Identification of Antibiotic Resistance Gene Hosts in Treatment Wetlands Using a Single-Cell Based High-Throughput Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling Regime
2.2. epicPCR
2.3. Illumina Sequencing and Data Analysis
3. Results and Discussion
3.1. AMR Gene Hosts along the Watercourse of the Treatment Wetlands
3.2. Potential Class 1 Integron Carriers in the Treatment Wetlands
3.3. Evidence for AMR Genes in Methanogenic Archaea
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collignon, P.; Beggs, J.J.; Walsh, T.R.; Gandra, S.; Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Buergmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Genet. 2021, 20, 257–269. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Woolhouse, M.E.J. Using sewage for surveillance of antimicrobial resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Genet. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2014, 9, 1269–1279. [Google Scholar] [CrossRef]
- Ma, L.; Li, A.-D.; Yin, X.-L.; Zhang, T. The Prevalence of Integrons as the Carrier of Antibiotic Resistance Genes in Natural and Man-Made Environments. Environ. Sci. Technol. 2017, 51, 5721–5728. [Google Scholar] [CrossRef]
- Zhang, A.N.; Li, L.-G.; Ma, L.; Gillings, M.R.; Tiedje, J.M.; Zhang, T. Conserved phylogenetic distribution and limited antibiotic resistance of class 1 integrons revealed by assessing the bacterial genome and plasmid collection. Microbiome 2018, 6, 130. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The Natural History of Integrons. Microorganisms 2021, 9, 2212. [Google Scholar] [CrossRef]
- Néron, B.; Littner, E.; Haudiquet, M.; Perrin, A.; Cury, J.; Rocha, E.P.C. IntegronFinder 2.0: Identification and Analysis of Integrons across Bacteria, with a Focus on Antibiotic Resistance in Klebsiella. Microorganisms 2022, 10, 700. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Genet. 2014, 13, 116–123. [Google Scholar] [CrossRef]
- Pärnänen, K.; Karkman, A.; Tamminen, M.; Lyra, C.; Hultman, J.; Paulin, L.; Virta, M. Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics. Sci. Rep. 2016, 6, 35790. [Google Scholar] [CrossRef]
- Stalder, T.; Press, M.O.; Sullivan, S.; Liachko, I.; Top, E.M. Linking the resistome and plasmidome to the microbiome. ISME J. 2019, 13, 2437–2446. [Google Scholar] [CrossRef]
- Spencer, S.J.; Tamminen, M.V.; Preheim, S.P.; Guo, M.T.; Briggs, A.W.; Brito, I.L.; Weitz, D.A.; Pitkänen, L.K.; Vigneault, F.; Virta, M.P.; et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2015, 10, 427–436. [Google Scholar] [CrossRef]
- Dai, S.; He, Q.; Han, Z.; Shen, W.; Deng, Y.; Wang, Y.; Qiao, W.; Yang, M.; Zhang, Y. Uncovering the diverse hosts of tigecycline resistance gene tet(X4) in anaerobic digestion systems treating swine manure by epicPCR. Water Res. X 2023, 19, 100174. [Google Scholar] [CrossRef]
- Hultman, J.; Tamminen, M.; Pärnänen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, fiy038. [Google Scholar] [CrossRef]
- Qi, Q.; Ghaly, T.M.; Penesyan, A.; Rajabal, V.; Stacey, J.A.; Tetu, S.G.; Gillings, M.R. Uncovering Bacterial Hosts of Class 1 Integrons in an Urban Coastal Aquatic Environment with a Single-Cell Fusion-Polymerase Chain Reaction Technology. Environ. Sci. Technol. 2023, 57, 4870–4879. [Google Scholar] [CrossRef]
- Wei, Z.; Feng, K.; Wang, Z.; Zhang, Y.; Yang, M.; Zhu, Y.-G.; Virta, M.P.J.; Deng, Y. High-Throughput Single-Cell Technology Reveals the Contribution of Horizontal Gene Transfer to Typical Antibiotic Resistance Gene Dissemination in Wastewater Treatment Plants. Environ. Sci. Technol. 2021, 55, 11824–11834. [Google Scholar] [CrossRef]
- Wei, Z.; Shen, W.; Feng, K.; Feng, Y.; He, Z.; Li, Y.; Jiang, C.; Liu, S.; Zhu, Y.-G.; Deng, Y. Organic fertilizer potentiates the transfer of typical antibiotic resistance gene among special bacterial species. J. Hazard. Mater. 2022, 435, 128985. [Google Scholar] [CrossRef] [PubMed]
- Diebold, P.J.; New, F.N.; Hovan, M.; Satlin, M.J.; Brito, I.L. Linking plasmid-based beta-lactamases to their bacterial hosts using single-cell fusion PCR. eLife 2021, 10, e66834. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.N.; Arias, C.A.; Brix, H. Constructed wetlands for water treatment: New developments. Water 2017, 9, 397. [Google Scholar] [CrossRef]
- Dotro, G.; Langergraber, G.; Molle, P.; Nivala, J.; Puigagut, J.; Stein, O.; von Sperling, M. Treatment Wetlands; IWA Publishing: London, UK, 2017. [Google Scholar]
- Matamoros, V.; García, J.; Bayona, J.M. Organic micropollutant removal in a full-scale surface flow constructed wetland fed with secondary effluent. Water Res. 2008, 42, 653–660. [Google Scholar] [CrossRef]
- Sossalla, N.A.; Nivala, J.; Reemtsma, T.; Schlichting, R.; König, M.; Forquet, N.; van Afferden, M.; Müller, R.A.; Escher, B.I. Removal of micropollutants and biological effects by conventional and intensified constructed wetlands treating municipal wastewater. Water Res. 2021, 201, 117349. [Google Scholar] [CrossRef]
- Wu, S.; Carvalho, P.N.; Müller, J.A.; Manoj, V.R.; Dong, R. Sanitation in constructed wetlands: A review on the removal of human pathogens and fecal indicators. Sci. Total Environ. 2016, 541, 8–22. [Google Scholar] [CrossRef]
- Knecht, C.A.; Krüger, M.; Kellmann, S.; Mäusezahl, I.; Möder, M.; Adelowo, O.O.; Vollmers, J.; Kaster, A.-K.; Nivala, J.; Müller, J.A. Cellular stress affects the fate of microbial resistance to folate inhibitors in treatment wetlands. Sci. Total Environ. 2022, 845, 157318. [Google Scholar] [CrossRef]
- Fluit, A.C.; Bayjanov, J.R.; Aguilar, M.D.; Cantón, R.; Tunney, M.M.; Elborn, J.S.; van Westreenen, M.; Ekkelenkamp, M.B. Characterization of clinical Ralstonia strains and their taxonomic position. Antonie van Leeuwenhoek 2021, 114, 1721–1733. [Google Scholar] [CrossRef]
- Haenelt, S.; Wang, G.; Kasmanas, J.C.; Musat, F.; Richnow, H.H.; da Rocha, U.N.; Müller, J.A.; Musat, N. The fate of sulfonamide resistance genes and anthropogenic pollution marker intI1 after discharge of wastewater into a pristine river stream. Front. Microbiol. 2023, 14, 1058350. [Google Scholar] [CrossRef]
- Lee, J.; Ju, F.; Maile-Moskowitz, A.; Beck, K.; Maccagnan, A.; McArdell, C.S.; Molin, M.D.; Fenicia, F.; Vikesland, P.J.; Pruden, A.; et al. Unraveling the riverine antibiotic resistome: The downstream fate of anthropogenic inputs. Water Res. 2021, 197, 117050. [Google Scholar] [CrossRef]
- Nivala, J.; Headley, T.; Wallace, S.; Bernhard, K.; Brix, H.; van Afferden, M.; Müller, R.A. Comparative analysis of constructed wetlands: The design and construction of the ecotechnology research facility in Langenreichenbach, Germany. Ecol. Eng. 2013, 61, 527–543. [Google Scholar] [CrossRef]
- Kahl, S.; Nivala, J.; van Afferden, M.; Müller, R.A.; Reemtsma, T. Effect of design and operational conditions on the performance of subsurface flow treatment wetlands: Emerging organic contaminants as indicators. Water Res. 2017, 125, 490–500. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Grape, M.; Motakefi, A.; Pavuluri, S.; Kahlmeter, G. Standard and real-time multiplex PCR methods for detection of trimethoprim resistance dfr genes in large collections of bacteria. Clin. Microbiol. Infect. 2007, 13, 1112–1118. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Wang, N.; Yang, X.; Jiao, S.; Zhang, J.; Ye, B.; Gao, S. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS ONE 2014, 9, e112626. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Vollmers, J.; Mäusezahl, I.; Kaster, A.-K.; Müller, J.A. Detection of the carbapenemase gene blaVIM-5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Sci. Rep. 2018, 8, 15116. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings. R package version 2.54.0; 2019. Available online: https://bioconductor.org/packages/release/bioc/html/Biostrings.html (accessed on 1 May 2023).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.2; 2018. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 1 May 2023).
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Roman, V.L.; Merlin, C.; Virta, M.P.J.; Bellanger, X. EpicPCR 2.0: Technical and Methodological Improvement of a Cutting-Edge Single-Cell Genomic Approach. Microorganisms 2021, 9, 1649. [Google Scholar] [CrossRef]
- Wang, Z.; Li, W.; Li, H.; Zheng, W.; Guo, F. Phylogenomics of Rhodocyclales and its distribution in wastewater treatment systems. Sci. Rep. 2020, 10, 3883. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.; Martínez-Lavanchy, P.M.; Goris, T.; Heider, J.; Boll, M.; Kaster, A.; Müller, J.A. IncP-type plasmids carrying genes for antibiotic resistance or for aromatic compound degradation are prevalent in sequenced Aromatoleum and Thauera strains. Environ. Microbiol. 2022, 24, 6411–6425. [Google Scholar] [CrossRef] [PubMed]
- Gerzova, L.; Videnska, P.; Faldynova, M.; Sedlar, K.; Provazník, I.; Cizek, A.; Rychlik, I. Characterization of Microbiota Composition and Presence of Selected Antibiotic Resistance Genes in Carriage Water of Ornamental Fish. PLoS ONE 2014, 9, e103865. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Osuntade, A.I. Class 1 Integron, Sulfonamide and Florfenicol Resistance Genes in Bacteria from Three Unsanitary Landfills, Ibadan, Nigeria. J. Microbiol. Infect. Dis. 2019, 9, 34–42. [Google Scholar] [CrossRef]
- Ryan, M.P.; Adley, C.C. Ralstonia spp.: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 33, 291–304. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Ubiquitous and persistent Proteobacteria and other Gram-negative bacteria in drinking water. Sci. Total Environ. 2017, 586, 1141–1149. [Google Scholar] [CrossRef]
- Wang, L.; Fu, L.; Liu, Z.; Guo, H.; Wang, L.; Feng, M.; He, Y.; Chen, Q.; Hu, J. Comparative Analysis of Antimicrobial Resistance, Integrons, and Virulence Genes Among Extended-Spectrum β-Lactamase-Positive Laribacter hongkongensis from Edible Frogs and Freshwater Fish. Microb. Drug Resist. 2019, 25, 855–864. [Google Scholar] [CrossRef]
- Woo, P.C.; Kuhnert, P.; Burnens, A.P.; Teng, J.L.; Lau, S.K.; Que, T.-L.; Yau, H.-H.; Yuen, K.-Y. Laribacter hongkongensis: A potential cause of infectious diarrhea. Diagn. Microbiol. Infect. Dis. 2003, 47, 551–556. [Google Scholar] [CrossRef]
- Atrouni, A.E.; Joly-Guillou, M.L.; Ehamze, M.; Ekempf, M. Reservoirs of Non-baumannii Acinetobacter Species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Álvarez, V.E.; Quiroga, M.P.; Galán, A.V.; Vilacoba, E.; Quiroga, C.; Ramírez, M.S.; Centrón, D. Crucial Role of the Accessory Genome in the Evolutionary Trajectory of Acinetobacter baumannii Global Clone 1. Front. Microbiol. 2020, 11, 342. [Google Scholar] [CrossRef]
- Lee, K.; Yum, J.H.; Yong, D.; Lee, H.M.; Kim, H.D.; Docquier, J.-D.; Rossolini, G.M.; Chong, Y. Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005, 49, 4485–4491. [Google Scholar] [CrossRef]
- Liu, C.-C.; Tang, C.Y.; Chang, K.-C.; Kuo, H.-Y.; Liou, M.-L. A comparative study of class 1 integrons in Acinetobacter baumannii. Gene 2014, 544, 75–82. [Google Scholar] [CrossRef]
- Ramírez, M.S.; Quiroga, C.; Centrón, D. Novel Rearrangement of a Class 2 Integron in Two Non-Epidemiologically Related Isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 5179–5181. [Google Scholar] [CrossRef]
- Garner, E.; Brown, C.L.; Schwake, D.O.; Rhoads, W.J.; Arango-Argoty, G.; Zhang, L.; Jospin, G.; Coil, D.A.; Eisen, J.A.; Edwards, M.A.; et al. Comparison of Whole-Genome Sequences of Legionella pneumophila in Tap Water and in Clinical Strains, Flint, Michigan, USA, 2016. Emerg. Infect. Dis. 2019, 25, 2013–2020. [Google Scholar] [CrossRef]
- Gordon, L.; Cloeckaert, A.; Doublet, B.; Schwarz, S.; Bouju-Albert, A.; Ganière, J.-P.; Le Bris, H.; Le Flèche-Matéos, A.; Giraud, E. Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J. Antimicrob. Chemother. 2008, 62, 65–71. [Google Scholar] [CrossRef]
- Schmidt, A.S.; Bruun, M.S.; Larsen, J.L.; Dalsgaard, I. Characterization of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicida isolates from various geographical areas. J. Antimicrob. Chemother. 2001, 47, 735–743. [Google Scholar] [CrossRef]
- Nguyen, P.M.; Arslan, M.; Kappelmeyer, U.; Mäusezahl, I.; Wiessner, A.; Müller, J.A. Spatial characterization of microbial sulfur cycling in horizontal-flow constructed wetland models. Chemosphere 2022, 309 Pt 1, 136605. [Google Scholar] [CrossRef]
- Lucey, B.; Crowley, D.; Moloney, P.; Cryan, B.; Daly, M.; O’Halloran, F.; Threlfall, E.J.; Fanning, S. Integronlike structures in Campylobacter spp. of human and animal origin. Emerg. Infect. Dis. 2000, 6, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Corno, G.; Ghaly, T.; Sabatino, R.; Eckert, E.M.; Galafassi, S.; Gillings, M.R.; Di Cesare, A. Class 1 integron and related antimicrobial resistance gene dynamics along a complex freshwater system affected by different anthropogenic pressures. Environ. Pollut. 2023, 316, 120601. [Google Scholar] [CrossRef] [PubMed]
- Buchenau, B.; Thauer, R.K. Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch. Microbiol. 2004, 182, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, A.D.; Assaf, R.; Aziz, R.K.; Brettin, T.; Bun, C.; Conrad, N.; Davis, J.J.; Dietrich, E.M.; Disz, T.; Gerdes, S.; et al. PATRIC as a unique resource for studying antimicrobial resistance. Briefings Bioinform. 2017, 20, 1094–1102. [Google Scholar] [CrossRef]
- Frigaard, N.-U.; Martinez, A.; Mincer, T.J.; DeLong, E.F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 2006, 439, 847–850. [Google Scholar] [CrossRef]
- Koretke, K.K.; Lupas, A.N.; Warren, P.V.; Rosenberg, M.; Brown, J.R. Evolution of Two-Component Signal Transduction. Mol. Biol. Evol. 2000, 17, 1956–1970. [Google Scholar] [CrossRef]
- Nelson, K.E.; Clayton, R.A.; Gill, S.R.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Nelson, W.C.; Ketchum, K.A.; et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 1999, 399, 323–329. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Tetu, S.G.; Penesyan, A.; Qi, Q.; Rajabal, V.; Gillings, M.R. Discovery of integrons in Archaea: Platforms for cross-domain gene transfer. Sci. Adv. 2022, 8, eabq6376. [Google Scholar] [CrossRef]
- Fonseca, D.R.; Halim, M.F.A.; Holten, M.P.; Costa, K.C. Type IV-Like Pili Facilitate Transformation in Naturally Competent Archaea. J. Bacteriol. 2020, 202, e00355-20. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Andremont, A.; Bengtsson-Palme, J.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Fick, J.; Flach, C.-F.; Gaze, W.H.; Kuroda, M.; et al. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018, 117, 132–138. [Google Scholar] [CrossRef]
- European Parliament. Strategic Approach to Pharmaceuticals in the Environment. 2020. Available online: https://www.europarl.europa.eu/doceo/document/TA-9-2020-0226_EN.pdf (accessed on 15 May 2023).
- UN Environment Programme. Frontiers 2017: Emerging Issues of Environmental Concern. 2017. Available online: https://www.unenvironment.org/resources/frontiers-2017-emerging-issues-environmental-concern (accessed on 1 May 2023).
- WHO. Technical Brief on Water, Sanitation, Hygiene (WASH) and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance (AMR). 2020. Available online: https://www.who.int/water_sanitation_health/publications/wash-wastewatermanagement-to-prevent-infections-and-reduce-amr/en/ (accessed on 1 May 2023).
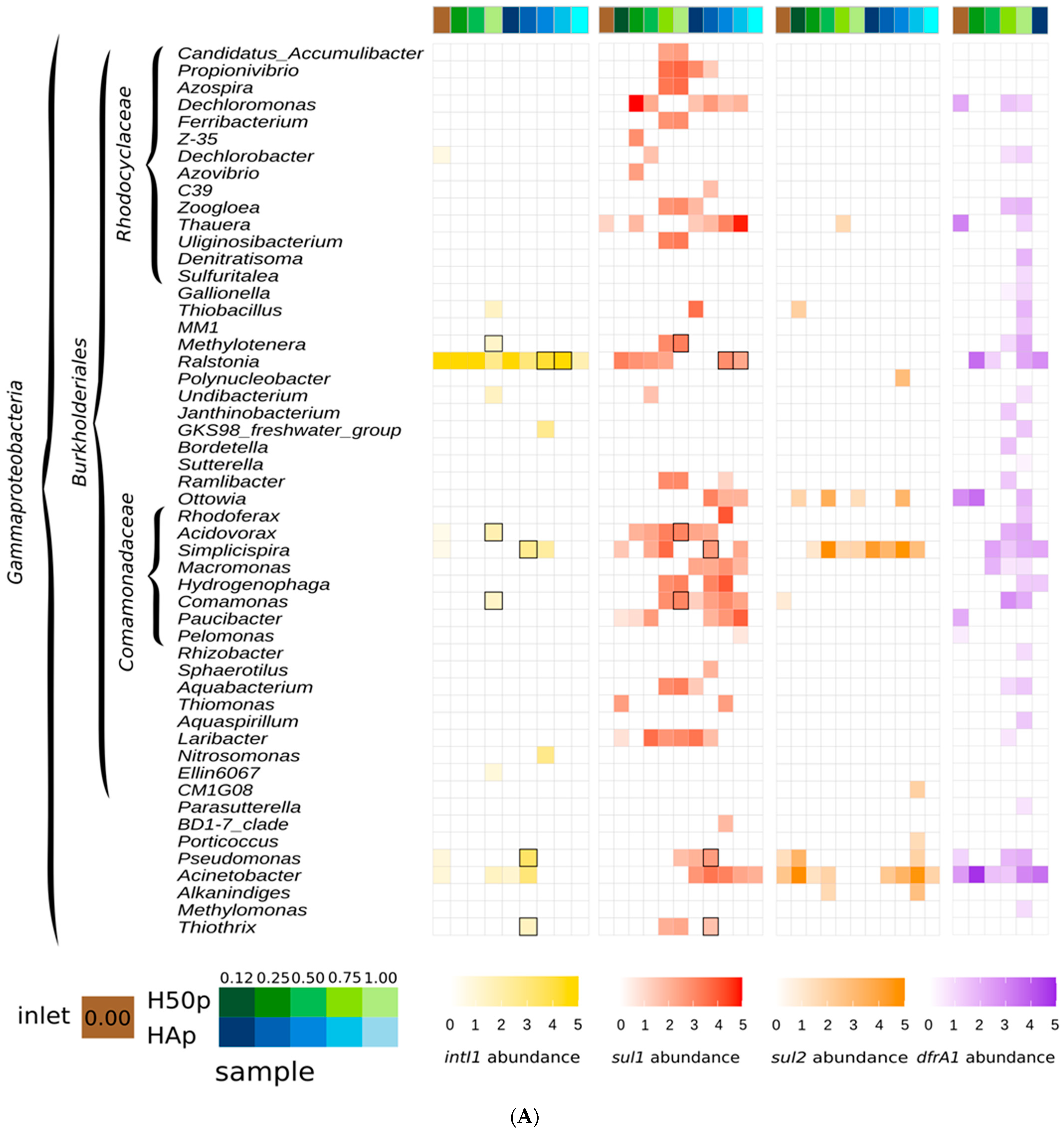
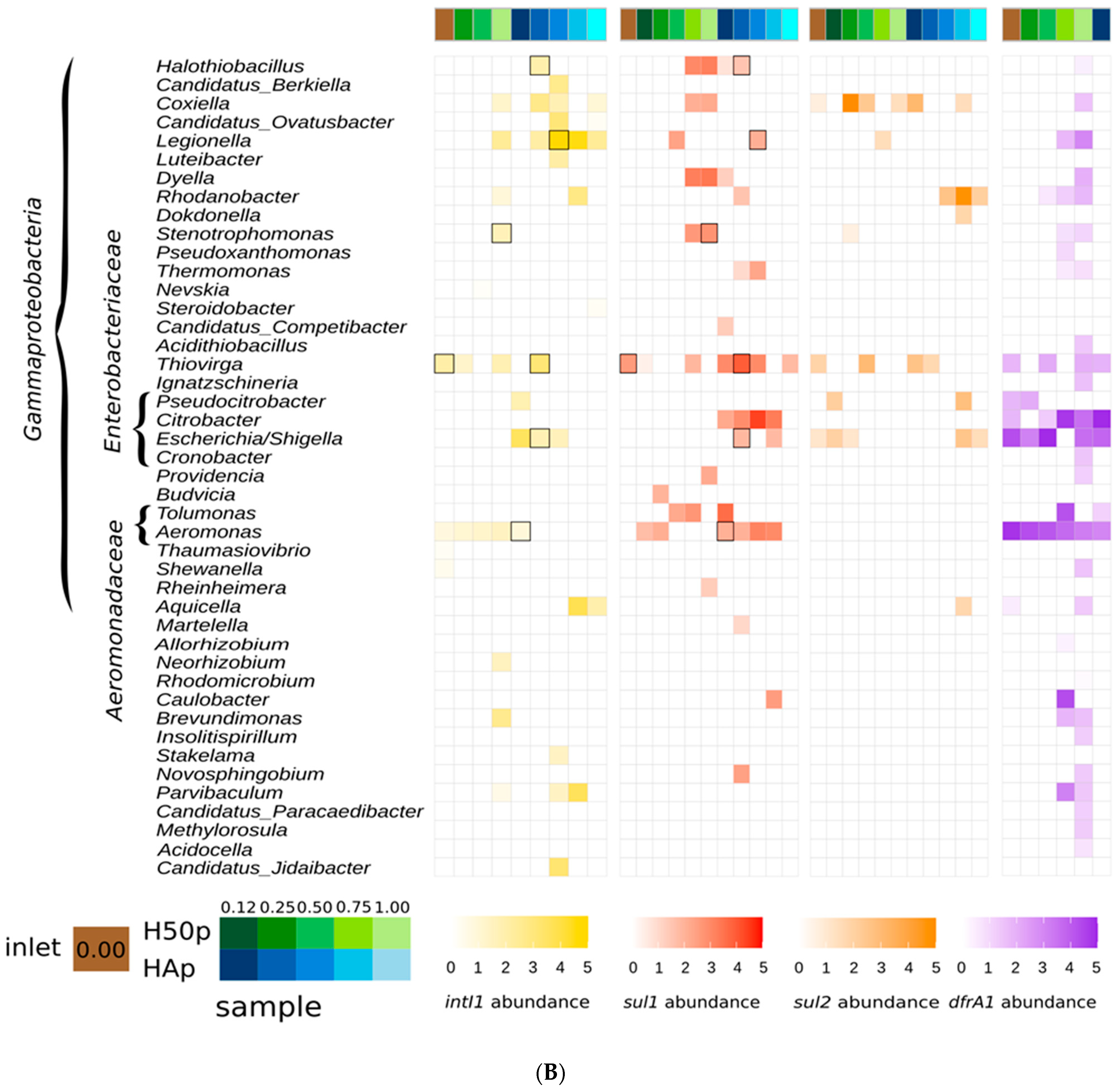

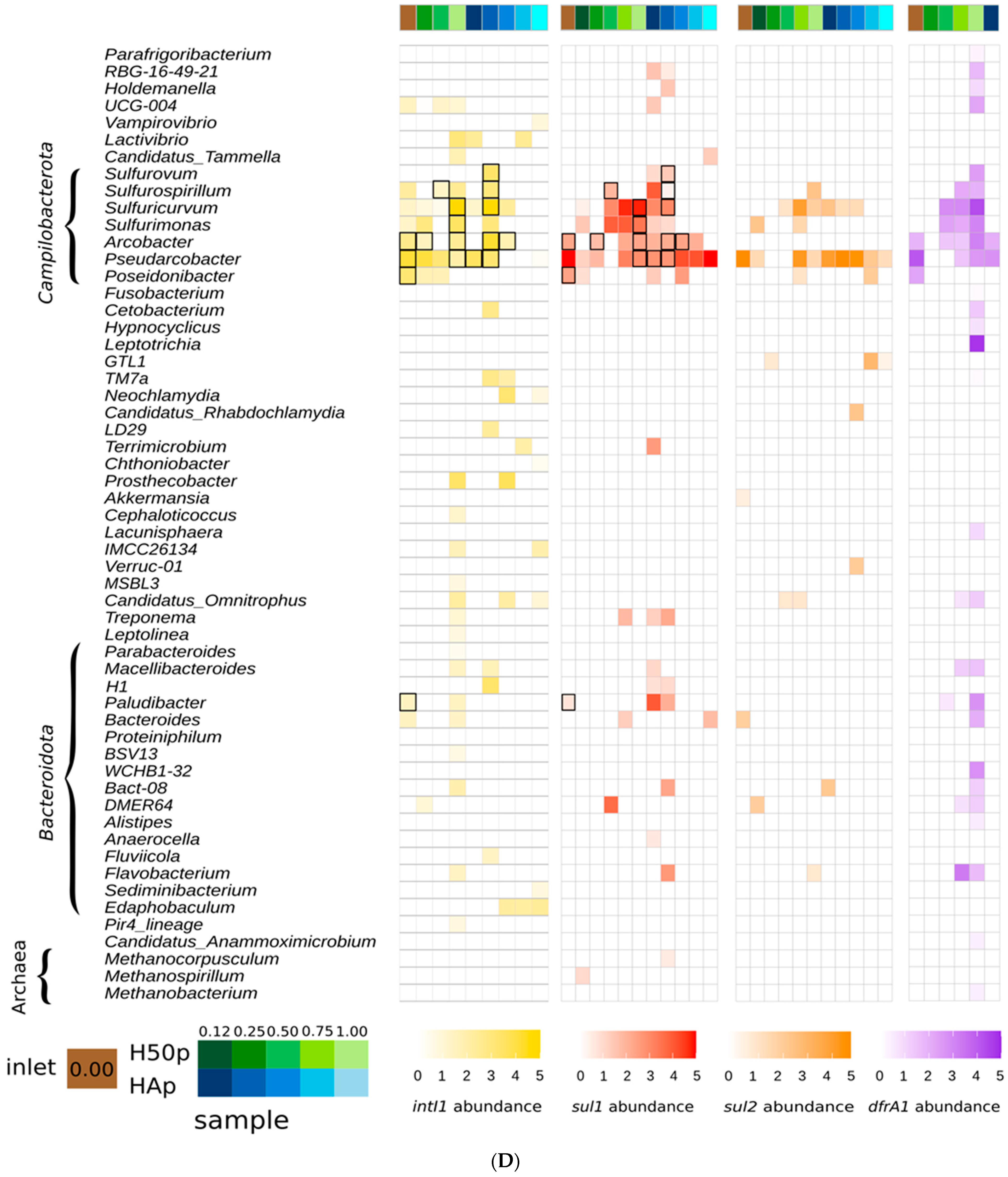
| intI1 | sul1 | sul2 | dfrA1 |
|---|---|---|---|
| Acidovorax | |||
| Acinetobacter | Acinetobacter | Acinetobacter | |
| Aeromonas | Aeromonas | Aeromonas | |
| Bdellovibrio | |||
| Citrobacter | |||
| Commamonas | |||
| Coxiella | Coxiella | ||
| Dechloromonas | |||
| Escherichia/Shigella | |||
| Laribacter | |||
| Legionella | |||
| Paucibacter | |||
| Protocatella | |||
| Ralstonia | Ralstonia | Ralstonia | |
| Simplicispira | Simplicispira | Simplicispira | |
| Subdoligranulum | Subdoligranulum | ||
| Thauera | |||
| Thiovirga | Thiovirga | Thiovirga | |
| Trichlorobacter | Trichlorobacter |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knecht, C.A.; Hinkel, M.; Mäusezahl, I.; Kaster, A.-K.; Nivala, J.; Müller, J.A. Identification of Antibiotic Resistance Gene Hosts in Treatment Wetlands Using a Single-Cell Based High-Throughput Approach. Water 2023, 15, 2432. https://doi.org/10.3390/w15132432
Knecht CA, Hinkel M, Mäusezahl I, Kaster A-K, Nivala J, Müller JA. Identification of Antibiotic Resistance Gene Hosts in Treatment Wetlands Using a Single-Cell Based High-Throughput Approach. Water. 2023; 15(13):2432. https://doi.org/10.3390/w15132432
Chicago/Turabian StyleKnecht, Camila A., Maja Hinkel, Ines Mäusezahl, Anne-Kristin Kaster, Jaime Nivala, and Jochen A. Müller. 2023. "Identification of Antibiotic Resistance Gene Hosts in Treatment Wetlands Using a Single-Cell Based High-Throughput Approach" Water 15, no. 13: 2432. https://doi.org/10.3390/w15132432
APA StyleKnecht, C. A., Hinkel, M., Mäusezahl, I., Kaster, A.-K., Nivala, J., & Müller, J. A. (2023). Identification of Antibiotic Resistance Gene Hosts in Treatment Wetlands Using a Single-Cell Based High-Throughput Approach. Water, 15(13), 2432. https://doi.org/10.3390/w15132432








