.2. Relation between AMR and Biofilm Formation Ability of Bacterial Isolates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | Antibiotic |
| AMR | Antimicrobial resistance |
| AMP | Ampicillin |
| ANOVA | One-way analysis of variance |
| ARB | Antibiotic resistant bacteria |
| ARGs | Antibiotic resistance genes |
| BF | Biofilm-formation ability |
| BI | Biofilm index |
| C | Chloramphenicol |
| CIP | Ciprofloxacin |
| CFU | Colony form unit |
| CTX | Cefotaxime |
| DW | Drinking water |
| DWDS | Drinking water distribution system |
| FF | Fosfomycin |
| FM | Nitrofurantoin |
| GEN | Gentamycin |
| HPC | Heterotrophic plate count |
| MDR | Multidrug resistance |
| OD | Optical density |
| ODc | Cutoff OD |
| P | Probability |
| S | Streptomycin |
| STX | Trimethoprim/sulfamethoxazole |
| Sul | sulphamethoxazole |
| TE | Tetracycline |
| TMP | Trimethoprim |
References
- Manaia, C.M.; Macedo, G.; Fatta-Kassinos, D.; Nunes, O.C. Antibiotic resistance in urban aquatic environments: Can it be controlled? Appl. Microbiol. Biotechnol. 2016, 100, 1543–1557. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marti, Е.; Jofre, J.; Balcazar, J.L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Bacterial diversity and antibiotic resistance in water habitats: Searching the links with the human microbiome. FEMS Microb. Rev. 2014, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, A. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Flemming, H.C.; Percival, S.I.; Walker, J.T. Contamination potential of biofilms in water distribution systems. Water Sci. Technol. 2002, 2, 271–280. [Google Scholar] [CrossRef]
- Wricke, B.; Korth, A.; Petzoldt, H.; Kruger, M. Change of bacterial water quality in drinking water distribution systems working with or without low chlorine residual. Water Sci. Technol. Water Supply 2002, 2, 275–281. [Google Scholar] [CrossRef]
- LeChevallier, M.W.; Cawthonm, C.D.; Leem, R.G. Inactivation of biofilm bacteria. Appl. Environ. Microbiol. 1988, 54, 2492–2499. [Google Scholar] [CrossRef] [Green Version]
- Codony, F.; Morato, J.; Mas, J. Role of discontinuous chlorination on microbial production by drinking water biofilms. Water Res. 2005, 39, 1896–1906. [Google Scholar] [CrossRef]
- Tsvetanova, Z. Quantification of the bacterial community of drinking water associated biofilms under different flow velocities and changing chlorination regimes. Appl. Water Sci. 2020, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef]
- Chen, X.; Stewart, P.S. Biofilm removal caused by chemical treatments. Water Res. 2000, 34, 4229–4233. [Google Scholar] [CrossRef]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbil. 2009, 75, 4093–4100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moskowitz, S.M.; Foster, J.M.; Emerson, J.; Burns, J.L. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 2004, 42, 1915–1922. [Google Scholar] [CrossRef] [Green Version]
- Eyoh, A.B.; Toukam, M.; Atashili, J.; Fokunang, C.; Gonsu, H.; Lyonga, E.E.; Mandi, H.; Ikomey, G.; Mukwele, B.; Mesembe, M.; et al. Relationship between multiple drug resistance and biofilm formation in Staphylococcus aureus isolated from medical and non-medical personnel in Yaounde, Cameroon. Pan Afr. Med. J. 2014, 17, 186. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Farkas, A.; Butiuc-Keul, A.; Ciatarâş, D.; Neamţu, C.; Crăciunaş, C.; Podar, D.; Drăgan-Bularda, M. Microbiological contamination and resistance genes in biofilms occurring during the drinking water treatment process. Sci. Total Environ. 2013, 443, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in waste-water, surface water and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Zhang, Y.; Marrs, C.F.; Ye, W.; Simon, C.; Foxman, B.; Nriagu, J. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl. Environ. Microbiol. 2009, 75, 5714–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narciso-da-Rocha, C.; Vaz-Moreira, I.; Svensson-Stadler, L.; Moore, E.R.B.; Manaia, C.M. Diversity and antibiotic resistance of Acinetobacter spp. in water from the source to the tap. Appl. Microbiol. Biotechnol. 2013, 97, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Beattie, T.K.; Knapp, C.W. Relationship between antibiotic- and disinfectant-resistance profiles in bacteria harvested from tap water. Chemosphere 2016, 152, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Destiаni, R.; Templeton, M.R. The antibiotic resistance of heterotrophic bacteria in tap waters in London. Water Supply 2019, 19, 179–190. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl. Environ. Microbiol. 2011, 77, 5697–5706. [Google Scholar] [CrossRef] [Green Version]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and antibiotic resistance in Pseudomonas spp. from drinking water. Sci. Total Environ. 2012, 426, 366–374. [Google Scholar] [CrossRef]
- Tsvetanova, Z.; Dimitrov, D.; Najdenski, H. Prevalence of antimicrobial resistance in a Bulgarian drinking water supply system. Water Supply 2022, submitted.
- Reasoner, D.J.; Geldreich, E. A new medium for enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation pf MICs and Zone Diameters. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 20 February 2022).
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 30, 2437. Available online: http://www.jove.com/details.php?id=2437 (accessed on 16 March 2022). [CrossRef]
- Simões, L.C.; Simões, M.; Vieira, M.J. Influence of the diversity of bacterial isolates from drinking water on resistance of biofilms to disinfection. Appl. Environ. Microbiol. 2010, 76, 6673–6679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boe-Hansen, R.; Albrechtsen, H.J.; Arvin, E.; Jørgensen, C. Dynamics of biofilm formation in a model drinking water distribution system. J. Water SRT Aqua 2002, 1, 399–406. [Google Scholar] [CrossRef]
- Kelly, J.J.; Minalt, N.; Culotti, A.; Pryor, M.; Packman, A. Temporal variations in the abundance and composition of biofilm communities colonizing drinking water distribution pipes. PLoS ONE 2014, 9, e98542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, C.D.; LeChevallier, M.W. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 2000, 66, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Tsvetanova, Z.; Najdenski, H. Biofilm formation potential of enteropathogenic bacteria and their survival in drinking water-associated biofilms. Acta Microbiol. Bulg. 2018, 34, 153–159. [Google Scholar]
- Zhu, Z.; Shan, L.; Hu, F.; Li, Z.; Zhong, D.; Yuan, Y.; Zhang, J. Biofilm formation potential and chlorine resistance of typical bacteria isolated from drinking water distribution systems. RSC Adv. 2020, 10, 31295. [Google Scholar] [CrossRef]
- Douterelo, I.; Jackson, M.; Solomon, C.; Boxall, J. Microbial analysis of in situ biofilm formation in drinking water distribution systems: Implications for monitoring and control of drinking water quality. Appl. Microbiol. Biotechnol. 2016, 100, 3301–3311. [Google Scholar] [CrossRef] [Green Version]
- Sahal, G.; Bilkay, I.S. Multi drug resistance in strong biofilm forming clinical isolates of Staphylococcus epidermidis. Braz. J. Microbiol. 2014, 45, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, M.; Simões, L.C.; Vieira, M.J. Species association increases biofilm resistance to chemical and mechanical treatment. Water Res. 2009, 43, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.W.K.; Periasamy, S.; Mukherjee, M.; Xie, C.; Kjelleberg, S.; Rice, S.A. Biofilm development and enhanced stress resistance of a model mixed-species community biofilm. SME J. 2014, 8, 894–907. [Google Scholar] [CrossRef] [PubMed]
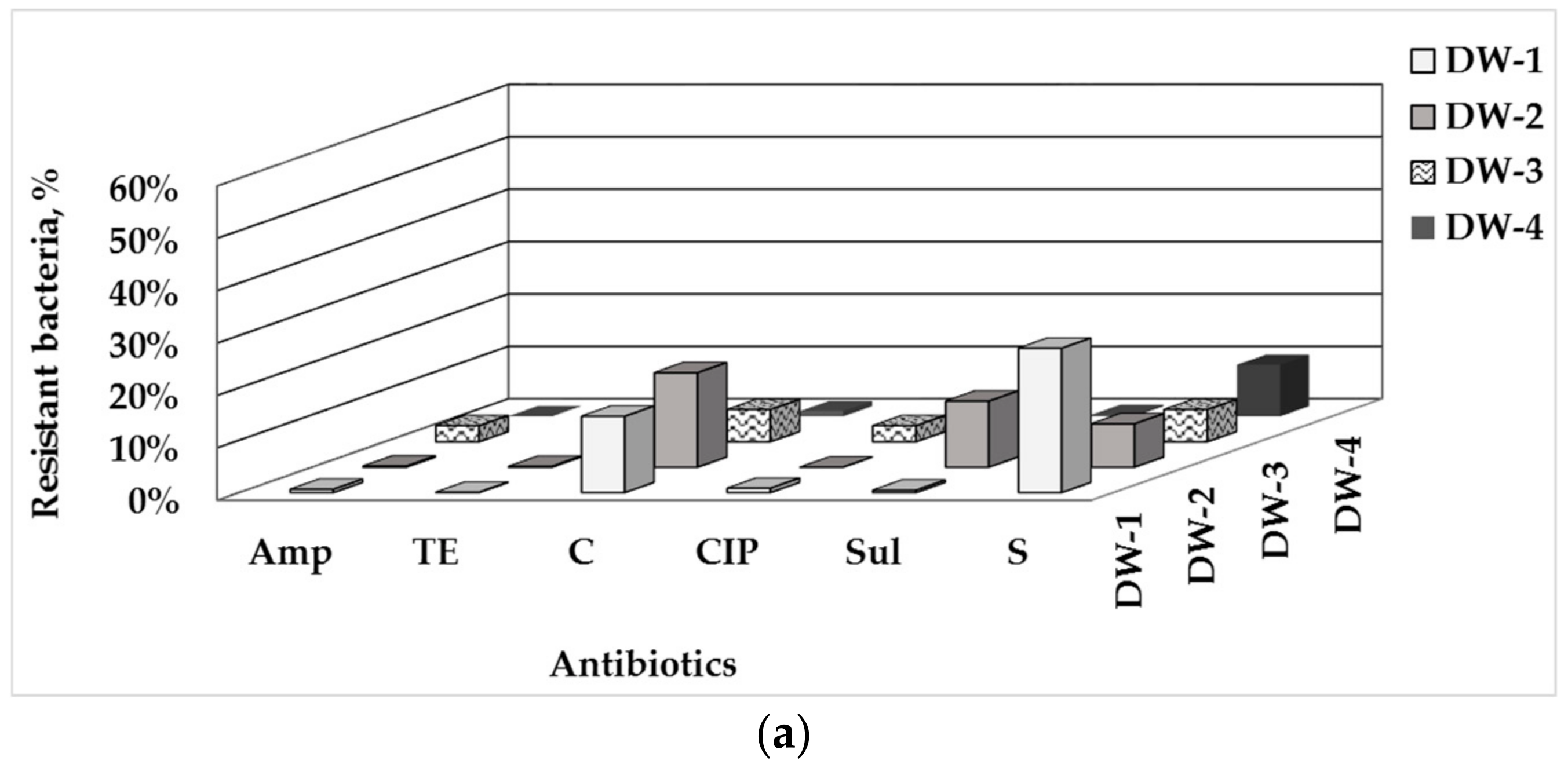
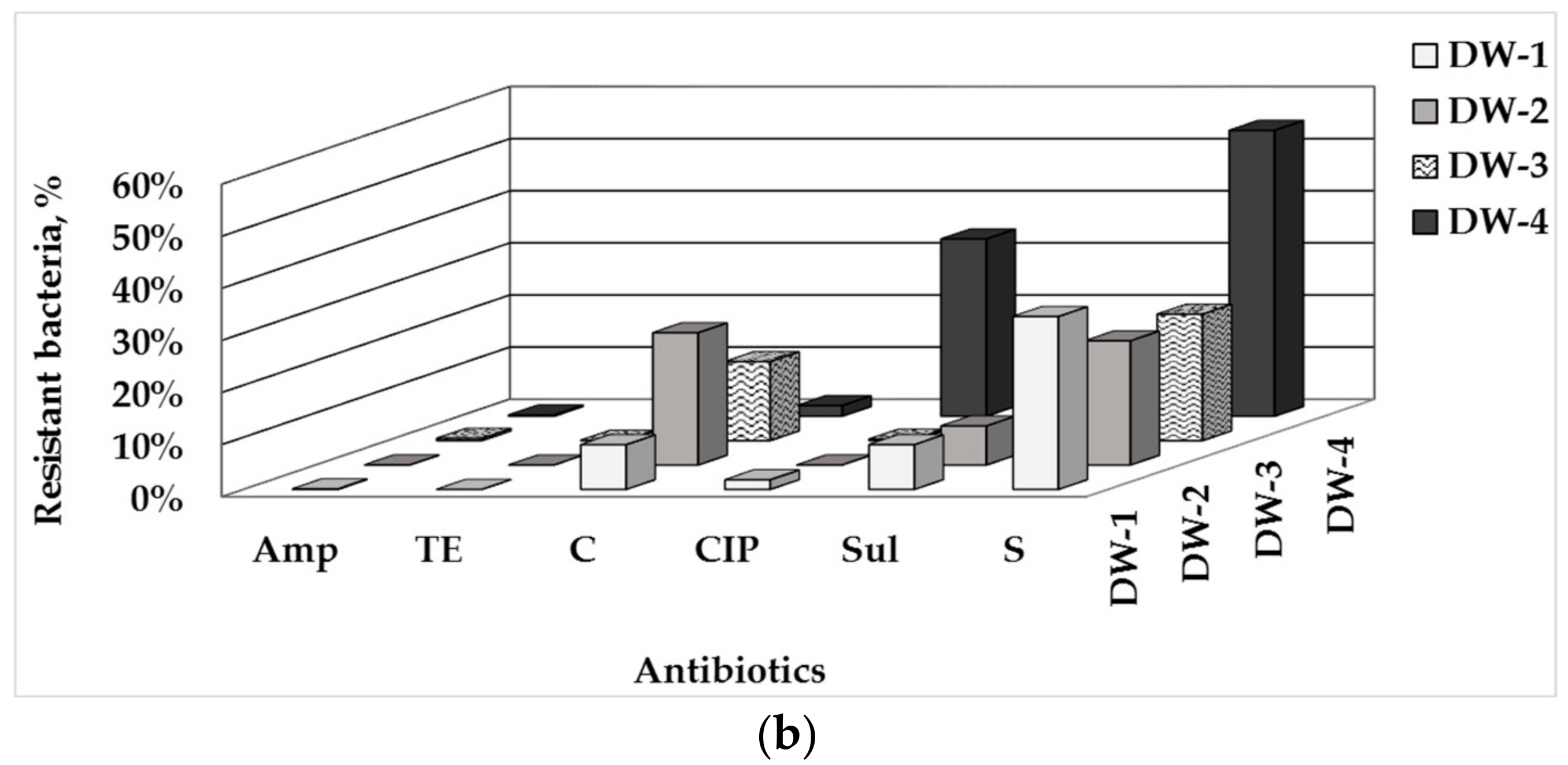
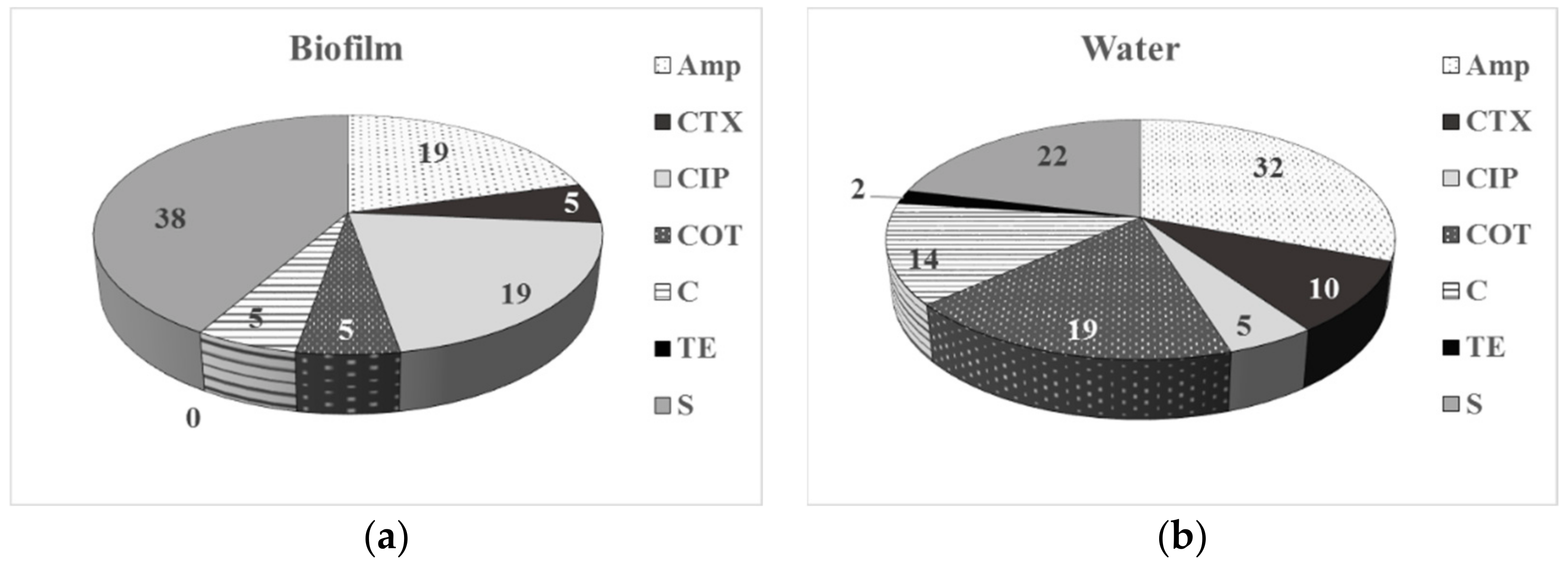

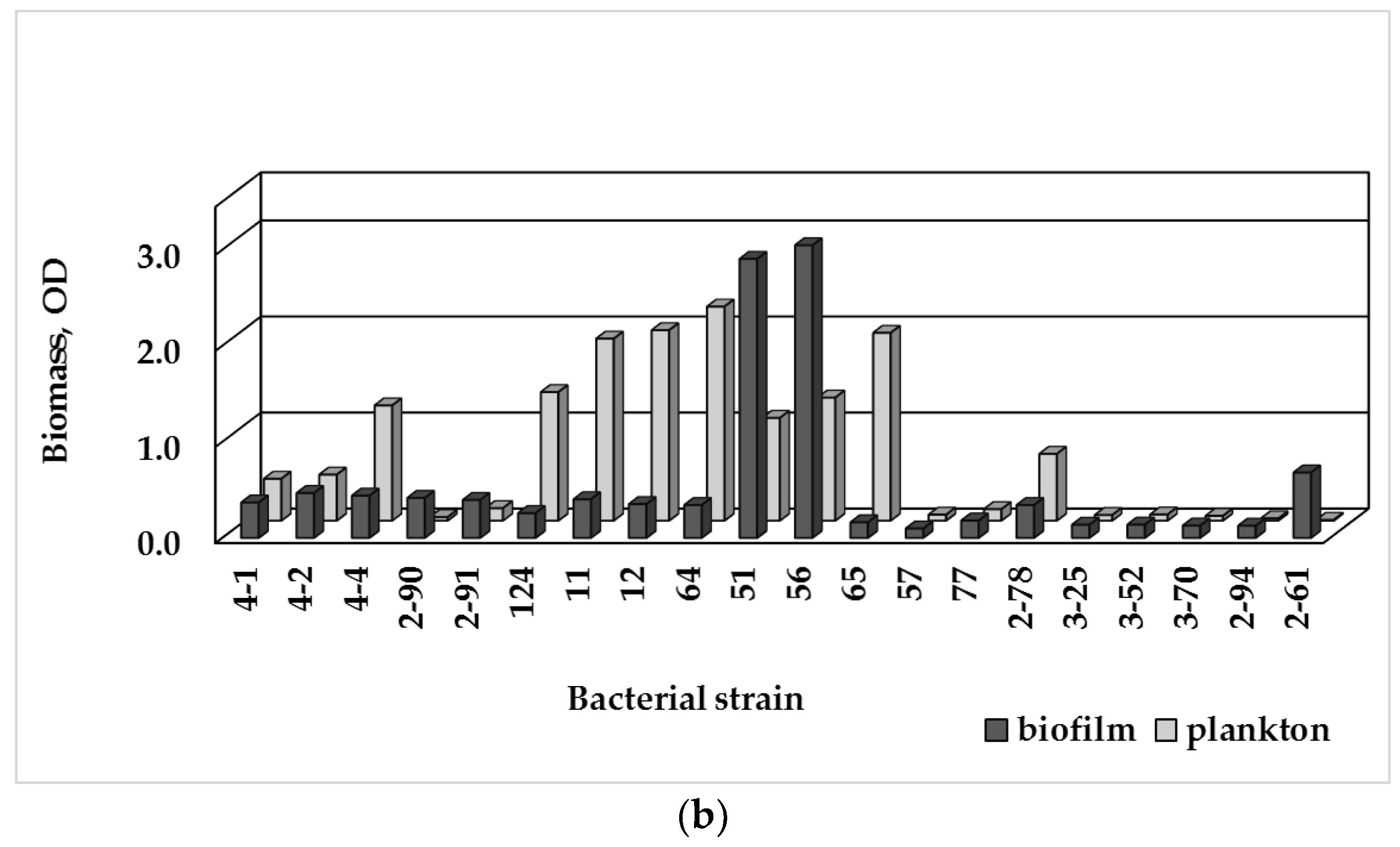
| Sampling Point | HPCs of Drinking Water, CFU/100 mL | HPC Density of the Biofilm Formed, CFU/cm2 |
|---|---|---|
| DW-1 | 2.9 (0.2) × 102 | 3.3 (2.4) × 104 |
| DW-2 | 7.9 (0.2) × 101 | 1.2 (1.2) × 104 |
| DW-3 | 1.3 (0.1) × 101 | 3.9 (2.1) × 105 |
| DW-4 | 1.6 (0.1) × 103 | 2.0 (0.9) × 105 |
| Strain № | Bacterial Taxon | AMR Phenotype | MDR, n | R2A | M63 | ||
|---|---|---|---|---|---|---|---|
| BI | BF | BI | BF | ||||
| 4-1 | Sphingomonas spp. | AMP, S | 2 | 0.8 | +++ | 0.8 | +++ |
| 4-2 | Sphingomonas spp. | AMP, S | 2 | 0.8 | +++ | 1.0 | +++ |
| 4-4 | Sphingomonas spp. | AMP, S | 2 | 0.9 | +++ | 0.4 | +++ |
| 124 | Sphingomonas paucimobilis | AMC, CZ, CN, CXM, FM | 2 | 0.1 | 0 | 0.2 | + |
| 2-90 | Embedobacter brevis | AMC, CZ, CN, CXM, FM | 2 | 7.9 | +++ | 11.3 | ++ |
| 2-91 | Embedobacter brevis | AMC, CZ, CN, CXM, FM | 2 | 3.1 | +++ | 3.0 | +++ |
| 11 | Pseudomonas spp. | AMP, AMC, CTX, CAZ, SXT, TE | 3 | 0.3 | ++ | 0.2 | ++ |
| 12 | Pseudomonas spp. | AMP, AMC, CTX, CAZ, SXT, TE | 3 | 0.5 | +++ | 0.2 | ++ |
| 64 | Pseudomonas spp. | AMP, CF, CTX, C, SXT | 3 | 0.1 | ++ | 0.2 | ++ |
| 56 | P. fluorescens | AMC, CN, CXM, CZ, FM | 2 | 0.9 | +++ | 2.4 | +++ |
| 51 | P. fluorescens | AMC, CN, CXM, CZ, FM | 2 | 1.0 | +++ | 2.7 | +++ |
| 65 | P. putida | AMP, AMC, CZ, CAZ, C, CIP, FM | 4 | 0.1 | ++ | 0.1 | ++ |
| 57 | Stenotrophomonas maltophilia | AMP, AMC, AN, ATM, CN, CTX, CXM, CZ, ETP, FF, FM, GEN, IPM, MEM, NN, TMP, TZP | 5 | 0.1 | + | 1.6 | ++ |
| 2-78 | Methylobacterium exorquens | sensitive | 0 | 1.6 | ++ | 0.5 | ++ |
| 77 | Methylobacterium spp. | SXT | 1 | 0.1 | + | 1.5 | + |
| 3-70 | Lysinibacillus sphaericus | S, NA | 2 | 0.2 | + | 2.6 | 0 |
| 3-25 | Bacillus spp. | Amp, CTX, CX, E | 2 | 0.4 | ++ | 2.3 | 0 |
| 3-52 | B. thuringiensis | AMP, AMC, CTX, CX, SXT | 2 | 0.1 | ++ | 2.2 | 0 |
| 2-94 | Staphylococcus cohnii | CIP, MXP | 2 | 0.2 | + | 5.4 | ++ |
| 2-61 | Micrococcus luteus | sensitive | 0 | 11.4 | +++ | 87.9 | +++ |
| BF or BI | Number of Strains in R2A (n) | Number of Strains in M63 (n) | ||
|---|---|---|---|---|
| MDR (5) | Non-MDR (15) | MDR (5) | Non-MDR (15) | |
| Strong (+++) | 1 | 8 | 0 | 7 |
| Weak (++; +) | 4 | 6 | 5 | 5 |
| Strong (BI > 1) | 0 | 4 | 1 | 10 |
| Weak (BI ≤ 1) | 5 | 11 | 4 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsvetanova, Z.; Tsvetkova, I.; Najdenski, H. Antimicrobial Resistance of Heterotrophic Bacteria in Drinking Water-Associated Biofilms. Water 2022, 14, 944. https://doi.org/10.3390/w14060944
Tsvetanova Z, Tsvetkova I, Najdenski H. Antimicrobial Resistance of Heterotrophic Bacteria in Drinking Water-Associated Biofilms. Water. 2022; 14(6):944. https://doi.org/10.3390/w14060944
Chicago/Turabian StyleTsvetanova, Zvezdimira, Iva Tsvetkova, and Hristo Najdenski. 2022. "Antimicrobial Resistance of Heterotrophic Bacteria in Drinking Water-Associated Biofilms" Water 14, no. 6: 944. https://doi.org/10.3390/w14060944
APA StyleTsvetanova, Z., Tsvetkova, I., & Najdenski, H. (2022). Antimicrobial Resistance of Heterotrophic Bacteria in Drinking Water-Associated Biofilms. Water, 14(6), 944. https://doi.org/10.3390/w14060944







