Human Oncogenic Epstein–Barr Virus in Water and Human Blood Infection of Communities in Phayao Province, Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Sampling Methods
2.2. Water Filtration
2.3. Specimens
2.4. DNA Extraction
2.4.1. DNA Extraction from Filter Membranes
2.4.2. DNA Extraction from Whole Blood Samples
2.5. EBV DNA Detection by PCR
2.6. EBV DNA Detection by qPCR
2.7. Metagenomics
2.8. Geographic Information System (GIS)
2.9. Statistical Analysis
3. Results
3.1. Water Quality Analysis
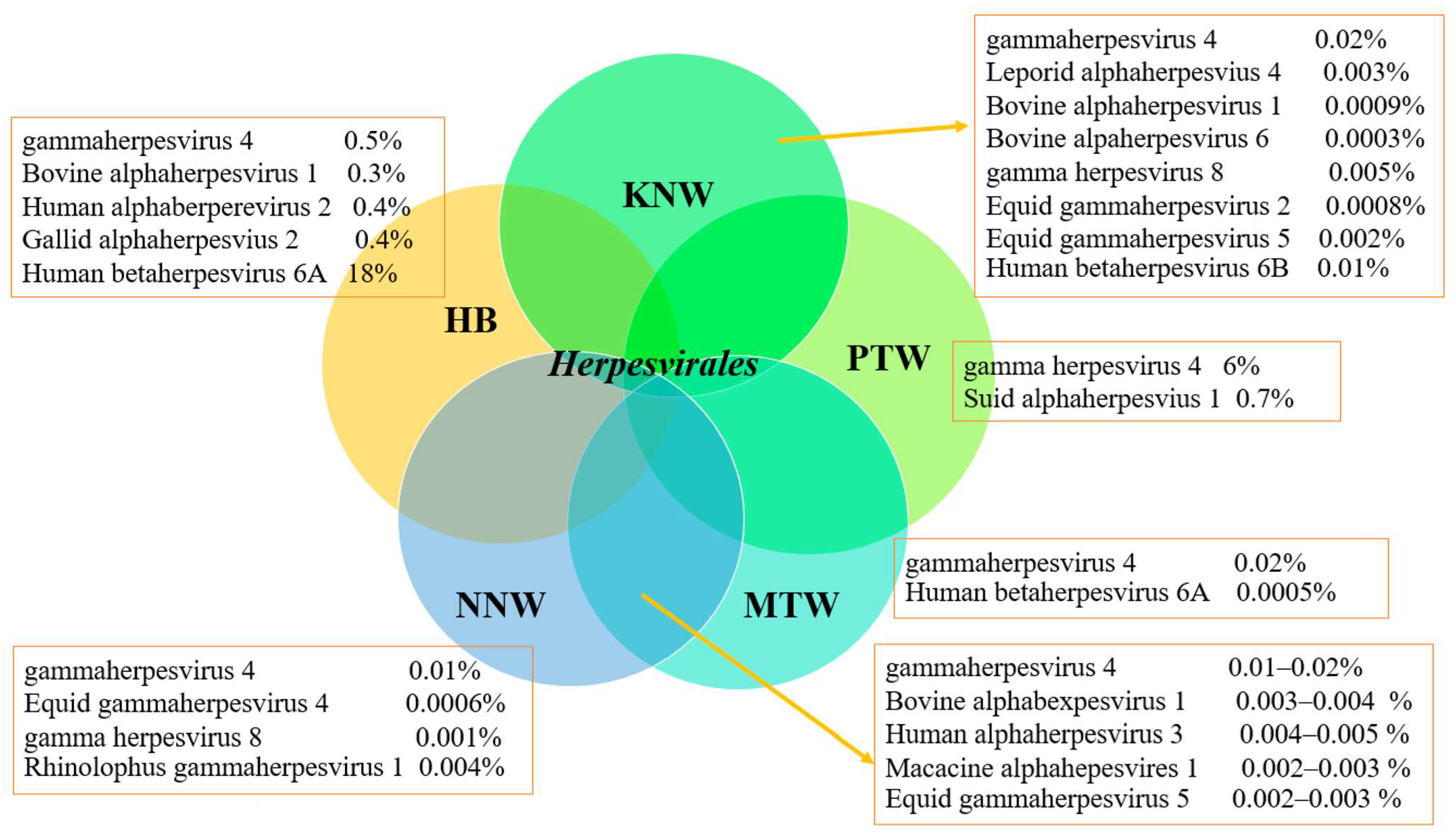
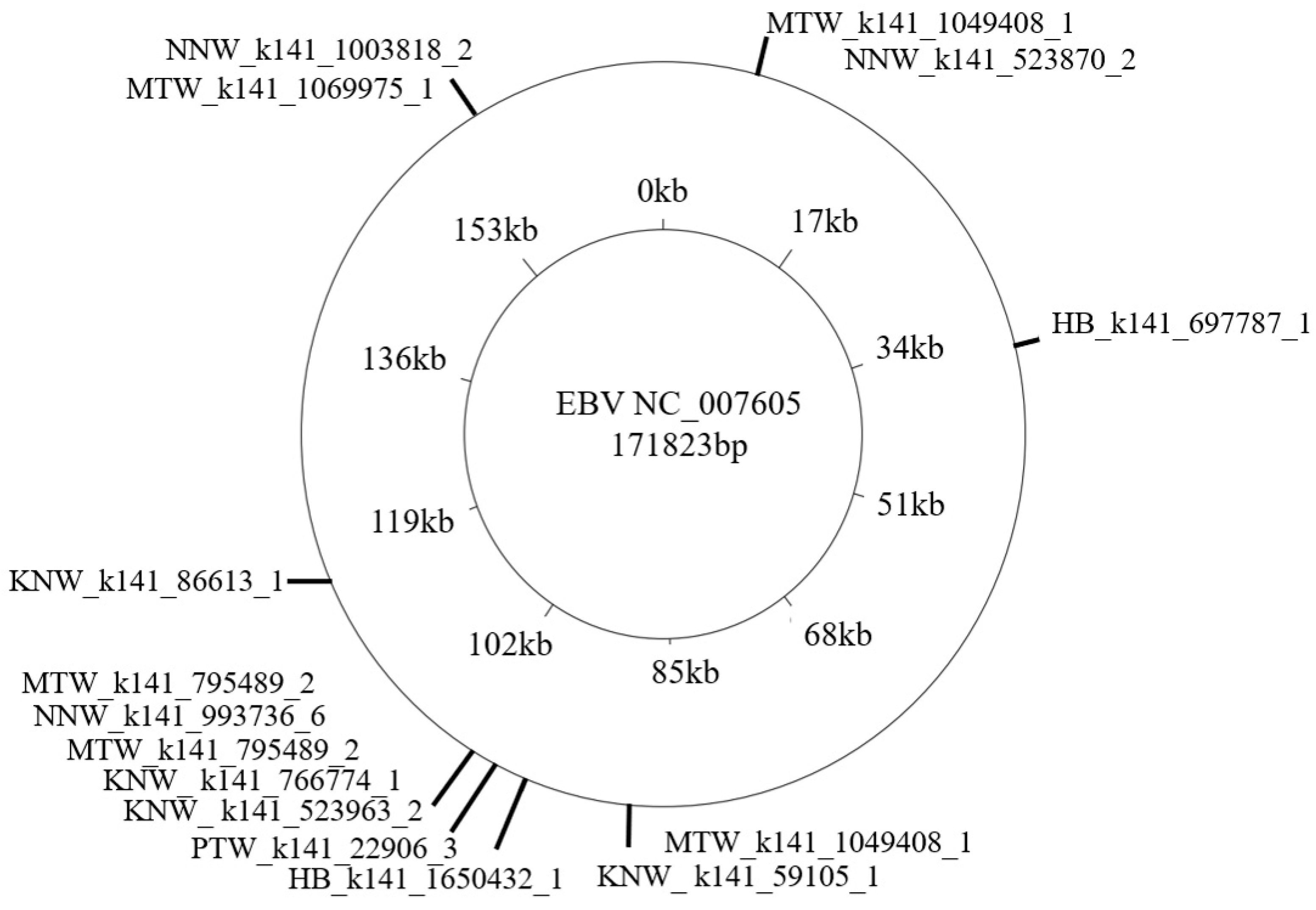
3.2. Metagenomics Analysis
3.3. EBV DNA Detection by PCR and qPCR and Environment Factors
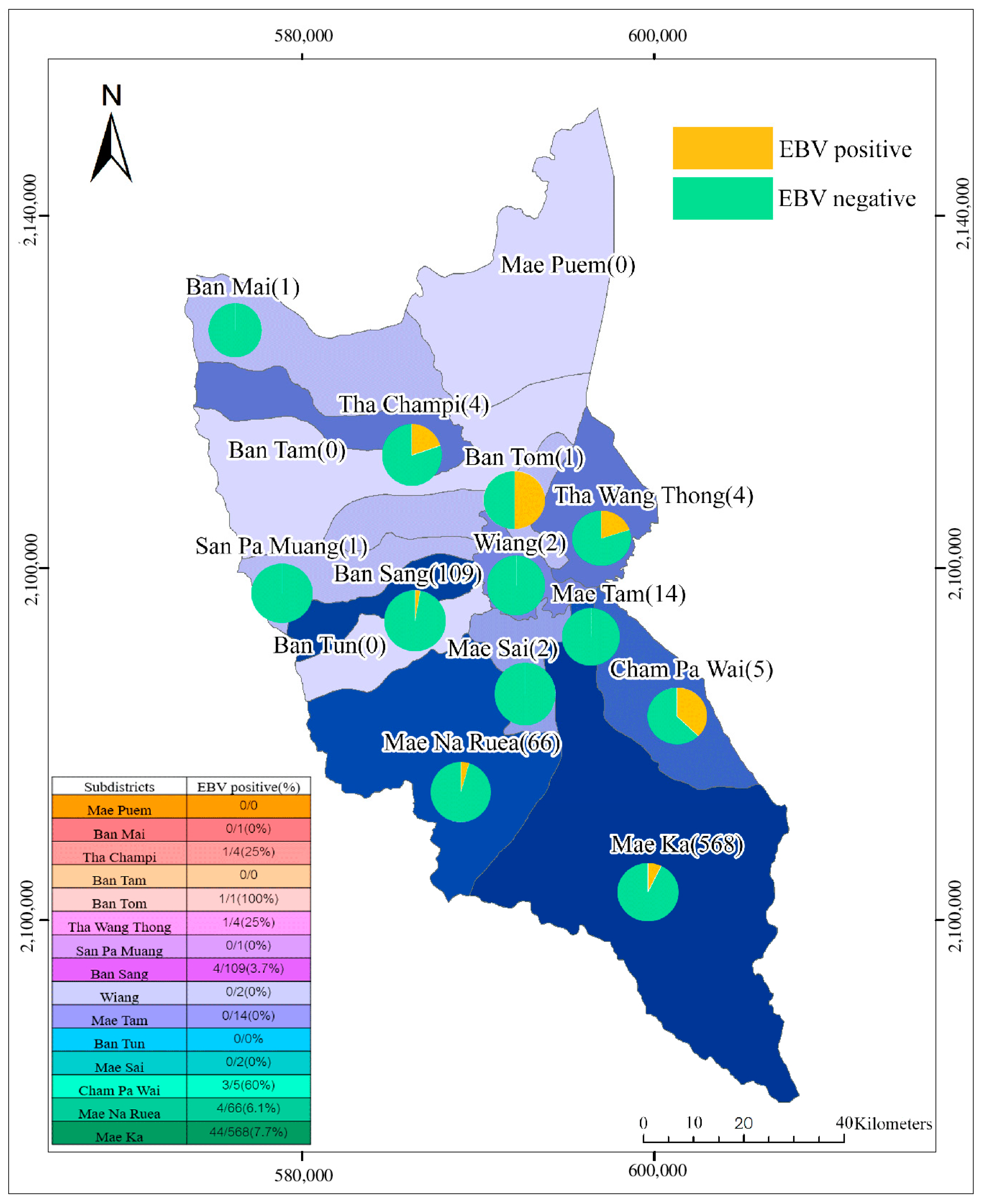
3.4. GIS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Billions of People Will Lack Access to Safe Water, Sanitation and Hygiene in 2030 Unless Progress Quadruples—Warn WHO, UNICEF. Available online: https://www.who.int/news/item/01-07-2021-billions-of-people-will-lack-access-to-safe-water-sanitation-and-hygiene-in-2030-unless-progress-quadruples-warn-who-unicef (accessed on 25 September 2022).
- Fan, L.; Liu, G.; Wang, F.; Geissen, V.; Ritsema, C.J. Factors Affecting Domestic Water Consumption in Rural Households upon Access to Improved Water Supply, Insights from the Wei River Basin, China. PLoS ONE 2013, 8, e71977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, J.; Bos, J.H.; de Vries, T.W.; de Jong-van den Berg, L.T. Use of antibiotics in rural and urban regions in the Netherlands: An observational drug utilization study. BMC Public Health 2014, 677, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, J.; Taiwo, O.O.; Ahluwalia, I.; Chungong, S. Disparities in Infectious Diseases among Women in Developing Countries. Emerg. Infect. Dis. 2004, 10, e24. [Google Scholar] [CrossRef]
- Singh, A.R.; Singh, S.A.; Shakuntala, A. Diseases of Poverty and Lifestyle, Well-Being and Human Development. Mens Sana Monogr. 2008, 6, 187–225. [Google Scholar] [CrossRef] [Green Version]
- Stewart, B.W. International Agency for Research on Cancer, Lyon, France. In World Cancer Report; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Di Bonito, P.; Iaconelli, M.; Gheit, T.; Tommasino, M.; Della Libera, S.; Bonadonna, L.; La Rosa, G. Detection of oncogenic viruses in water environments by a Luminex-based multiplex platform for high throughput screening of infectious agents. Water Res. 2017, 123, 549–555. [Google Scholar] [CrossRef]
- Al-Jamaei, A.; van Dijk, B.; Helder, M.; Forouzanfar, T.; Leemans, C.; de Visscher, J. A population-based study of the epidemiology of oral squamous cell carcinoma in the Netherlands with emphasis on young adults. Int. J. Oral Maxillofac. Surg. 2018, 51, 18–26. [Google Scholar] [CrossRef]
- Oliver, R.J.; Dearing, J.; Hindle, I. Oral cancer in young adults, report of three cases and review of the literature. Br. Dent. J. 2000, 188, 362–365. [Google Scholar] [CrossRef] [Green Version]
- di Martino, E.; Smith, L.; Bradley, S.H.; Hemphill, S.; Wright, J.; Renzi, C.; Bergin, R.; Emery, J.; Neal, R.D. Incidence trends for twelve cancers in younger adults-a rapid review. Br. J. Cancer 2022, 126, 1374–1386. [Google Scholar] [CrossRef]
- Brunner, J.L.; Yarber, C.M. Evaluating the Importance of Environmental Persistence for Ranavirus Transmission and Epidemiology. Adv. Virus Res. 2018, 101, 129–148. [Google Scholar]
- Wyn-Jones, P. Chapter 9 The Detection of Waterborne Viruses. Perspect. Med. Virol. 2007, 17, 177–204. [Google Scholar]
- Shoham, D.; Jahangir, A.; Ruenphet, S.; Takehara, K. Persistence of avian influenza viruses in various artificially frozen environmental water types. Influenza Res. Treat. 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gall, A.M.; Mariñas, B.J.; Lu, Y.; Shisler, J.L. Waterborne Viruses, A Barrier to Safe Drinking Water. PLoS Pathog. 2015, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Adefisoye, M.A.; Nwodo, U.U.; Green, E.; Okoh, A.I. Quantitative PCR Detection and Characterisation of Human Adenovirus, Rotavirus and Hepatitis A Virus in Discharged Effluents of Two Wastewater Treatment Facilities in the Eastern Cape, South Africa. Food Environ. Virol. 2016, 8, 262–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haramoto, E.; Katayama, H.; Ohgaki, S. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 2004, 70, 2154–2160. [Google Scholar] [CrossRef] [Green Version]
- Vecchia, A.D.; Kluge, M. Presence of Torque teno virus (TTV) in tap water in public schools from Southern Brazil. Food Environ. Virol. 2013, 5, 41–45. [Google Scholar] [CrossRef]
- Rashid, M.; Khan, M.N.; Jalbani, N. Detection of Human Adenovirus, Rotavirus, and Enterovirus in Tap Water and Their Association with the Overall Quality of Water in Karachi, Pakistan. Food Environ. Virol. 2021, 13, 44–52. [Google Scholar] [CrossRef]
- Botzenhart, K. Viruses in drinking water. Bundesgesundheitsblatt Gesundh. Gesundheitsschutz. 2007, 50, 296–301. [Google Scholar] [CrossRef]
- Hao, W.; Inger, K.; Per, S. Hepatitis E virus genotype 3 strains and a plethora of other viruses detected in raw and still in tap water. Water Res. 2020, 168, 115141. [Google Scholar]
- Martin-Latil, S.; Hennechart-Collette, C.; Guillier, L.; Perelle, S. Duplex RT-qPCR for the detection of hepatitis E virus in water using a process control. Int. J. Food Microbiol. 2012, 157, 167–173. [Google Scholar] [CrossRef]
- Breitbart, M.; Rohwer, F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 2005, 39, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Bezerra, R.S.; Bitencourt, H.T.; Covas, D.T.; Kashima, S.; Slavov, S.N. Metagenomic identification of human Gemykibivirus-2 (HuGkV-2) in parenterally infected blood donors from the Brazilian Amazon. Int. J. Infect. Dis. 2020, 98, 249–251. [Google Scholar] [CrossRef]
- Lau, P.; Cordey, S.; Brito, F.; Tirefort, D.; Petty, T.J.; Turin, L.; Guichebaron, A.; Docquier, M.; Zdobnov, E.M.; Waldvogel-Abramowski, S.; et al. Metagenomics analysis of red blood cell and fresh-frozen plasma units. Transfusion 2017, 57, 1787–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summary of Excreted and Waterborne Viruses. Available online: https://www.waterpathogens.org/book/summary-of-excreted-and-waterborne-viruses (accessed on 25 May 2022).
- Maliha, G.; Jintao, L.; Lei, Z.; Khodahemmati, S.; Wang, M.; Wang, Y.; Zhao, L.; Jia, R.; Chen, S.; Zeng, Y. Water Carcinogenicity and Prevalence of HPV Infection in Esophageal Cancer Patients in Huaihe River Basin, China. Gastroenterol. Res. Pract. 2018, 6, 2028986. [Google Scholar]
- Uzoma, I.C.; Taiwo, I.A.; Granai, M.; Di Stefano, G.; Sorrentino, E.; Mannucci, S.; Durosinmi, M.A.; Lazzi, S.; Leoncini, L.; Akinloye, O. Distinct pattern of lymphoid neoplasms characterizations according to the WHO classification (2016) and prevalence of associated Epstein-Barr virus infection in Nigeria population. Infect. Agent Cancer 2021, 16, 36. [Google Scholar] [CrossRef]
- Spacek, M.; Hubacek, P.; Markova, J.; Zajac, M.; Vernerova, Z.; Kamaradova, K.; Stuchly, J.; Kozak, T. Plasma EBV-DNA monitoring in Epstein-Barr virus-positive Hodgkin lymphoma patients. APMIS 2011, 119, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, S.Y. Epstein-Barr Virus (Herpesviridae) General Features. In Encyclopedia of Virology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 487–494. [Google Scholar]
- Stevens, S.J.; Verkuijlen, S.A.; Hariwiyanto, B.; Harijadi; Fachiroh, J.; Paramita, D.K.; Tan, I.B.; Haryana, S.M.; Middeldorp, J.M. Diagnostic value of measuring Epstein-Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV immunoglobulin A (IgA) and IgG antibody levels in blood of nasopharyngeal carcinoma patients from Indonesia. J. Clin. Microbiol. 2005, 43, 3066–3073. [Google Scholar] [CrossRef] [Green Version]
- Zanella, M.; Cordey, S.; Kaiser, L. Beyond Cytomegalovirus and Epstein-Barr Virus: A Review of Viruses Composing the Blood Virome of Solid Organ Transplant and Hematopoietic Stem Cell Transplant Recipients. Clin. Microbiol. Rev. 2020, 33, e00027-20. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.K.; Al-Sadeq, D.W.; Ali, N.H.; Pintus, G.; Abou-Saleh, H.; Nasrallah, G.K. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population. Front Oncol. 2018, 13, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.H.; Tam, J.S.; Peiris, J.S. Epstein-Barr virus (EBV) infection in infancy. J. Clin. Virol. 2001, 21, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Meyohas, M.C.; Maréchal, V.; Desire, N.; Bouillie, J.; Frottier, J.; Nicolas, J.C. Study of mother-to-child Epstein-Barr virus transmission by means of nested PCRs. J. Virol. 1996, 70, 6816–6819. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.M.; Lee, C.Y.; Chang, M.H.; Wang, J.D.; Hsu, C.Y. Primary infections of Epstein-Barr virus, cytomegalovirus, and human herpesvirus-6. Arch. Dis. Child. 1993, 68, 408–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaz, B.H. Transfusion Transmitted Diseases. Transfus. Med. Hemost. 2009, 361–371. [Google Scholar] [CrossRef]
- Odumade, O.A.; Hogquist, K.A.; Balfour, H.H.J. Progress and problems in understanding and managing primary Epstein-Barr virus infections. Clin. Microbiol. Rev. 2011, 24, 193–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umakanthan, S.; Bukelo, M.M. Molecular Genetics in Epstein-Barr Virus-Associated Malignancies. Life 2021, 11, 593. [Google Scholar] [CrossRef]
- Houen, G.; Trier, N.H. Epstein-Barr Virus and Systemic Autoimmune Diseases. Front. Immunol. 2021, 11, 587380. [Google Scholar] [CrossRef] [PubMed]
- Suntornlohanakul, R.; Wanlapakorn, N.; Vongpunsawad, S.; Thongmee, T.; Chansaenroj, J.; Poovorawan, Y. Seroprevalence of Anti-EBV IgG among Various Age Groups from Khon Kaen Province, Thailand. Asian Pac. J. Cancer Prev. 2015, 16, 7583–7587. [Google Scholar] [CrossRef] [Green Version]
- Thammaborvorn, R.; Mungmee, V.; Thammachotruja, L.; Kowitdamrong, E.; Bhattarakosol, P. Prevalence of viral infections in clinical specimens at Virology Laboratory Unit during the Year 1998 to 2004. Chula Med. J. 2007, 51, 229–239. [Google Scholar]
- De Paschale, M.; Clerici, P. Serological diagnosis of Epstein-Barr virus infection: Problems and solutions. World J. Virol. 2012, 12, 31–43. [Google Scholar] [CrossRef]
- Pan, R.; Liu, X.; Zhou, S.; Ning, Z.; Zheng, H.; Gao, M.; Ding, Y.; Yao, W.; Liao, X.; He, N. Differential prevalence and correlates of whole blood Epstein-Barr virus DNA between HIV-positive and HIV-negative men who have sex with men in Shanghai, China. Epidemiol. Infect. 2017, 145, 2330–2340. [Google Scholar] [CrossRef] [Green Version]
- Gupta, I.; Nasrallah, G.K.; Sharma, A.; Jabeen, A.; Smatti, M.K.; Al-Thawadi, H.A.; Sultan, A.A.; Alkhalaf, M.; Vranic, S.; Moustafa, A.E.A. Prevalence of human Papillomaviruses (HPV) and Epstein-Barr virus (EBV) in healthy blood donors from diverse nationalities in Qatar. Cancer Cell Int. 2020, 3, 107. [Google Scholar] [CrossRef] [Green Version]
- Smatti, M.K.; Yassine, H.M.; AbuOdeh, R.; AlMarawani, A.; Taleb, S.A.; Althani, A.A.; Nasrallah, G.K. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE 2017, 12, e0189033. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.-H.; Lu, T.-X.; Tian, T.; Wang, L.; Fan, L.; Xu, J.; Zhang, R.; Gong, Q.-X.; Zhang, Z.-H.; Li, J.-Y.; et al. Epstein–Barr virus (EBV) DNA in whole blood as a superior prognostic and monitoring factor than EBV-encoded small RNA in situ hybridization in diffuse large B-cell lymphoma. Clin. Microbiol. Infect. 2015, 21, 596–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traore, L.; Tao, I.; Bisseye, C.; Diarra, B.; Compaore, T.R.; Nebie, Y.; Asshi, M.; Ouedraogo, A.; Zohoncon, T.; Djigma, F.; et al. Molecular diagnostic of cytomegalovirus, Epstein Barr virus and Herpes virus 6 infections among blood donors by multiplex real-time PCR in Ouagadougou, Burkina Faso. Pan Afr. Med. J. 2016, 99, 298. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Phusingha, P.; Vatanasapt, P.; Promthet, S.; Eklaksananan, T.; Pientong, C. Prevalence of Epstein-Barr virus (EBV) in Oral Exfoliated Cells: A Pilot Prospective Study of Oral Squamous Cell Carcinoma. Srinagarind Med. J. 2013, 28, 286–290. [Google Scholar]
- Gulley, M.L.; Tang, W. Laboratory assays for Epstein-Barr virus-related disease. J. Mol. Diagn. 2008, 10, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Okano, M.; Thiele, G.M.; Davis, J.R.; Grierson, H.L.; Purtilo, D.T. Epstein-Barr virus and human diseases: Recent advances in diagnosis. Clin. Microbiol. Rev. 1988, 1, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Ito, Y.; Suzuki, R. Epstein-Barr virus (EBV) load: The significance and application for each EBV-associated disease. Rev. Med. Virol. 2008, 18, 305–319. [Google Scholar] [CrossRef]
- Lanamtieng, T.; Teawtrakul, N.; Ungarreevittaya, P. Prevalence and clinical characteristics of EBV-positive diffuse large B-cell lymphoma in adult patients: A single center study in Thailand. Leuk Lymphoma 2020, 61, 2262–2264. [Google Scholar] [CrossRef]
- Ok, C.Y.; Papathomas, T.G.; Medeiros, L.J.; Young, K.H. EBV-positive diffuse large B-cell lymphoma of the elderly. Blood 2013, 122, 328–340. [Google Scholar] [CrossRef] [Green Version]
- Fafi-Kremer, S.; Brengel-Pesce, K.; Barguès, G.; Bourgeat, M.J.; Genoulaz, O.; Seigneurin, J.M.; Morand, P. Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma. J. Clin. Virol. 2014, 30, 157–164. [Google Scholar] [CrossRef]
- Kedi, W.; Dongjiang, X.; Zhi, L.; Yan, G.; Kun, J.; Jianrong, S. The rational specimen for the quantitative detection of Epstein-Barr virus DNA load. Clin. Chem. Lab. Med. 2019, 57, 759–765. [Google Scholar] [CrossRef]
- Louten, J. Virus Structure and Classification. Essent. Hum. Virol. 2016, 19–29. [Google Scholar] [CrossRef]
- Amon, W.; Farrell, P.J. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 2005, 15, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Laidler, J.R.; Stedman, K.M. Virus Silicification under Simulated Hot Spring Conditions. Astrobiology 2010, 10, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parimal, P.; Frenkel, V.S. Advances in Membrane Technologies for Water Treatment Materials. Processes and Applications; Woodhead Publishing Series in Energy; Elsevies: Amsterdam, The Netherlands, 2015; pp. 329–347. [Google Scholar]
- Duangjit, S.; Somsuwan, B.; Inpeng, S.; Buddhisa, S.; Bumrungthai, S. The Specific Properties of Phusang Hot Spring Water, Safety and Benefits. Cosmetics 2022, 9, 89. [Google Scholar] [CrossRef]
- Junter, G.A.; Lebrun, L. Cellulose-based virus-retentive filters, a review. Rev. Environ. Sci. Biotechnol. 2017, 16, 455–489. [Google Scholar] [CrossRef]
- Hofscheier, A.; Ponciano, A.; Bonzheim, I.; Adam, P.; Lome-Maldonado, C.; Vela, T.; Cortes, E.; Ortiz-Hidalgo, C.; Fend, F.; Quintanilla-Martinez, L. Geographic variation in the prevalence of Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly, a comparative analysis of a Mexican and a German population. Mod. Pathol. 2011, 24, 1046–1054. [Google Scholar] [CrossRef] [Green Version]
- Bon, M.A.; van Oeveren-Dybicz, A.; van den Bergh, F.A. Genotyping of HLA-B27 by real-time PCR without hybridization probes. Clin. Chem. 2000, 46, 1000–1002. [Google Scholar] [CrossRef]
- Lao, T.D.; Nguyen, D.; Nguyen, T.M.; Le, T.A.H. Molecular Screening for Epstein-Barr virus (EBV), Detection of Genomic EBNA-1, EBNA-2, LMP-1, LMP-2 Among Vietnamese Patients with Nasopharyngeal Brush Samples. Asian Pac. J. Cancer Prev. 2017, 18, 675–1679. [Google Scholar]
- Guideline Value of Provincial Waterworks Authority. 2017. Available online: https://www.pwa.co.th/download/pwastandard50-1.pdf (accessed on 25 May 2022).
- Guideline Value of Thai Metropolitan Waterworks Authority. 2017. Available online: https://web.mwa.co.th/more_news.php?cid=1491 (accessed on 25 May 2022).
- Dayaram, A.; Franz, M.; Schattschneider, A.; Damiani, A.M.; Bischofberger, S.; Osterrieder, N.; Greenwood, A.D. Long term stability and infectivity of herpesviruses in water. Sci. Rep. 2017, 7, 46559. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.J. Gamma herpesviruses and Pulmonary Fibrosis: Evidence from Humans, Horses, and Rodents. Vet. Pathol. 2014, 51, 372–384. [Google Scholar] [CrossRef]
- Nerurkar, L.S.; West, F.; May, M.; Madden, D.L.; Sever, J.L. Survival of herpes simplex virus in water specimens collected from hot tubs in spa facilities and on plastic surfaces. JAMA 1983, 9, 3081–3083. [Google Scholar] [CrossRef]
- Virani, S.; Bilheem, S.; Chansaard, W.; Chitapanarux, I.; Daoprasert, K.; Khuanchana, S.; Leklob, A.; Pongnikorn, D.; Rozek, L.S.; Siriarechakul, S.; et al. National and Subnational Population-Based Incidence of Cancer in Thailand: Assessing Cancers with the Highest Burdens. Cancers 2018, 9, 108. [Google Scholar] [CrossRef] [Green Version]
- Draborg, A.H.; Duus, K.; Houen, G. Epstein-Barr virus in systemic autoimmune diseases. Clin. Dev. Immunol. 2013, 2013, 535738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lünemann, A.; Rowe, M.; Nadal, D. Innate Immune Recognition of EBV. Epstein Barr Virus 2015, 2, 265–287. [Google Scholar]
- Frappier, L. The Epstein-Barr Virus EBNA1 Protein. Scientifica 2012, 2012, 438204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, P.D.; Hsieh, J.J.; Ruf, I.K.; Rawlins, D.R.; Hayward, S.D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 1994, 168, 5375–5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.M.; Lee, K.-H.; Farrell, C.J.; Ling, P.D.; Kempkes, B.; Park, J.H.; Hayward, S.D. EBNA2 is required for protection of latently Epstein-Barr virus-infected B cells against specific apoptotic stimuli. J. Virol. 2004, 78, 12694–12697. [Google Scholar] [CrossRef]
- Blanco, A.; Guix, S.; Fuster, N.; Fuentes, C.; Bartolomé, R.; Cornejo, T.; Pintó, R.M.; Bosch, A. Norovirus in Bottled Water Associated with Gastroenteritis Outbreak, Spain, 2016. Emerg. Infect. Dis. 2017, 23, 1531–1534. [Google Scholar] [CrossRef]
- Curutiu, C.; Iordache, F.; Gurban, P.; Lazar, V.; Chifiriuc, M.C. Main Microbiological Pollutants of Bottled Waters and Beverages. Bottled Packaged Water 2019, 403–422. [Google Scholar] [CrossRef]
- Hjalgrim, H.; Ekstrom-Smedby, K.; Rostgaard, K.; Amini, R.M.; Molin, D.; Hamilton-Dutoit, S.; Schollkopf, C.; Chang, E.T.; Ralfkiaer, E.; Adami, H.O.; et al. Cigarette smoking and risk of Hodgkin lymphoma: A population-based case-control study. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1561–1566. [Google Scholar] [CrossRef] [Green Version]
- Willett, E.V.; O’Connor, S.; Smith, A.G.; Roman, E. Does smoking or alcohol modify the risk of Epstein-Barr virus-positive or -negative Hodgkin lymphoma? Epidemiology 2007, 18, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Koriyama, C.; Matsuo, K.; Kim, W.H.; Herrera-Goepfert, R.; Liao, L.M.; Eurgast-EPIC Group; Yu, J.; Carrasquilla, G.; Sung, J.J.; et al. Case-case comparison of smoking and alcohol risk associations with Epstein-Barr virus-positive gastric cancer. Int. J. Cancer 2014, 134, 948–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Liang, K.Y.; Chang, Y.S.; Wang, Y.F.; Hsieh, T.; Hsu, M.M.; Chen, J.Y.; Liu, M.Y. Multiple risk factors of nasopharyngeal carcinoma: Epstein-Barr virus, malarial infection, cigarette smoking and familial tendency. Anticancer. Res. 1990, 10, 547–553. [Google Scholar] [PubMed]
- Ruan, H.-L.; Xu, F.-H.; Liu, W.-S.; Feng, Q.-S.; Chen, L.-Z.; Zeng, Y.-X.; Jia, W.-H. Alcohol and tea consumption in relation to the risk of nasopharyngeal carcinoma in Guangdong, China. Front. Med. China 2010, 4, 448–456. [Google Scholar] [CrossRef]
- Schwartz, S.M.; Doody, D.; Fitzgibbons, E.D.; Ricks, S.; Porter, P.L.; Chen, C. Oral squamous cell cancer risk in relation to alcohol consumption and alcohol dehydrogenase-3 genotypes. Cancer Epidemiol. Biomark. Prev. 2001, 10, 1137–1144. [Google Scholar]
- Wang, H.; Wang, X.; Xu, J.; Wang, L.; Liu, C. The role of cigarette smoking and alcohol consumption in the differentiation of oral squamous cell carcinoma for the males in China. J. Cancer Res. Ther. 2015, 11, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Bakkalci, D.; Jia, Y.; Winter, J.R.; Lewis, J.E.; Taylor, G.S.; Stagg, H.R. Risk factors for Epstein Barr virus-associated cancers: A systematic review, critical appraisal, and mapping of the epidemiological evidence. J. Glob. Health 2020, 10, 010405. [Google Scholar] [CrossRef] [PubMed]
- Hashim, N.A.N.; Ramzi, N.H.; Velapasamy, S.; Alex, L.; Chahil, J.K.; Lye, S.H.; Munretnam, K.; Haron, M.R.; Ler, L.W. Identification of Genetic and Non-genetic Risk Factors for Nasopharyngeal Carcinoma in a Southeast Asian Population. Asian Pac. J. Cancer Prev. 2012, 13, 6005–6010. [Google Scholar] [CrossRef]
- Yu, M.C.; Ho, J.H.; Lai, S.H.; Henderson, B.E. Cantonese-style salted fish as a cause of nasopharyngeal carcinoma: Report of a case-control study in Hong Kong. Cancer Res. 1986, 46, 956–961. [Google Scholar]
- Keegan, T.H.; Glaser, S.L.; Clarke, C.A.; Dorfman, R.F.; Mann, R.B.; DiGiuseppe, J.A.; Chang, E.T.; Ambinder, R.F. Body size, physical activity, and risk of Hodgkin’s lymphoma in women. Cancer Epidemiol. Biomark. Prev. 2016, 15, 1095–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.-F.; Liu, W.-S.; Qin, H.-D.; Xu, Y.-F.; Yu, D.-D.; Feng, Q.-S.; Chen, L.-Z.; Shu, X.-O.; Zeng, Y.-X.; Jia, W.-H. Effect of family history of cancers and environmental factors on risk of nasopharyngeal carcinoma in Guangdong, China. Cancer Epidemiol. 2010, 34, 419–424. [Google Scholar] [CrossRef] [PubMed]

| Property | Standard * | Mae Ka (No. 1) | Wiang | Unit | |
|---|---|---|---|---|---|
| Physical | Color | 15 | 0.480 | 0.700 | Pt.Co unit |
| Turbidity | 5 | 17.235 | 1.575 | NTU | |
| pH | 6.5–8.5 | 7.97 | 7.63 | ||
| Chemical | Total dissolved solids | 1000 | 205.00 | 135.75 | mg/L |
| Fe | 0.3 | 0.330 | 0.062 | mg/L | |
| Mn | 0.1 | 0.135 | 0.030 | mg/L | |
| Cu | 2.0 | 0.010 | 0.003 | mg/L | |
| Zn | 3.0 | 0.018 | 0.010 | mg/L | |
| Total Hardness (CaCO3) | 300 | 99.5 | 71.0 | mg/L | |
| SO42− | 6.525 | 12.695 | mg/L | ||
| Cl− | 250 | Not detected | 16.715 | mg/L | |
| F− | 1.5 | 0.24 | 0.34 | mg/L | |
| NO3− | 50 | 1.77 | Not detected | mg/L | |
| NO2− | 3 | Not detected | Not detected | mg/L | |
| Microbiological | Total coliform bacteria | - | 23.0 | 1.1 | MPN-100 mL |
| Escherichia coli | - | Detected | Not detected | per 100 mL | |
| Chemical Poison | Hg | 1 | Not detected | Not detected | mg/L |
| Pb | 10 | 0.001 | Not detected | mg/L | |
| As | 10 | 0.002 | 0.001 | mg/L | |
| Cr | 50 | Not detected | Not detected | mg/L | |
| Cd | 3 | Not detected | Not detected | mg/L |
| Demographical Factor | All Three Methods (%) | p-Value | |
|---|---|---|---|
| Positive | Negative | ||
| Sex | |||
| Male | 17 (7.36) | 214 (92.64) | 0.944 |
| Female | 42 (7.22) | 540 (92.78) | |
| Age (years) | |||
| Mean | 44.37 | 52.61 | 0.000 |
| SD | 16.05 | 18.71 | |
| Age Groups (years) | |||
| 1–10 | 0 (0) | 21 (100) | |
| 11–20 | 7 (12.28) | 50 (87.72) | |
| 21–30 | 6 (11.54) | 46 (88.46) | |
| 31–40 | 5 (9.62) | 47 (90.38) | |
| 41–50 | 20 (16.81) | 99 (83.19) | |
| 51–60 | 15 (7.54) | 184 (92.46) | |
| 61–70 | 2 (0.93) | 214 (99.07) | |
| 71–80 | 4 (5.19) | 73 (94.81) | |
| 81–90 | 0 (0) | 20 (100) | |
| Congenital Disease | |||
| Yes | 23 (5.96) | 363 (94.04) | 0.175 |
| No | 36 (8.43) | 391 (91.57) | |
| Family History of Cancer | |||
| Yes | 16 (8.70) | 168 (91.30) | 0.392 |
| No | 43 (6.84) | 586 (93.16) | |
| Exercise | |||
| Yes | 48 (7.35) | 605 (92.65) | 0.835 |
| No | 11 (6.87) | 149 (93.13) | |
| Boiled/Filtered Drinking Water | |||
| Yes | 41 (7.65) | 495 (92.35) | 0.549 |
| No | 18 (6.50) | 259 (93.50) | |
| Bottled Drinking Water | |||
| No | 8 (12.70) | 55 (87.30) | 0.083 |
| Yes | 51 (6.80) | 699 (93.20) | |
| Cleaning of Water for Consumption | |||
| Yes | 46 (6.73) | 638 (93.27) | 0.178 |
| No | 13 (10.08) | 116 (89.92) | |
| Tap Water Used for Brushing Teeth | |||
| No a | 14 (12.17) | 101 (87.83) | 0.028 |
| Yes | 45 (6.45) | 653 (93.55) | |
| Alcohol Consumption | |||
| Yes | 35 (11.40) | 272 (88.60) | 0.000 |
| No | 24 (4.74) | 482 (95.26) | |
| Serving Spoon | |||
| No | 13 (10.16) | 115 (89.84) | 0.168 |
| Yes | 46 (6.72) | 639 (93.28) | |
| Smoking Status | |||
| Yes | 13 (10.74) | 108 (89.26) | 0.109 |
| No | 46 (6.65) | 646 (93.35) | |
| Secondhand Smoke Status | |||
| Yes | 20 (11.76) | 150 (88.24) | 0.011 |
| No | 39 (6.07) | 604 (93.93) | |
| Eat Fresh Fruit (Per Week) | |||
| No | 6 (19.35) | 25 (80.65) | 0.019 |
| 1–2 times | 8 (9.64) | 75 (90.36) | |
| 3–4 times | 17 (8.50) | 183 (91.50) | |
| 5–7 times | 28 (5.61) | 471 (94.39) | |
| Eat Vegetables (per Week) | |||
| No | 3 (8.57) | 32 (91.43) | 0.236 |
| 1–2 times | 6 (10.17) | 53 (89.83) | |
| 3–4 times | 15 (10.49) | 128 (89.51) | |
| 5–7 times | 35 (6.08) | 541 (93.92) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pongpakdeesakul, S.; Ekalaksananan, T.; Pientong, C.; Iamchuen, N.; Buddhisa, S.; Mahingsa, K.; Pingyod, A.; Sangsrijun, W.; Passorn, S.; Chopjitt, P.; et al. Human Oncogenic Epstein–Barr Virus in Water and Human Blood Infection of Communities in Phayao Province, Thailand. Water 2023, 15, 323. https://doi.org/10.3390/w15020323
Pongpakdeesakul S, Ekalaksananan T, Pientong C, Iamchuen N, Buddhisa S, Mahingsa K, Pingyod A, Sangsrijun W, Passorn S, Chopjitt P, et al. Human Oncogenic Epstein–Barr Virus in Water and Human Blood Infection of Communities in Phayao Province, Thailand. Water. 2023; 15(2):323. https://doi.org/10.3390/w15020323
Chicago/Turabian StylePongpakdeesakul, Sutida, Tipaya Ekalaksananan, Chamsai Pientong, Niti Iamchuen, Surachat Buddhisa, Khwanruedee Mahingsa, Arunee Pingyod, Wanwipa Sangsrijun, Supaporn Passorn, Peechanika Chopjitt, and et al. 2023. "Human Oncogenic Epstein–Barr Virus in Water and Human Blood Infection of Communities in Phayao Province, Thailand" Water 15, no. 2: 323. https://doi.org/10.3390/w15020323
APA StylePongpakdeesakul, S., Ekalaksananan, T., Pientong, C., Iamchuen, N., Buddhisa, S., Mahingsa, K., Pingyod, A., Sangsrijun, W., Passorn, S., Chopjitt, P., Duangjit, S., & Bumrungthai, S. (2023). Human Oncogenic Epstein–Barr Virus in Water and Human Blood Infection of Communities in Phayao Province, Thailand. Water, 15(2), 323. https://doi.org/10.3390/w15020323






