Past and Present Environmental Factors Differentially Influence Genetic and Morphological Traits of Italian Barbels (Pisces: Cyprinidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Barbel Samples
2.2. Genetic Data
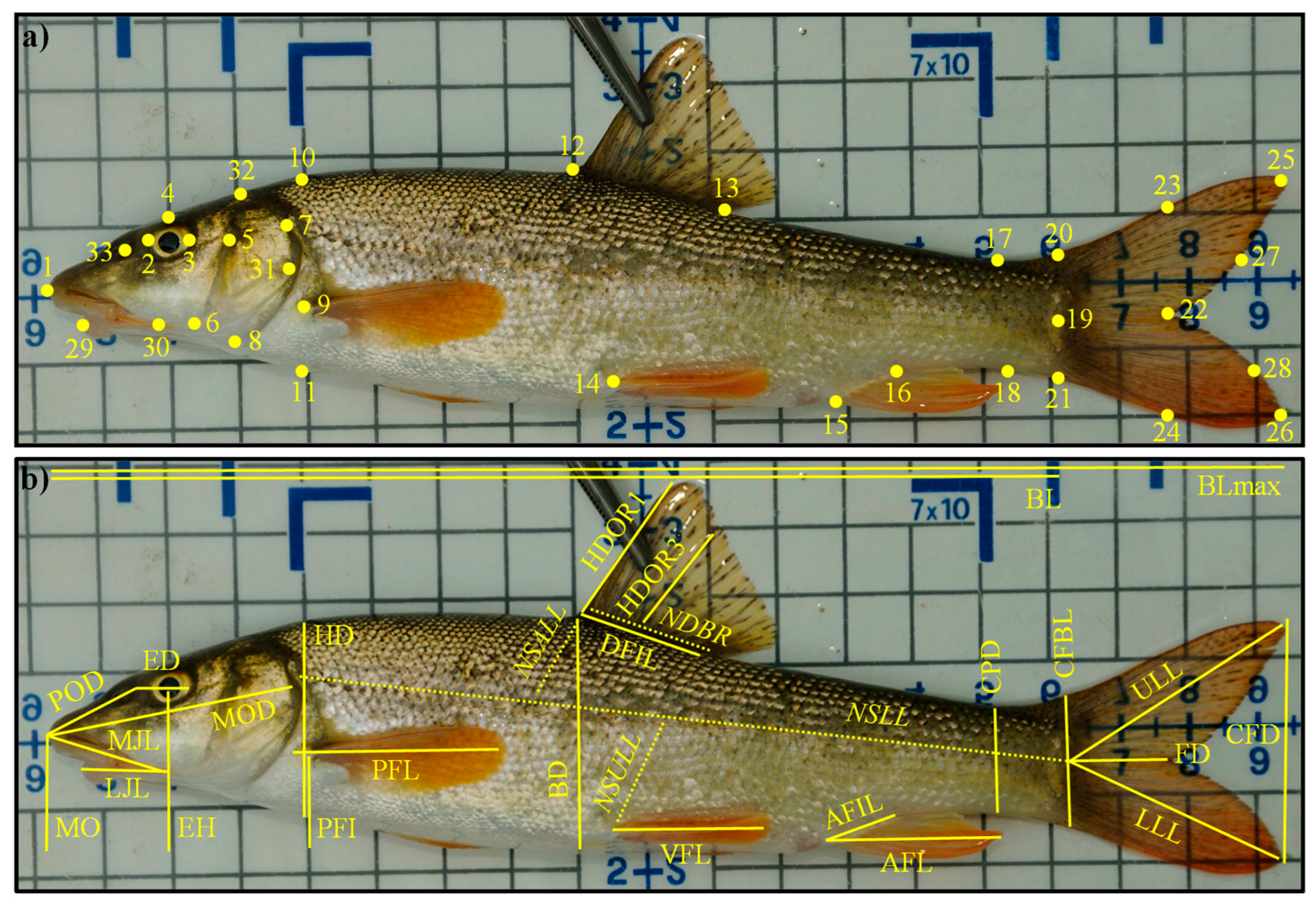
2.3. Morphological Data
2.4. Environmental Data
2.5. Data Analysis
2.5.1. Genetic Data
2.5.2. Morphological Data
2.5.3. Environmental Data
3. Results
3.1. Genetics of Barbel Populations along a Latitudinal Gradient
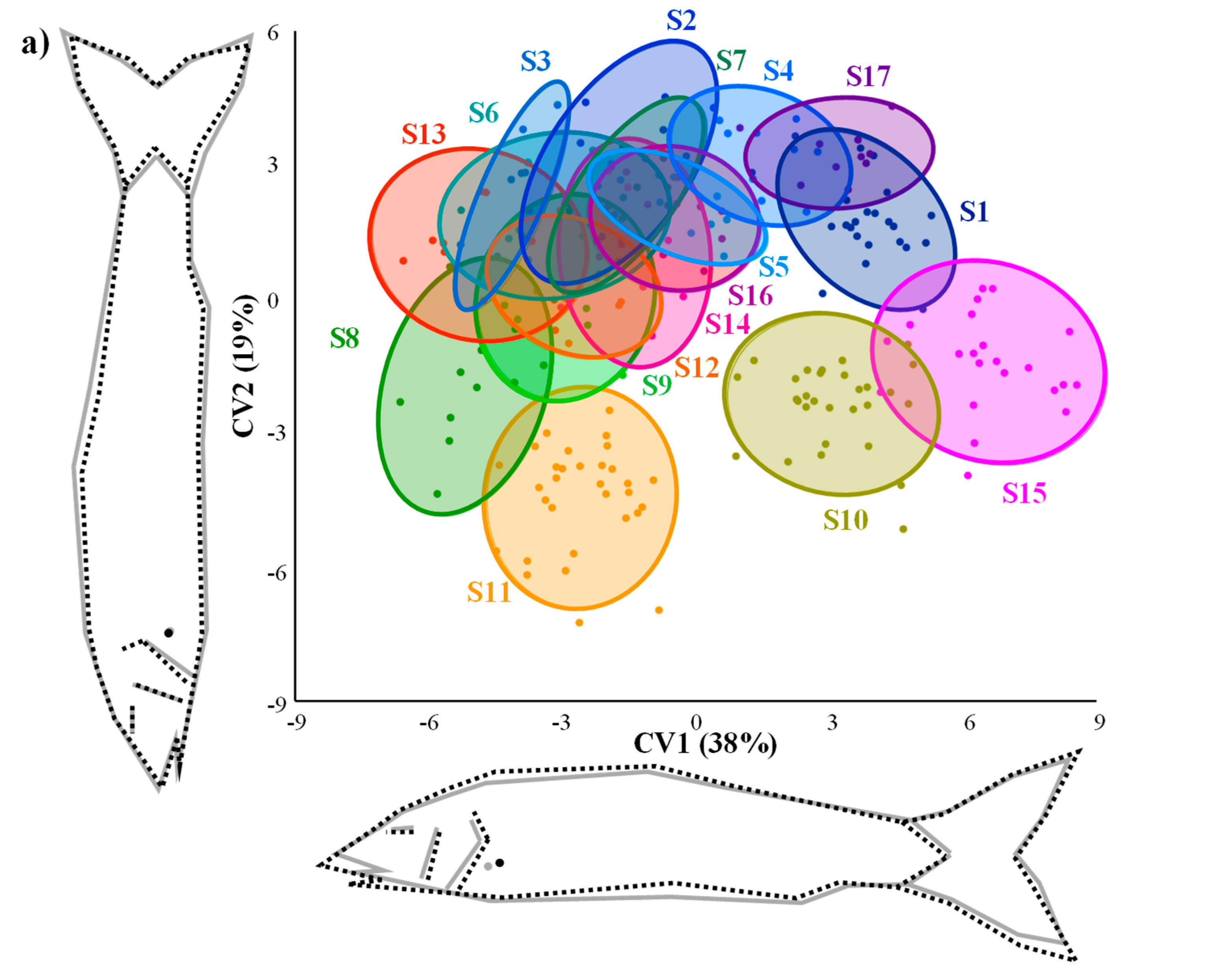
3.2. Morphology of Barbel Populations along a Latitudinal Gradient
3.3. Environmental Factors Influencing Barbel Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sagnes, P.; Statzner, B. Hydrodynamic abilities of riverine fish: A functional link between morphology and velocity use. Aquat. Living Resour. 2009, 22, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Franssen, N.R.; Stewart, L.K.; Schaefer, J.F. Morphological divergence and flow induced phenotypic plasticity in a native fish from anthropogenically altered stream habitats. Ecol. Evol. 2013, 3, 4648–4657. [Google Scholar] [CrossRef] [PubMed]
- Samways, K.M.; Leavitt, P.R.; Magnan, P.; Rodríguez, M.A.; Peres-Neto, P.R. Convergent polymorphism between stream and lake habitats: The case of brook char. Can. J. Fishe. Aquat. Sci. 2015, 72, 1406–1414. [Google Scholar] [CrossRef]
- Thacker, C.E.; Gkenas, C. Morphometric convergence among European sand gobies in freshwater (Gobiiformes: Gobionellidae). Ecol. Evol. 2019, 9, 8087–8103. [Google Scholar] [CrossRef] [PubMed]
- Van Valen, L. Morphological variation and width of ecological niche. Am. Nat. 1965, 99, 377–390. [Google Scholar] [CrossRef]
- Bolnick, D.; Svanbäck, R.; Araújo, M.; Persson, L. Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proc. Natl. Acad. Sci. USA 2007, 104, 10075–10079. [Google Scholar] [CrossRef] [Green Version]
- Snowberg, L.K.; Hendrix, K.M.; Bolnick, D.I. Covarying variances: More morphologically variable populations also exhibit more diet variation. Oecologia 2015, 178, 89–101. [Google Scholar] [CrossRef]
- Li, W.; Zhai, D.; Wang, C.; Gao, X.; Liu, H.; Cao, W. Relationships Among Trophic Niche Width, Morphological Variation, and Genetic Diversity of Hemiculter leucisculus in China. Front. Ecol. Evol. 2021, 9, 691218. [Google Scholar] [CrossRef]
- Yedier, S.; Bostanci, D. Intra-and Inter-species Discriminations: Scorpaena species from the Black Sea, Aegean Sea, Sea of Marmara and Mediterranean Sea. Sci. Mar. 2021, 85, 197–209. [Google Scholar] [CrossRef]
- Scheiner, S.M. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 1993, 24, 35–68. [Google Scholar] [CrossRef]
- Price, T.D.; Qvarnstrom, A.; Irwin, D.E. The role of phenotypic plasticity in driving genetic evolution. Proc. Biol. Sci. 2003, 270, 1433–1440. [Google Scholar] [CrossRef] [Green Version]
- Bower, L.M.; Saenz, D.E.; Winemiller, K.O. Widespread convergence in stream fishes. Biol. J. Linn. Soc. 2021, 133, 863–879. [Google Scholar] [CrossRef]
- Kirk, M.A.; Rahel, F.J.; Laughlin, D.C. Environmental filters of freshwater fish community assembly along elevation and latitudinal gradients. Glob. Ecol. Biogeogr. 2022, 31, 470–485. [Google Scholar] [CrossRef]
- Bower, L.M.; Winemiller, K.O. Fish assemblage convergence along stream environmental gradients: An intercontinental analysis. Ecography 2019, 42, 1691–1702. [Google Scholar] [CrossRef] [Green Version]
- Higham, T.E.; Ferry, L.A.; Schmitz, L.; Irschick, D.J.; Starko, S.; Anderson, P.S.; Bergmann, P.J.; Jamniczky, H.A.; Monteiro, L.R.; Navon, D.; et al. Linking ecomechanical models and functional traits to understand phenotypic diversity. Trends Ecol. Evol. 2021, 36, 860–873. [Google Scholar] [CrossRef]
- Viganò, G.; Confortola, G.; Fornaroli, R.; Cabrini, R.; Canobbio, S.; Mezzanotte, V.; Bocchiola, D. Effects of future climate change on a river habitat in an Italian alpine catchment. J. Hydrol. Eng. 2016, 21, 04015063. [Google Scholar] [CrossRef]
- Fuso, F.; Stucchi, L.; Bonacina, L.; Fornaroli, R.; Bocchiola, D. Evaluation of water temperature under changing climate and its effect on river habitat in a regulated Alpine catchment. J. Hydrol. 2022, 616, 128816. [Google Scholar] [CrossRef]
- Mantua, N.; Tohver, I.; Hamlet, A. Climate change impacts on streamflow extremes and summertime stream temperature and their possible consequences for freshwater salmon habitat in Washington State. Clim. Chang. 2010, 102, 187–223. [Google Scholar] [CrossRef]
- Isaak, D.J.; Wollrab, S.; Horan, D.; Chandler, G. Climate change effects on stream and river temperatures across the northwest US from 1980-2009 and implications for salmonid fishes. Clim. Chang. 2012, 113, 499–524. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.; Laizé, C.L.R.; Acreman, M.C.; Florke, M. How will climate change modify river flow regimes in Europe? Hydrol. Earth Syst. Sci. 2013, 17, 325–339. [Google Scholar] [CrossRef]
- van Vliet, M.T.; Franssen, W.H.; Yearsley, J.R.; Ludwig, F.; Haddeland, I.; Lettenmaier, D.P.; Kabat, P. Global river discharge and water temperature under climate change. Glob. Environ. Chang. 2013, 23, 450–464. [Google Scholar] [CrossRef]
- Ye, F.; Kameyama, S. Long-term nationwide spatiotemporal changes of freshwater temperature in Japan during 1982–2016. J. Environ. Manage. 2021, 281, 111866. [Google Scholar] [CrossRef] [PubMed]
- Benejam, L.; Alcaraz, C.; Sasal, P.; Simon-Levert, G.; García-Berthou, E. Life history and parasites of the invasive mosquitofish (Gambusia holbrooki) along a latitudinal gradient. Biol. Invasions 2009, 11, 2265–2277. [Google Scholar] [CrossRef]
- Parra, I.; Nicola, G.G.; Vøllestad, L.A.; Elvira, B.; Almodóvar, A. Latitude and altitude differentially shape life history trajectories between the sexes in non-anadromous brown trout. Evol. Ecol. 2014, 28, 707–720. [Google Scholar] [CrossRef]
- Riesch, R.; Martin, R.A.; Diamond, S.E.; Jourdan, J.; Plath, M.; Brian Langerhans, R. Thermal regime drives a latitudinal gradient in morphology and life history in a livebearing fish. Biol. J. Linn. Soc. 2018, 125, 126–141. [Google Scholar] [CrossRef] [Green Version]
- Rohde, K. Latitudinal gradients in species diversity: The search for the primary cause. Oikos 1992, 65, 514–527. [Google Scholar] [CrossRef] [Green Version]
- Barlow, G.W. Causes and significance of morphological variation in fishes. Syst. Zool. 1961, 10, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Osborne, M.J.; Perkin, J.S.; Gido, K.B.; Turner, T.F. Comparative riverscape genetics reveals reservoirs of genetic diversity for conservation and restoration of Great Plains fishes. Mol. Ecol. 2014, 23, 5663–5679. [Google Scholar] [CrossRef] [Green Version]
- De Santis, V.; Quadroni, S.; Britton, R.J.; Carosi, A.; Gutmann Roberts, C.; Lorenzoni, M.; Crosa, G.; Zaccara, S. Biological and trophic consequences of genetic introgression between endemic and invasive Barbus fishes. Biol. Invasions 2021, 23, 3351–3368. [Google Scholar] [CrossRef]
- Prunier, J.G.; Dubut, V.; Loot, G.; Tudesque, L.; Blanchet, S. The relative contribution of river network structure and anthropogenic stressors to spatial patterns of genetic diversity in two freshwater fishes: A multiple-stressors approach. Freshwater Biol. 2018, 63, 6–21. [Google Scholar] [CrossRef]
- Radojković, N.; Marinović, Z.; Milošković, A.; Radenković, M.; Đuretanović, S.; Lujić, J.; Simić, V. Effects of stream damming on morphological variability of fish: Case study on large spot barbel Barbus balcanicus. Turk. J. Fish. Aquat. Sci. 2018, 19, 231–239. [Google Scholar] [CrossRef]
- Machado, C.B.; Braga-Silva, A.; Freitas, P.D.; Galetti Jr, P.M. Damming shapes genetic patterns and may affect the persistence of freshwater fish populations. Freshwater Biol. 2022, 67, 603–618. [Google Scholar] [CrossRef]
- Berbel-Filho, W.M.; Martinez, P.A.; Ramos, T.; Torres, R.A.; Lima, S.M. Inter-and intra-basin phenotypic variation in two riverine cichlids from northeastern Brazil: Potential eco-evolutionary damages of Sao Francisco interbasin water transfer. Hydrobiologia 2016, 766, 43–56. [Google Scholar] [CrossRef]
- Schmitt, T.; Fritz, U.; Delfino, M.; Ulrich, W.; Habel, J.C. Biogeography of Italy revisited: Genetic lineages confirm major phylogeographic patterns and a pre-Pleistocene origin of its biota. Front. Zool. 2021, 18, 34. [Google Scholar] [CrossRef]
- Gumiero, B.; Maiolini, B.; Comiti, F.; Moroni, F. The Italian Rivers. In Rivers of Europe, 2nd ed.; Tockner, K., Zarfl, C., Robinson, C.T., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 657–685. [Google Scholar] [CrossRef]
- Skoulikidis, N.T.; Sabater, S.; Datry, T.; Morais, M.M.; Buffagni, A.; Dörflinger, G.; Zogaris, S.; Sánchez-Montoya, M.D.M.; Bonada, N.; Kalogianni, E.; et al. Non-perennial Mediterranean rivers in Europe: Status, pressures, and challenges for research and management. Sci. Total Environ. 2017, 577, 1–18. [Google Scholar] [CrossRef]
- Darvini, G.; Memmola, F. Assessment of the impact of climate variability and human activities on the runoff in five catchments of the Adriatic Coast of south-central Italy. J. Hydrol. Reg. Stud. 2020, 31, 100712. [Google Scholar] [CrossRef]
- Lorenzoni, M.; Carosi, A.; Quadroni, S.; De Santis, V.; Vanetti, I.; Delmastro, G.B.; Zaccara, S. Cryptic diversity within endemic Italian barbels: Revalidation and description of new Barbus species (Teleostei: Cyprinidae). J. Fish Biol. 2021, 98, 1433–1449. [Google Scholar] [CrossRef]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Publications Kottelat: Cornol, Switzerland; Berlin, Germany, 2007. [Google Scholar]
- Buonerba, L.; Zaccara, S.; Delmastro, G.B.; Lorenzoni, M.; Salzburger, W.; Gante, H.F. Intrinsic and extrinsic factors act at different spatial and temporal scales to shape population structure, distribution and speciation in Italian Barbus (Osteichthyes: Cyprinidae). Mol. Phylogenet. Evol. 2015, 89, 115–129. [Google Scholar] [CrossRef]
- Zaccara, S.; Quadroni, S.; De Santis, V.; Vanetti, I.; Carosi, A.; Britton, R.; Lorenzoni, M. Genetic and morphological analysis reveal a complex biogeographic pattern in the endemic barbel populations of the southern Italian peninsula. Ecol. Evol. 2019, 9, 10185–10197. [Google Scholar] [CrossRef]
- Bianco, P.G. Potential role of the palaeohistory of the Mediterranean and Paratethys basins on the early dispersal of Euro-Mediterranean freshwater fishes. Ichthyol. Explor. Fres. 1990, 1, 167–184. [Google Scholar]
- Livi, S.; De Innocentiis, S.; Longobardi, A.; Cataudella, S.; Tancioni, L.; Rampacci, M.; Marino, G. Genetic structure of Barbus spp. populations in the Marche Region of central Italy and its relevance to conservation actions. J. Fish Biol. 2013, 82, 806–826. [Google Scholar] [CrossRef] [PubMed]
- Geiger, M.F.; Schreiner, C.; Delmastro, G.B.; Herder, F. Combining geometric morphometrics with molecular genetics to investigate a putative hybrid complex: A case study with barbels Barbus spp. (Teleostei: Cyprinidae). J. Fish Biol. 2016, 88, 1038–1055. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Quadroni, S.; Vanetti, I.; Carosi, A.; La Porta, G.; Crosa, G.; Britton, R.; Lorenzoni, M. Morphologic and genetic variability in the Barbus fishes (Teleostei, Cyprinidae) of Central Italy. Zool. Scr. 2019, 48, 289–301. [Google Scholar] [CrossRef]
- Bianco, P.G. A revision of the Italian Barbus species (Cypriniformes: Cyprinidae). Ichthyol. Explor. Fres. 1995, 6, 305–324. [Google Scholar]
- Lorenzoni, M.; Carosi, A.; Angeli, V.; Bicchi, A.; Pedicillo, G.; Viali, P. Individuazione e Riconoscimento dei Barbi Autoctoni nel Bacino del Fiume Paglia. Provincia di Terni-Assessorato alla Programmazione Faunistica; Arti grafiche Tiezzi: Terni, Italy, 2006. [Google Scholar]
- Meraner, A.; Venturi, A.; Ficetola, G.F.; Rossi, S.; Candiotto, A.; Gandolfi, A. Massive invasion of alien Barbus barbus and introgressive hybridization with endemic Barbus plebejus in Northern Italy: Where, how and why? Mol. Ecol. 2013, 22, 5295–5312. [Google Scholar] [CrossRef]
- Carosi, A.; Ghetti, L.; La Porta, G.; Lorenzoni, M. Ecological effects of the European barbel Barbus barbus (L., 1758) (Cyprinidae) invasion on native barbel populations in the Tiber River basin (Italy). Eur. Zool. J. 2017, 84, 420–435. [Google Scholar] [CrossRef] [Green Version]
- Piccoli, F.; Burgazzi, G.; Laini, A.; Ferrari, C.; Voccia, A.; Filonzi, L.; Bolpagni, R.; Nonnis Marzano, F. Barbel species arrangement in a regional Natura 2000 network (Emilia Romagna, northern Italy): An altitudinal perspective. J. Limnol. 2017, 76, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Zaccara, S.; Quadroni, S.; De Santis, V.; Vanetti, I.; Carosi, A.; Crosa, G.; Britton, R.J.; Lorenzoni, M. Genetic and phenotypic displacement of an endemic Barbus complex by invasive European barbel Barbus barbus in Central Italy. Biol. Invasions 2020, 23, 521–535. [Google Scholar] [CrossRef]
- Bagenal, T. Methods for Assessment of Fish Production in Fresh Waters; Blackwell Scientific Publications: Oxford, UK, 1978. [Google Scholar]
- Vilizzi, L.; Copp, G.H.; Britton, J.R. Age and growth of European barbel Barbus barbus (Cyprinidae) in the small, mesotrophic River Lee and relative to other populations in England. Knowl. Managt. Aquatic Ecosyst. 2013, 409, 9. [Google Scholar] [CrossRef] [Green Version]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Briolay, J.; Galtier, N.; Brito, R.M.; Bouvet, Y. Molecular phylogeny of Cyprinidae inferred from cytochrome b DNA sequences. Mol. Phylogenet. Evol. 1998, 9, 100–108. [Google Scholar] [CrossRef]
- Rossi, G.; Zuffi, G.; Gandolfi, G.; Marchi, A.; Rinaldi, M.; Valli, M. Analisi della Distribuzione delle Specie del Genere Barbus Cuvier, 1871 nei Bacini Idrografici della Regione Abruzzo; Dipartimento di Scienze Biologiche, Geologiche e Ambientali dell’Università di Bologna: Bologna, Italy, 2013. [Google Scholar]
- Antognazza, C.M.; Andreou, D.; Zaccara, S.; Britton, R.J. Loss of genetic integrity and biological invasions result from stocking and introductions of Barbus barbus: Insights from rivers in England. Ecol. Evol. 2016, 6, 1280–1292. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 6 May 2021).
- Adams, D.C.; Collyer, M.L.; Kaliontzopoulou, A.; Geomorph: Software for Geometric Morphometric Analysis. R Package Version 3.0.6. 2018. Available online: https://cran.r-project.org/package=geomorph (accessed on 6 May 2021).
- Beacham, T.D. Meristic and morphometric variation in pink salmon (Oncorhynchus gorbuscha) in southern British Columbia and Puget Sound. Can. J. Zool. 1985, 63, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Xiang, T.; Dong, X.; Grenouillet, G. Ecological and biological traits of non-native freshwater fish species differentiate them from native species in China. Ecol. Indic. 2021, 131, 108218. [Google Scholar] [CrossRef]
- Olden, J.D.; Jackson, D.A.; Peres-Neto, P.R. Predictive models of fish species distributions: A note on proper validation and chance predictions. T. Am. Fish. Soc. 2002, 131, 329–336. [Google Scholar] [CrossRef]
- Ha, L.M.; Iguchi, K.I. Geographical continuity and discontinuity in the meristic characteristics of ayus of the southern subspecies Plecoglossus altivelis ryukyuensis. Ichthyol. Res. 2021, 68, 177–181. [Google Scholar] [CrossRef]
- Toussaint, A.; Charpin, N.; Brosse, S.; Villéger, S. Global functional diversity of freshwater fish is concentrated in the Neotropics while functional vulnerability is widespread. Sci. Rep. 2016, 6, 22125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reecht, Y.; Rochet, M.-J.; Trenkel, V.M.; Jennings, S.; Pinnegar, J.K. Use of morphological characteristics to define functional groups of predatory fishes in the Celtic Sea. J. Fish Biol. 2013, 83, 355–377. [Google Scholar] [CrossRef] [Green Version]
- Winemiller, K.O. Ecomorphological diversification in lowland freshwater fish assemblages from five biotic regions. Ecol. Monogr. 1991, 61, 343–365. [Google Scholar] [CrossRef]
- Boyle, K.S.; Horn, M.H. Comparison of feeding guild structure and ecomorphology of intertidal fish assemblages from central California and central Chile. Mar. Ecol. Prog. Ser. 2006, 319, 65–84. [Google Scholar] [CrossRef]
- Dumay, O.; Tari, P.S.; Tomasini, J.A.; Mouillot, D. Functional groups of lagoon fish species in Languedoc Roussillon, southern France. J. Fish Biol. 2004, 64, 970–983. [Google Scholar] [CrossRef]
- Lefcheck, J.S.; Buchheister, A.; Laumann, K.M.; Stratton, M.A.; Sobocinski, K.L.; Chak, S.T.C.; Clardy, T.R.; Reynolds, P.L.; Latour, R.J.; Duffy, J.E. Dimensions of biodiversity in Chesapeake Bay demersal fishes: Patterns and drivers through space and time. Ecosphere 2014, 5, art14. [Google Scholar] [CrossRef]
- Fulton, C.J.; Bellwood, D.R.; Wainwright, P.C. The relationship between swimming ability and habitat use in wrasses (Labridae). Mar. Biol. 2001, 139, 25–33. [Google Scholar] [CrossRef]
- Webb, P.W. Body form, locomotion and foraging in aquatic vertebrates. Am. Zool. 1984, 24, 107–120. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. QGIS Association. 2022. Available online: http://www.qgis.or (accessed on 4 June 2021).
- Available online: http://wms.pcn.minambiente.it/ogc?map=/ms_ogc/WMS_v1.3/Vettoriali/Carta_geolitologica.map (accessed on 4 June 2021).
- Available online: http://wms.pcn.minambiente.it/ogc?map=/ms_ogc/WMS_v1.3/Vettoriali/Corine_Land_Cover2012.map (accessed on 4 June 2021).
- Available online: http://www.scia.isprambiente.it (accessed on 4 October 2021).
- DECRETO 8 Novembre 2010, n. 260. Regolamento Recante i Criteri Tecnici per la Classificazione dello Stato dei Corpi Idrici Superficiali, per la Modifica delle Norme Tecniche del Decreto Legislativo 3 Aprile 2006, n. 152, Recante Norme in Materia Ambientale, Predisposto ai Sensi dell’articolo 75, Comma 3, del Medesimo Decreto Legislativo. (11G0035) (GU Serie Generale n.30 del 07-02-2011-Suppl. Ordinario n. 31). Available online: https://www.gazzettaufficiale.it/eli/id/2011/02/07/011G0035/sg (accessed on 9 July 2021).
- Buffagni, A.; Erba, S. Intercalibrazione e classificazione di qualità ecologica dei fiumi per la 2000/60/EC (WFD): l’indice STAR-ICMi. IRSA-CNR Not. Dei Metod. Anal. 2007, 1, 94–100. [Google Scholar]
- Available online: https://www.arpa.marche.it/images/PUBBLICAZIONI/BACINI_2015-2017/Relazione%20fiumi%202015-2017_senza_accorp_lug2019_revfeb2020.pdf (accessed on 6 October 2021).
- Available online: https://www.artaabruzzo.it/acque-superficiali.php?id_page=1 (accessed on 6 October 2021).
- Available online: https://www.arpamolise.it/Acque/Super/rapporto_2018.pdf (accessed on 6 October 2021).
- Available online: https://www.arpa.puglia.it/pagina3381_stato-ecologico-delle-acque-superficiali-interne.html (accessed on 6 October 2021).
- Available online:. Available online: https://www.regione.marche.it/Regione-Utile/Ambiente/Tutela-delle-acque/PTA#Documentazione (accessed on 8 October 2021).
- Available online: https://www.regione.abruzzo.it/content/piano-tutela-delle-acque (accessed on 8 October 2021).
- Available online: https://www.regione.molise.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/13780 (accessed on 8 October 2021).
- Available online: http://www.adb.basilicata.it/adb/pStralcio/distretto_bas.asp (accessed on 8 October 2021).
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Data from: PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef]
- Won, H.; Jeon, H.B.; Suk, H.Y. Evidence of an ancient connectivity and biogeodispersal of a bitterling species, Rhodeus notatus, across the Korean Peninsula. Sci. Rep. 2020, 10, 1011. [Google Scholar] [CrossRef] [Green Version]
- Gido, K.B.; Whitney, J.E.; Perkin, J.S.; Turner, T.F. Fragmentation, connectivity and fish species persistence in freshwater ecosystems. In Conservation of Freshwater Fishes; Closs, G.P., Krkosek, M., Olden, J.D., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 292–323. [Google Scholar]
- Jordan, D.S. The Days of a Man: Being Memories of a Naturalist, Teacher, and Minor Prophet of Democracy (Vol. 2); World Book Company: Chicago, IL, USA, 1922. [Google Scholar]
- Hubbs, C.L. Variations in the number of vertebrae and other meristic characters of fishes correlated with the temperature of water during development. Am. Nat. 1922, 56, 360–372. [Google Scholar] [CrossRef] [Green Version]
- McDowall, R. Jordan’s and other ecogeographical rules, and the vertebral number in fishes. J. Biogeogr. 2008, 35, 501–508. [Google Scholar] [CrossRef]
- Cal, L.; Suarez-Bregua, P.; Cerdá-Reverter, J.M.; Braasch, I.; Rotllant, J. Fish pigmentation and the melanocortin system. Comp. Biochem. Phys. A 2017, 211, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Nüsslein-Volhard, C.; Singh, A.P. How fish color their skin: A paradigm for development and evolution of adult patterns: Multipotency, plasticity, and cell competition regulate proliferation and spreading of pigment cells in zebrafish coloration. BioEssays 2017, 39, 1600231. [Google Scholar] [CrossRef] [Green Version]
- Mizusawa, K.; Yamamura, Y.; Kasagi, S.; Cerdá-Reverter, J.M.; Takahashi, A. Expression of genes for melanotropic peptides and their receptors for morphological color change in goldfish Carassius auratus. Gen. Comp. Endocr. 2018, 264, 138–150. [Google Scholar] [CrossRef]
- Luo, M.; Lu, G.; Yin, H.; Wang, L.; Atuganile, M.; Dong, Z. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquacult. 2021, 13, 2395–2412. [Google Scholar] [CrossRef]
- Chavin, W. Fundamental aspects of morphological melanin color changes in vertebrate skin. Am. Zool. 1969, 9, 505–520. [Google Scholar] [CrossRef]
- Sugimoto, M. Morphological color changes in fish: Regulation of pigment cell density and morphology. Microsc. Res. Techniq. 2002, 58, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Henning, F.; Jones, J.C.; Franchini, P.; Meyer, A. Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genom. 2013, 14, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashidian, G.; Rainis, S.; Prokić, M.D.; Faggio, C. Effects of different levels of carotenoids and light sources on swordtail fish (Xiphophorus helleri) growth, survival rate and reproductive parameters. Nat. Prod. Res. 2020, 35, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luo, M.; Yin, H.; Zhu, W.; Fu, J.; Dong, Z. Effects of background adaptation on the skin color of Malaysian red tilapia. Aquaculture 2020, 521, 735061. [Google Scholar] [CrossRef]
- Aspengren, S.; Sköld, H.; Wallin, M. Different strategies for color change. Cell. Mol. Life Sci. 2009, 66, 187–191. [Google Scholar] [CrossRef]
- Donnelly, W.A.; Dill, L.M. Evidence for crypsis in coho salmon, Oncorhynchus kisutch (Walbaum), parr: Substrate colour preference and achromatic reflectance. J. Fish Biol. 1984, 25, 183–195. [Google Scholar] [CrossRef]
- Atkinson, D. Temperature and organism size –a biological law for ectotherms. Adv. Ecol. Res. 1994, 25, 1–58. [Google Scholar] [CrossRef]
- Wootton, H.F.; Morrongiello, J.R.; Schmitt, T.; Audzijonyte, A. Smaller adult fish size in warmer water is not explained by elevated metabolism. Ecol. Lett. 2022, 25, 1177–1188. [Google Scholar] [CrossRef]
- Rypel, A.L. The cold-water connection: Bergmann’s rule in north American freshwater fishes. Am. Nat. 2014, 183, 147–156. [Google Scholar] [CrossRef]
- Blanck, A.; Lamouroux, N. Large-scale intraspecific variation in life-history traits of European freshwater fish. J. Biogeogr. 2007, 34, 862–875. [Google Scholar] [CrossRef]
- Crozier, L.G.; Hutchings, J.A. Plastic and evolutionary responses to climate change in fish. Evol. Appl. 2014, 7, 68–87. [Google Scholar] [CrossRef]
- Rahel, F.J.; Hubert, W.A. Fish assemblages and habitat gradients in a Rocky Mountain-Great Plains stream: Biotic zonation and additive patterns of community change. Trans. Am. Fish. Soc. 1991, 120, 319–332. [Google Scholar] [CrossRef]
- Olinger, C.T.; Peoples, B.K.; Frimpong, E.A. Reproductive life history of Heterandria bimaculata (Heckel, 1848) (Poeciliinae: Poeciliidae) in the Honduran interior highlands: Trait variation along an elevational gradient. Neotrop. Ichthyol. 2016, 14, e150050. [Google Scholar] [CrossRef] [Green Version]
- Michel, M.J.; Chien, H.; Beachum, C.E.; Bennett, M.G.; Knouft, J.H. Climate change, hydrology, and fish morphology: Predictions using phenotype-environment associations. Climatic Chang. 2017, 140, 563–576. [Google Scholar] [CrossRef]
- Rudolfsen, T.; Watkinson, D.A.; Poesch, M. Morphological divergence of the threatened Rocky Mountain sculpin (Cottus sp.) is driven by biogeography and flow regime: Implications for mitigating altered flow regime to freshwater fishes. Aquat. Conserv. 2018, 28, 78–86. [Google Scholar] [CrossRef]
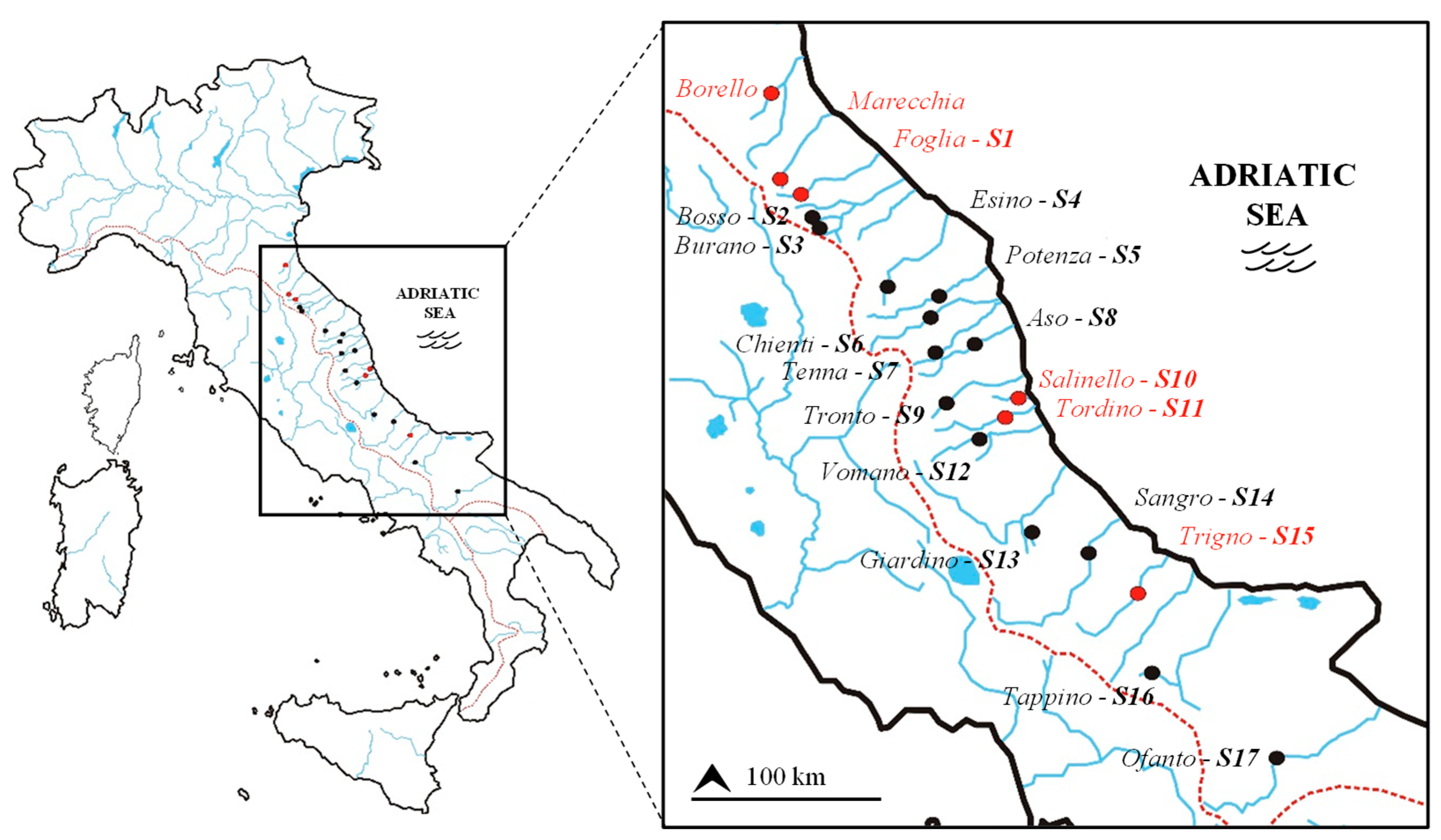


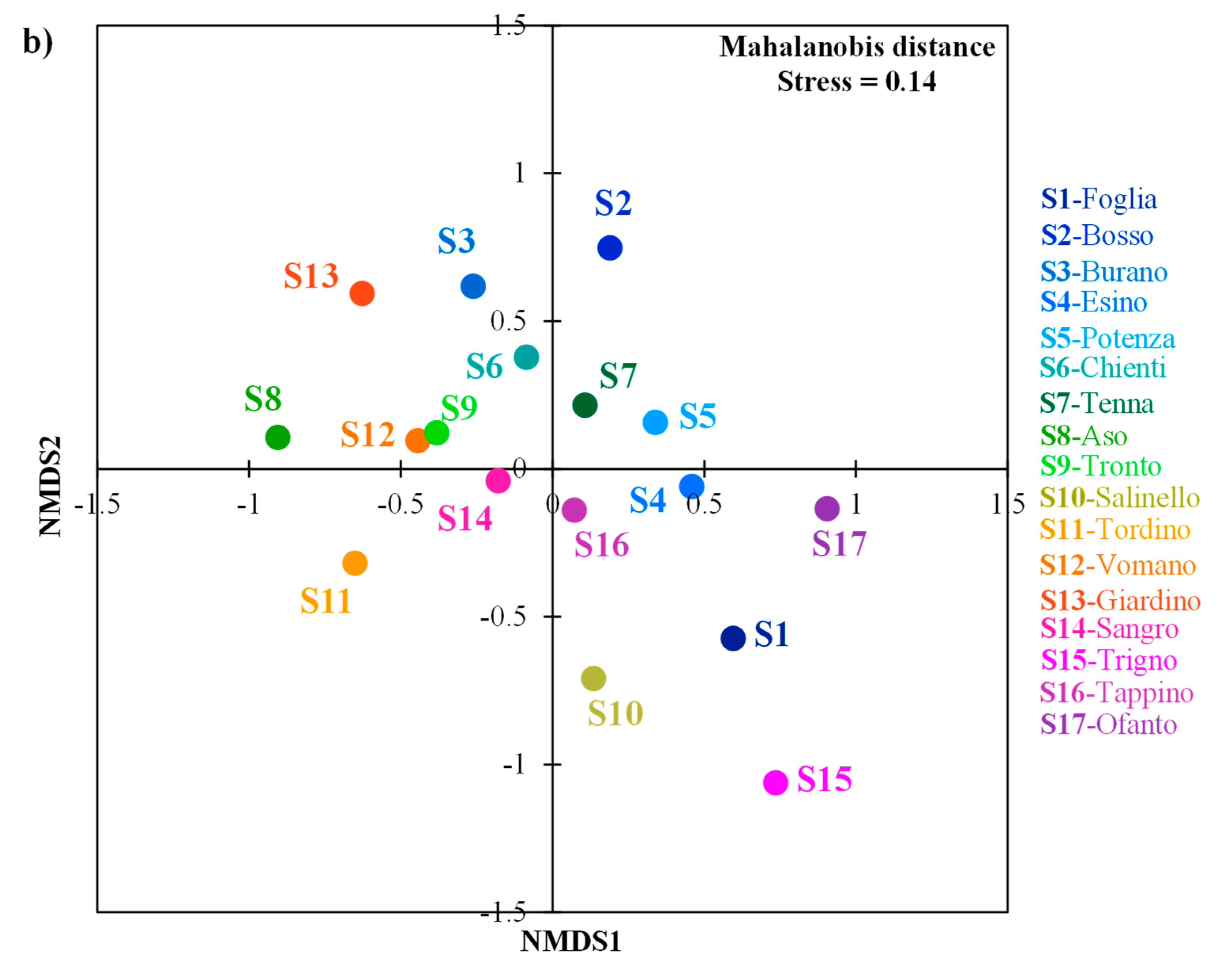

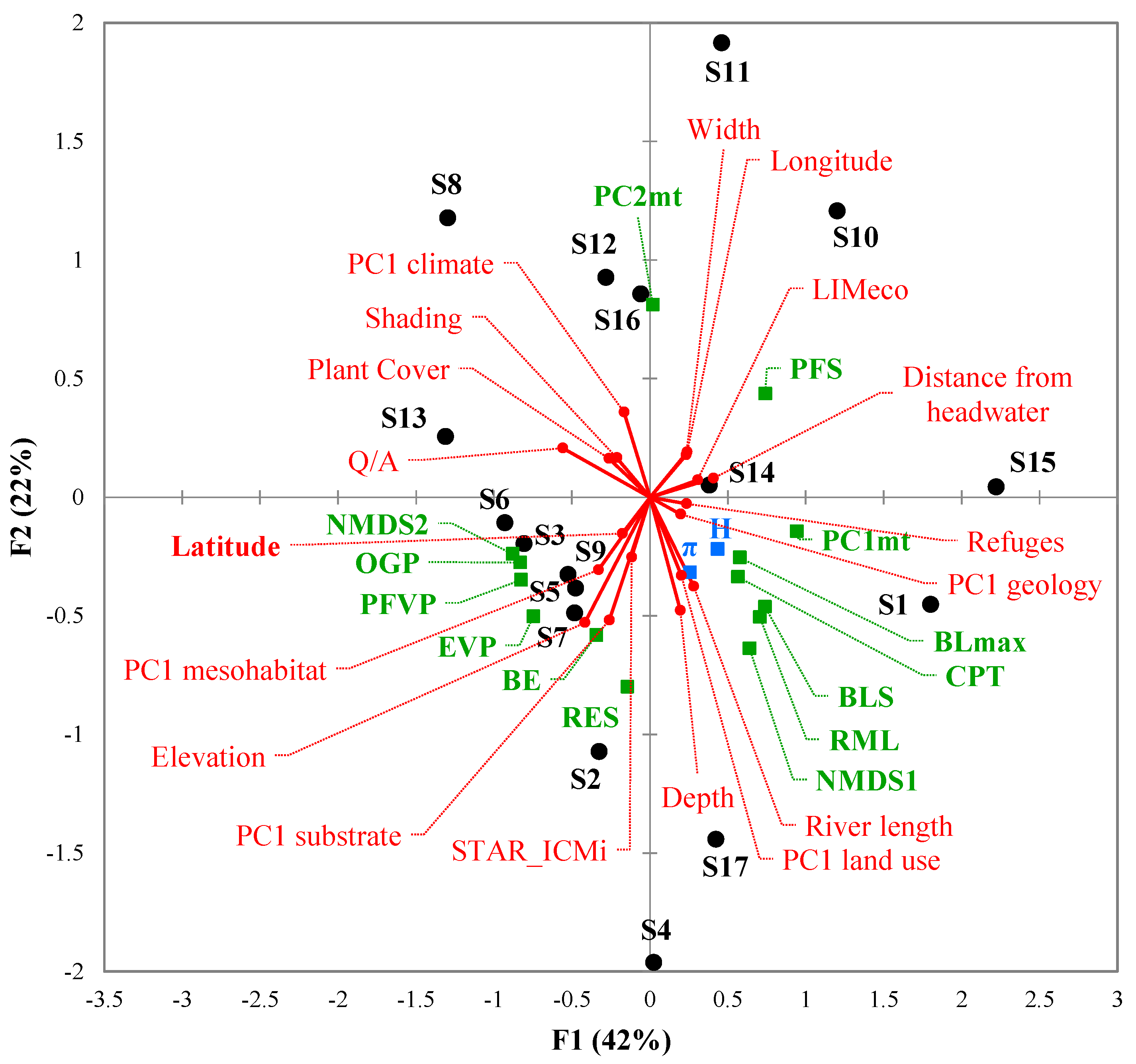
| Watercourse | Basin | Latitude (N) | Longitude (E) | Elevation (m) | Distance from headwater (km) | Species | ID | Ng | Nm | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Borello | Savio | 44°3′8.1″ | 12°10′47″ | 77 | 30 | No purebred | - | - | - | This study |
| Marecchia | Marecchia | 43°49′35.2″ | 12°15′2.4″ | 335 | 27 | No purebred | - | - | - | This study |
| Foglia | Foglia | 43°45′1.2″ | 12°27′13.3″ | 300 | 25 | B. plebejus | S1 | 13 | 23 | This study |
| Bosso | Metauro | 43°30′49.8″ | 12°33′42.8″ | 398 | 10 | B. plebejus | S2 | 43 | 10 | [45] |
| Burano | Metauro | 43°28′ 48.3″ | 12°37′16.4″ | 341 | 14 | B. plebejus | S3 | 14 | 10 | [45] |
| Esino | Esino | 43°24′24.4″ | 12°58′47″ | 237 | 19 | B. plebejus | S4 | 26 | 10 | [45] |
| Potenza | Potenza | 43°16′59.8″ | 13°20′5.1″ | 150 | 45 | B. plebejus | S5 | 25 | 10 | [45] |
| Chienti | Chienti | 43°9′ 35.4″ | 13°14′37″ | 268 | 40 | B. plebejus | S6 | 34 | 10 | [45] |
| Tenna | Tenna | 43°1′10.6″ | 13° 25′ 34″ | 363 | 14 | B. plebejus | S7 | 19 | 10 | [45] |
| Aso | Aso | 43°1′25.5″ | 13°37′20.1″ | 185 | 38 | B. plebejus | S8 | 27 | 10 | [45] |
| Tronto | Tronto | 42°46′12.2″ | 13°24′29.4″ | 370 | 39 | B. plebejus | S9 | 28 | 10 | [45] |
| Salinello | Salinello | 42°47′17″ | 13°53′28.2″ | 28 | 39 | B. plebejus | S10 | 21 | 29 | This study |
| Tordino | Tordino | 42°42′8″ | 13°49′49.9″ | 97 | 44 | B. plebejus | S11 | 19 | 31 | This study |
| Vomano | Vomano | 42°35′21.6″ | 13°40′51.4″ | 203 | 33 | B. samniticus | S12 | 25 | 10 | [45] |
| Giardino | Aterno-Pescara | 42°10′25.8″ | 13°49′51.4″ | 247 | 0.5 | B. samniticus | S13 | 24 | 10 | [41] |
| Sangro | Sangro | 42°3′10.7″ | 14°20′51.3″ | 116 | 93 | B. samniticus | S14 | 23 | 10 | [41] |
| Trigno | Trigno | 41° 54′ 14.2″ | 14°38′43.7″ | 121 | 61 | B. samniticus | S15 | 18 | 23 | This study |
| Tappino | Fortore | 41°33′13.2″ | 14°52′33.9″ | 239 | 30 | B. fucini | S16 | 27 | 10 | [41] |
| Ofanto | Ofanto | 41°4′26.1″ | 15°32′46.4″ | 219 | 74 | B. fucini | S17 | 20 | 10 | [41] |
| Functional Trait | Code | Formula | Potential Link with Fish Functions | References |
|---|---|---|---|---|
| Maximum body length | BLmax | BLmax | Size is linked to metabolism, trophic impacts, locomotion ability, nutrient cycling | [66] |
| Body elongation | BE | BL/BD | Hydrodynamism | [67] |
| Eye vertical position | EVP | EH/BD | Position of fish and/or of its prey in the water column | [68] |
| Relative eye size | RES | ED/HD | Visual acuity | [69] |
| Oral gape position | OGP | MO/BD | Feeding position in the water column | [70,71] |
| Relative maxillary length | RML | MJL/HD | Size of mouth and strength of jaw | [66] |
| Body lateral shape | BLS | HD/BD | Hydrodynamism and head size | [66] |
| Pectoral fin vertical position | PFVP | PFI/BD | Pectoral fin use for swimming | [70] |
| Pectoral fin size | PFS | PFL/BL | Pectoral fin use for swimming | [72] |
| Caudal peduncle throttling | CPT | CFD/CPD | Caudal propulsion efficiency through reduction of drag | [73] |
| Species | Watercourse | ID | n | PSh | S | π | H | |
|---|---|---|---|---|---|---|---|---|
| B. plebejus | Foglia | S1 | 7 | 0.3 | 8 | 0.002 ± 0.001 | 0.79 ± 0.11 | |
| Bosso | S2 | 7 | 0.4 | 6 | 0.002 ± 0.001 | 0.87 ± 0.06 | ||
| Burano | S3 | 4 | 0.3 | 5 | 0.002 ± 0.001 | 0.68 ± 0.07 | ||
| Esino | S4 | 11 | 0.5 | 11 | 0.002 ± 0.001 | 0.85 ± 0.05 | ||
| Potenza | S5 | 4 | 0.0 | 3 | 0.0006 ± 0.0006 | 0.46 ± 0.10 | ||
| Chienti | S6 | 4 | 0.3 | 4 | 0.001 ± 0.001 | 0.6 ± 0.07 | ||
| Tenna | S7 | 2 | 1.0 | 1 | 0.0001 ± 0.0002 | 0.10 ± 0.09 | ||
| Aso | S8 | 3 | 1.0 | 4 | 0.001 ± 0.001 | 0.55 ± 0.08 | ||
| Tronto | S9 | 4 | 0.5 | 4 | 0.001 ± 0.001 | 0.54 ± 0.09 | ||
| Salinello | S10 | 4 | 0.5 | 6 | 0.001 ± 0.001 | 0.61 ± 0.09 | ||
| Tordino | S11 | 6 | 0.5 | 5 | 0.001 ± 0.001 | 0.70 ± 0.10 | ||
| B. samniticus | Vomano | S12 | 5 | 1.0 | 6 | 0.0005 ± 0.0005 | 0.30 ± 0.12 | |
| Giardino | S13 | 3 | 0.7 | 3 | 0.0003 ± 0.0004 | 0.16 ± 0.10 | ||
| Sangro | S14 | 2 | 0.5 | 3 | 0.002 ± 0.001 | 0.52 ± 0.03 | ||
| Trigno | S15 | 5 | 0.8 | 4 | 0.001 ± 0.001 | 0.76 ± 0.06 | ||
| B. fucini | Tappino | S16 | 3 | 1.0 | 4 | 0.001 ± 0.0008 | 0.32 ± 0.10 | |
| Ofanto | S17 | 3 | 1.0 | 3 | 0.0007 ± 0.0006 | 0.43 ± 0.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quadroni, S.; De Santis, V.; Carosi, A.; Vanetti, I.; Zaccara, S.; Lorenzoni, M. Past and Present Environmental Factors Differentially Influence Genetic and Morphological Traits of Italian Barbels (Pisces: Cyprinidae). Water 2023, 15, 325. https://doi.org/10.3390/w15020325
Quadroni S, De Santis V, Carosi A, Vanetti I, Zaccara S, Lorenzoni M. Past and Present Environmental Factors Differentially Influence Genetic and Morphological Traits of Italian Barbels (Pisces: Cyprinidae). Water. 2023; 15(2):325. https://doi.org/10.3390/w15020325
Chicago/Turabian StyleQuadroni, Silvia, Vanessa De Santis, Antonella Carosi, Isabella Vanetti, Serena Zaccara, and Massimo Lorenzoni. 2023. "Past and Present Environmental Factors Differentially Influence Genetic and Morphological Traits of Italian Barbels (Pisces: Cyprinidae)" Water 15, no. 2: 325. https://doi.org/10.3390/w15020325
APA StyleQuadroni, S., De Santis, V., Carosi, A., Vanetti, I., Zaccara, S., & Lorenzoni, M. (2023). Past and Present Environmental Factors Differentially Influence Genetic and Morphological Traits of Italian Barbels (Pisces: Cyprinidae). Water, 15(2), 325. https://doi.org/10.3390/w15020325