SARS-CoV-2 Wastewater Monitoring in Thuringia, Germany: Analytical Aspects and Normalization of Results
Abstract
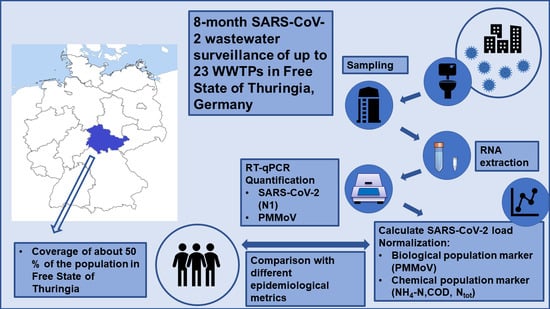
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites, Wastewater Sampling, and Transport
2.2. Physicochemical Standard Parameters
2.3. RNA Extraction from Wastewater and Quantification using RT-qPCR
2.4. Normalization Parameters
- Inhabitant-weighted (COD-based) 24 h virus load per WWTP [GC SARS-CoV-2/inhabitantCOD and 24 h];
- Inhabitant-weighted (nitrogen-based) 24 h virus load per WWTP [GC SARS-CoV-2/inhabitantNtot and 24 h].
- Average concentration per sampling day [GC SARS-CoV-2/L] (flow-normalized viral load of all sampled WWTPs):
- Average inhabitant-weighted 24 h virus load per sampling day (population marker X = NH4-N, COD, Ntot) [GC SARS-CoV-2/inhabitantx and 24 h]:
- Average relative signal (PMMoV) per sampling day [(GC SARS-CoV-2/24 h)/(GC PMMoV/24 h)]:
2.5. Epidemiological Metrics
- 7-day incidences per 100,000 inhabitants on the federal state level and county/urban district level;
- 7-day-hospitalized COVID-19 cases per 100,000 inhabitants on the federal-state level;
- The relative proportion of positive tests (positive test rate) [57]: At the time of our study, the collection of testing data in Germany was based on data from voluntarily participating laboratories in this part of the surveillance system. Testing data from this survey were reported on the federal-state level weekly. The report covers both detection using PCR and serological diagnostics using antibody detection. At the time of our study, two laboratories in Thuringia participated voluntarily and reported 20,429,600 testing data to the Robert Koch Institute. We used these data as a proxy for the positive test rate in the Free State of Thuringia.
2.6. Data Analysis
3. Results
3.1. Performance of the 4S Method for the SARS-CoV-2 Monitoring Program
3.2. Description of SARS-CoV-2 Concentration and Normalization Parameters
3.3. Virus Signal in Wastewater and Epidemiological Metrics
3.3.1. Virus Signal in the Pooled Wastewater Data Set and Epidemiological Metrics on the Federal Level
3.3.2. Virus Signal Per WWTP and Epidemiological Metrics on the Federal Level
3.4. Correlation between Epidemiological Metrics and Pooled Viral Signals in the Wastewater
4. Discussion
4.1. Performance of the 4S Method for the SARS-CoV-2 Monitoring Program
4.2. Normalization Parameters
4.3. Correlation between Virus Signals in the Wastewater and Epidemiological Metrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perez-Zabaleta, M.; Archer, A.; Khatami, K.; Jafferali, M.H.; Nandy, P.; Atasoy, M.; Birgersson, M.; Williams, C.; Cetecioglu, Z. Long-term SARS-CoV-2 surveillance in the wastewater of Stockholm: What lessons can be learned from the Swedish perspective? Sci. Total Environ. 2023, 858 Pt 3, 160023. [Google Scholar] [CrossRef]
- World Health Organization. Environmental Surveillance for SARS-COV-2 to Complement Public Health Surveillance–Interim Guidance: WHO/HEP/ECH/WSH/2022.1; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, J.; Guo, L.; Yao, H.; Wang, L.; Xia, X.; Zhang, W. Fecal viral shedding in COVID-19 patients: Clinical significance, viral load dynamics and survival analysis. Virus Res. 2020, 289, 198147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, P.M.; Graber, T.E.; Mercier, E.; Montpetit, D.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021, 770, 145319. [Google Scholar] [CrossRef] [PubMed]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada): Comparing twelve SARS-CoV-2 normalization approaches. Sci. Total Environ. 2023, 856 Pt 1, 158964. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Sherchan, S.; Orive, G.; Lertxundi, U.; Haramoto, E.; Honda, R.; Kumar, M.; Arora, S.; Kitajima, M.; et al. Correlation between SARS-CoV-2 RNA concentration in wastewater and COVID-19 cases in community: A systematic review and meta-analysis. J. Hazard. Mater. 2022, 441, 129848. [Google Scholar] [CrossRef] [PubMed]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: Findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021. [Google Scholar] [CrossRef]
- Bertels, X.; Demeyer, P.; van den Bogaert, S.; Boogaerts, T.; van Nuijs, A.L.N.; Delputte, P.; Lahousse, L. Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: A systematic review. Sci. Total Environ. 2022, 820, 153290. [Google Scholar] [CrossRef]
- Augusto, M.R.; Claro, I.C.M.; Siqueira, A.K.; Sousa, G.S.; Caldereiro, C.R.; Duran, A.F.A.; de Miranda, T.B.; Bomediano Camillo, L.d.M.; Cabral, A.D.; de Freitas Bueno, R. Sampling strategies for wastewater surveillance: Evaluating the variability of SARS-COV-2 RNA concentration in composite and grab samples. J. Environ. Chem. Eng. 2022, 10, 107478. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Bertsch, P.M.; Bibby, K.; Choi, P.M.; Farkas, K.; Gyawali, P.; Hamilton, K.A.; Haramoto, E.; Kitajima, M.; et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health 2020, 17, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Whitney, O.N.; Kennedy, L.C.; Fan, V.B.; Hinkle, A.; Kantor, R.; Greenwald, H.; Crits-Christoph, A.; Al-Shayeb, B.; Chaplin, M.; Maurer, A.C.; et al. Sewage, Salt, Silica, and SARS-CoV-2 (4S): An Economical Kit-Free Method for Direct Capture of SARS-CoV-2 RNA from Wastewater. Environ. Sci. Technol. 2021, 55, 4880–4888. [Google Scholar] [CrossRef] [PubMed]
- Hubert, C.R.J.; Acosta, N.; Waddell, B.J.M.; Hasing, M.E.; Qiu, Y.; Fuzzen, M.; Harper, N.B.J.; Bautista, M.A.; Gao, T.; Papparis, C.; et al. Tracking Emergence and Spread of SARS-CoV-2 Omicron Variant in Large and Small Communities by Wastewater Monitoring in Alberta, Canada. Emerg. Infect. Dis. 2022, 28, 1770–1776. [Google Scholar] [CrossRef]
- Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; Whitney, O.N.; Fan, V.B.; Crits-Christoph, A.; Harris-Lovett, S.; Flamholz, A.I.; Al-Shayeb, B.; Liao, L.D.; et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X 2021, 12, 100111. [Google Scholar] [CrossRef] [PubMed]
- Schill, R.; Nelson, K.L.; Harris-Lovett, S.; Kantor, R.S. The dynamic relationship between COVID-19 cases and SARS-CoV-2 wastewater concentrations across time and space: Considerations for model training data sets. Sci. Total Environ. 2023, 162069. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, A.; Schoth, J.; Meinert-Berning, C.; Agrawal, S.; Bastian, D.; Orschler, L.; Ciesek, S.; Teichgräber, B.; Wintgens, T.; Lackner, S.; et al. Wastewater surveillance allows early detection of SARS-CoV-2 omicron in North Rhine-Westphalia, Germany. Sci. Total Environ. 2022, 846, 157375. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Rainey, A.L.; Loeb, J.C.; Robinson, S.E.; Lednicky, J.A.; McPherson, J.; Colson, S.; Allen, M.; Coker, E.S.; Sabo-Attwood, T.; Maurelli, A.T.; et al. Wastewater surveillance for SARS-CoV-2 in a small coastal community: Effects of tourism on viral presence and variant identification among low prevalence populations. Environ. Res. 2022, 208, 112496. [Google Scholar] [CrossRef]
- Mazumder, P.; Dash, S.; Honda, R.; Sonne, C.; Kumar, M. Sewage surveillance for SARS-CoV-2: Molecular detection, quantification, and normalization factors. Curr. Opin. Environ. Sci. Health 2022, 28, 100363. [Google Scholar] [CrossRef]
- Kumar, M.; Jiang, G.; Kumar Thakur, A.; Chatterjee, S.; Bhattacharya, T.; Mohapatra, S.; Chaminda, T.; Kumar Tyagi, V.; Vithanage, M.; Bhattacharya, P.; et al. Lead time of early warning by wastewater surveillance for COVID-19: Geographical variations and impacting factors. Chem. Eng. J. 2022, 441, 135936. [Google Scholar] [CrossRef]
- Bayati, M.; Hsieh, H.-Y.; Hsu, S.-Y.; Li, C.; Rogers, E.; Belenchia, A.; Zemmer, S.A.; Blanc, T.; LePage, C.; Klutts, J.; et al. Identification and quantification of bioactive compounds suppressing SARS-CoV-2 signals in wastewater-based epidemiology surveillance. Water Res. 2022, 221, 118824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Duran, S.S.F.; Lim, W.Y.S.; Tan, C.K.I.; Cheong, W.C.D.; Suwardi, A.; Loh, X.J. SARS-CoV-2 in wastewater: From detection to evaluation. Mater. Today Adv. 2022, 13, 100211. [Google Scholar] [CrossRef]
- Oloye, F.F.; Xie, Y.; Challis, J.K.; Femi-Oloye, O.P.; Brinkmann, M.; McPhedran, K.N.; Jones, P.D.; Servos, M.R.; Giesy, J.P. Understanding common population markers for SARS-CoV-2 RNA normalization in wastewater—A review. Chemosphere 2023, 333, 138682. [Google Scholar] [CrossRef]
- Langeveld, J.; Schilperoort, R.; Heijnen, L.; Elsinga, G.; Schapendonk, C.E.M.; Fanoy, E.; Schepper, E.I.T.; de Koopmans, M.P.G.; de Graaf, M.; Medema, G.; et al. Normalisation of SARS-CoV-2 concentrations in wastewater: The use of flow, electrical conductivity and crAssphage. Sci. Total Environ. 2023, 865, 161196. [Google Scholar] [CrossRef] [PubMed]
- Masri, N.Z.; Card, K.G.; Caws, E.A.; Babcock, A.; Powell, R.; Lowe, C.J.; Donovan, S.; Norum, S.; Lyons, S.; de Pol, S.; et al. Testing specificity and sensitivity of wastewater-based epidemiology for detecting SARS-CoV-2 in four communities on Vancouver Island, Canada. Environ. Adv. 2022, 9, 100310. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, S.F.; Rossi, G.; Low, A.S.; McAteer, S.P.; O’Keefe, B.; Findlay, D.; Cameron, G.J.; Pollard, P.; Singleton, P.T.R.; Ponton, G.; et al. Site Specific Relationships between COVID-19 Cases and SARS-CoV-2 Viral Load in Wastewater Treatment Plant Influent. Environ. Sci. Technol. 2021, 55, 15276–15286. [Google Scholar] [CrossRef]
- Alamin, M.; Tsuji, S.; Hata, A.; Hara-Yamamura, H.; Honda, R. Selection of surrogate viruses for process control in detection of SARS-CoV-2 in wastewater. Sci. Total Environ. 2022, 823, 153737. [Google Scholar] [CrossRef]
- Mitranescu, A.; Uchaikina, A.; Kau, A.-S.; Stange, C.; Ho, J.; Tiehm, A.; Wurzbacher, C.; EDrewes, J. Wastewater-Based Epidemiology for SARS-CoV-2 Biomarkers: Evaluation of Normalization Methods in Small and Large Communities in Southern Germany. ACS EST Water 2022, 2, 2460–2470. [Google Scholar] [CrossRef]
- Markt, R.; Stillebacher, F.; Nägele, F.; Kammerer, A.; Peer, N.; Payr, M.; Scheffknecht, C.; Dria, S.; Draxl-Weiskopf, S.; Mayr, M.; et al. Expanding the Pathogen Panel in Wastewater Epidemiology to Influenza and Norovirus. Viruses 2023, 15, 263. [Google Scholar] [CrossRef]
- Graham, K.E.; Loeb, S.K.; Wolfe, M.K.; Catoe, D.; Sinnott-Armstrong, N.; Kim, S.; Yamahara, K.M.; Sassoubre, L.M.; Mendoza Grijalva, L.M.; Roldan-Hernandez, L.; et al. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2021, 55, 488–498. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities. ACS EST Water 2021, 1, 1955–1965. [Google Scholar] [CrossRef]
- Zheng, X.; Li, S.; Deng, Y.; Xu, X.; Ding, J.; Lau, F.T.K.; Yau, C.I.; Poon, L.L.M.; Tun, H.M.; Zhang, T.; et al. Quantification of SARS-CoV-2 RNA in wastewater treatment plants mirrors the pandemic trend in Hong Kong. Sci. Total Environ. 2022, 844, 157121. [Google Scholar] [CrossRef] [PubMed]
- Scobie, H.M.; Panaggio, M.; Binder, A.M.; Gallagher, M.E.; Duck, W.M.; Graff, P.; Silk, B.J. Correlations and Timeliness of COVID-19 Surveillance Data Sources and Indicators―United States, October 1, 2020–March 22, 2023. Morb. Mortal. Wkly. Rep. MMWR 2023, 72, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Dumke, R.; Geissler, M.; Skupin, A.; Helm, B.; Mayer, R.; Schubert, S.; Oertel, R.; Renner, B.; Dalpke, A.H. Simultaneous Detection of SARS-CoV-2 and Influenza Virus in Wastewater of Two Cities in Southeastern Germany, January to May 2022. Int. J. Environ. Res. Public Health 2022, 19, 3374. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Orschler, L.; Lackner, S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci Rep 2021, 11, 5372. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Weber, F.-A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751, 141750. [Google Scholar] [CrossRef] [PubMed]
- Breulmann, M.; Kallies, R.; Bernhard, K.; Gasch, A.; Müller, R.A.; Harms, H.; Chatzinotas, A.; van Afferden, M. A long-term passive sampling approach for wastewater-based monitoring of SARS-CoV-2 in Leipzig, Germany. Sci. Total Environ. 2023, 887, 164143. [Google Scholar] [CrossRef]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 wastewater surveillance in Germany: Long-term RT-digital droplet PCR monitoring, suitability of primer/probe combinations and biomarker stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef]
- Gawlik, B.; Tavazzi, S.; Mariani, G.; Skejo, H.; Sponar, M.; Higgins, T.; Medema, G.; Wintgens, T. SARS-CoV-2 Surveillance Employing Sewage-Towards a Sentinel System; JRC Technical Report; Publications Office of the European Union: Luxembourg, 2021; ISBN 978-92-76-36888-5. [Google Scholar]
- Wilhelm, A.; Agrawal, S.; Schoth, J.; Meinert-Berning, C.; Bastian, D.; Orschler, L.; Ciesek, S.; Teichgräber, B.; Wintgens, T.; Lackner, S.; et al. Early Detection of SARS-CoV-2 Omicron BA.4 and BA.5 in German Wastewater. Viruses 2022, 14, 1876. [Google Scholar] [CrossRef]
- DIN 38409-44:1992; German Standardised Method for the Analysis of Water, Waste Water and Sludge; Summarised Effect and Substance Parameters (Group H); Determination of the Chemical Oxygen Demand (COD) in the Range 5 to 50 mg/l (H 44). DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 1992.
- DIN ISO 15705:2002; Water Quality-Determination of Chemical Oxygen Demand (ST-COD): Cuvette Test (ISO 15705:2002). DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 2003.
- ISO 6060:1989; Water Quality; Determination of Chemical Oxygen Consumption. DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 1989.
- DIN 38405-9:2011; German Standardised Methods for the Analysis of Water, Waste Water and Sludge- Anions (Group D)-Part 9: Photometric Determination of Nitrate (D 9). DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 2011.
- DIN EN ISO 11905-1:1998; Water Quality-Determination of Nitrogen: Part 1: Determination of Nitrogen by Oxidative Digestion with Peroxodisulfate (ISO 11905-1:1997). DIN German Institute for Standardization; German Version EN ISO 11905-1:1998; Beuth Verlag GmbH: Berlin, Germany, 1998.
- DIN 38406-5:1983; German Standardised Method for the Analysis of Water, Waste Water and Sludge; Cations (group E); Determination of Ammonium Nitrogen (E 5). DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 1983.
- ISO 7150-1:1984; Water quality; Determination of ammonium; Part 1: Manual spectrometric method. DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 1984.
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- ISO 10705-1:1995; Detection and Enumeration of Bacteriophages: Part 1: Enumeration of F-Specific RNA Bacteriophages. DIN German Institute for Standardization; Beuth Verlag GmbH: Berlin, Germany, 2002.
- Mondal, S.; Feirer, N.; Brockman, M.; Preston, M.A.; Teter, S.J.; Ma, D.; Goueli, S.A.; Moorji, S.; Saul, B.; Cali, J.J.; et al. A direct capture method for purification and detection of viral nucleic acid enables epidemiological surveillance of SARS-CoV-2. Sci. Total Environ. 2021, 795, 148834. [Google Scholar] [CrossRef] [PubMed]
- Kantor, R.S.; Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; Harris-Lovett, S.; Metzger, M.; Thornton, M.M.; Paluba, J.M.; Nelson, K.L. Operationalizing a routine wastewater monitoring laboratory for SARS-CoV-2. PLoS Water 2022, 1, e0000007. [Google Scholar] [CrossRef]
- ATV-DVWK—Rules, ATV-DVWK-A 198: Vereinheitlichung und Herleitung von Bemessungswerten für Abwasseranlagen; Deutsche Vereinigung für Wasserwirtschaft, Abwasser und Abfall e.V.: Hennef, Germany, 2003; ISBN 3-924063-48-6.
- Been, F.; Rossi, L.; Ort, C.; Rudaz, S.; Delémont, O.; Esseiva, P. Population normalization with ammonium in wastewater-based epidemiology: Application to illicit drug monitoring. Environ. Sci. Technol. 2014, 48, 8162–8169. [Google Scholar] [CrossRef] [PubMed]
- Arabzadeh, R.; Grünbacher, D.M.; Insam, H.; Kreuzinger, N.; Markt, R.; Rauch, W. Data filtering methods for SARS-CoV-2 wastewater surveillance. Water Sci. Technol. 2021, 84, 1324–1339. [Google Scholar] [CrossRef] [PubMed]
- Robert Koch-Institute (RKI). 7-Day Incidences by State and County. Available online: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Daten/Fallzahlen_Inzidenz_aktualisiert.html (accessed on 20 October 2022).
- Robert Koch-Institute (RKI). Laboratory-Based SARS-CoV-2 Surveillance Data Sheets: Federal State Thueringen. Available online: https://ars.rki.de/Content/COVID19/Reports.aspx (accessed on 20 October 2022).
- von Sperling, M.; Verbyla, M.E.; Oliveira, S.M.A.C. (Eds.) Assessment of Treatment Plant Performance and Water Quality Data: A Guide for Students, Researchers and Practitioners, 1st ed.; IWA Publishing: London, UK, 2020; ISBN 9781780409320. [Google Scholar]
- Blainey, P.; Krzywinski, M.; Altman, N. Points of significance: Replication. Nat. Methods 2014, 11, 879–880. [Google Scholar] [CrossRef] [PubMed]
- Ontario Clean Water Agency, Ministry of the Environment, Conservation and Parks, MECP Wastewater Surveillance Initiative: Protocol for Evaluations of RT-qPCR Performance Characteristics: Technical Guidance; Ministry of the Environment, Conservation and Parks (MECP): Toronto, ON, Canada, 2022; ISBN 978-1-4868-5481-3.
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandão, J.; Choi, P.M.; et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef]
- Kantor, R.S.; Nelson, K.L.; Greenwald, H.D.; Kennedy, L.C. Challenges in Measuring the Recovery of SARS-CoV-2 from Wastewater. Environ. Sci. Technol. 2021, 55, 3514–3519. [Google Scholar] [CrossRef]
- Balboa, S.; Mauricio-Iglesias, M.; Rodriguez, S.; Martínez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021, 772, 145268. [Google Scholar] [CrossRef]
- Forés, E.; Bofill-Mas, S.; Itarte, M.; Martínez-Puchol, S.; Hundesa, A.; Calvo, M.; Borrego, C.M.; Corominas, L.L.; Girones, R.; Rusiñol, M.; et al. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021, 768, 144786. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Iker, B.C.; Pepper, I.L.; Gerba, C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes--identification of potential viral indicators. Sci. Total Environ. 2014, 488, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, L.; Kontou, A.; Strati, A.; Galani, A.; Kostakis, M.; Kapes, V.; Lianidou, E.; Thomaidis, N.; Markou, A. Evaluation of viral concentration and extraction methods for SARS-CoV-2 recovery from wastewater using droplet digital and quantitative RT-PCR. Case Stud. Chem. Environ. Eng. 2022, 6, 100224. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kennedy, L.C.; Wolfe, M.K.; Criddle, C.S.; Duong, D.H.; Topol, A.; White, B.J.; Kantor, R.S.; Nelson, K.L.; Steele, J.A.; et al. SARS-CoV-2 RNA is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works. Environ. Sci. Water Res. Technol. 2022, 8, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Lucansky, V.; Samec, M.; Burjanivova, T.; Lukacova, E.; Kolkova, Z.; Holubekova, V.; Turyova, E.; Hornakova, A.; Zaborsky, T.; Podlesniy, P.; et al. Comparison of the methods for isolation and detection of SARS-CoV-2 RNA in municipal wastewater. Front. Public Health 2023, 11, 1116636. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Waldman, P.; Ferrier-Rembert, A.; Frenois-Veyrat, G.; Mouchel, J.M.; Boni, M.; Maday, Y.; Marechal, V.; Moulin, L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 2021, 198, 117183. [Google Scholar] [CrossRef] [PubMed]
- Breadner, P.R.; Dhiyebi, H.A.; Fattahi, A.; Srikanthan, N.; Hayat, S.; Aucoin, M.G.; Boegel, S.J.; Bragg, L.M.; Craig, P.M.; Xie, Y.; et al. A comparative analysis of the partitioning behaviour of SARS-CoV-2 RNA in liquid and solid fractions of wastewater. Sci. Total Environ. 2023, 895, 165095. [Google Scholar] [CrossRef]
- Sweetapple, C.; Wade, M.J.; Melville-Shreeve, P.; Chen, A.S.; Lilley, C.; Irving, J.; Grimsley, J.M.; Bunce, J.T. Dynamic population normalisation in wastewater-based epidemiology for improved understanding of the SARS-CoV-2 prevalence: A multi-site study. J. Water Health 2023, 21, 625–642. [Google Scholar] [CrossRef]
- Schenk, H.; Heidinger, P.; Insam, H.; Kreuzinger, N.; Markt, R.; Nägele, F.; Oberacher, H.; Scheffknecht, C.; Steinlechner, M.; Vogl, G.; et al. Prediction of hospitalisations based on wastewater-based SARS-CoV-2 epidemiology. Sci. Total Environ. 2023, 873, 162149. [Google Scholar] [CrossRef]
- Rosario, K.; Symonds, E.M.; Sinigalliano, C.; Stewart, J.; Breitbart, M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009, 75, 7261–7267. [Google Scholar] [CrossRef]
- Hamza, I.A.; Jurzik, L.; Uberla, K.; Wilhelm, M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011, 45, 1358–1368. [Google Scholar] [CrossRef]
- Kitajima, M.; Sassi, H.P.; Torrey, J.R. Pepper mild mottle virus as a water quality indicator. NPJ Clean Water 2018, 1, 19. [Google Scholar] [CrossRef]
- Dhiyebi, H.A.; Abu Farah, J.; Ikert, H.; Srikanthan, N.; Hayat, S.; Bragg, L.M.; Qasim, A.; Payne, M.; Kaleis, L.; Paget, C.; et al. Assessment of seasonality and normalization techniques for wastewater-based surveillance in Ontario, Canada. Front. Public Health 2023, 11, 1186525. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Murakami, M.; Ahmed, W.; Iwamoto, R.; Okabe, S.; Kitajima, M. Wastewater-based prediction of COVID-19 cases using a highly sensitive SARS-CoV-2 RNA detection method combined with mathematical modeling. Environ. Int. 2023, 107743. [Google Scholar] [CrossRef] [PubMed]
- Burnet, J.-B.; Cauchie, H.-M.; Walczak, C.; Goeders, N.; Ogorzaly, L. Persistence of endogenous RNA biomarkers of SARS-CoV-2 and PMMoV in raw wastewater: Impact of temperature and implications for wastewater-based epidemiology. Sci. Total Environ. 2023, 857, 159401. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; North, D.; Wu, Z.; Shaffer, M.; Ahmed, W.; Bibby, K. Within- and between-Day Variability of SARS-CoV-2 RNA in Municipal Wastewater during Periods of Varying COVID-19 Prevalence and Positivity. ACS EST Water 2021, 1, 2097–2108. [Google Scholar] [CrossRef]
- Baldwin, W.M.; Dayton, R.D.; Bivins, A.W.; Scott, R.S.; Yurochko, A.D.; Vanchiere, J.A.; Davis, T.; Arnold, C.L.; Asuncion, J.E.T.; Bhuiyan, M.A.N.; et al. Highly socially vulnerable communities exhibit disproportionately increased viral loads as measured in community wastewater. Environ. Res. 2023, 222, 115351. [Google Scholar] [CrossRef]
- Water Environment Federation. Summary Report: Pilot Program for Onsite Testing of SARS-CoV-2 in Correctional Facility Wastewater; Water Environment Federation: Alexandria, VA, USA, 2022. [Google Scholar]
- Gonçalves, J.; Torres-Franco, A.; Rodriguéz, E.; Diaz, I.; Koritnik, T.; Da Silva, P.G.; Mesquita, J.R.; Trkov, M.; Paragi, M.; Muñoz, R.; et al. Centralized and decentralized wastewater-based epidemiology to infer COVID-19 transmission - A brief review. One Health 2022, 15, 100405. [Google Scholar] [CrossRef]
- Haeusser, S.; Weber, M.; Mauer, C.; Linnemann, V.; Pfannstiel, A.; Pinnekamp, J.; Wintgens, T.; Klümper, C.; Beier, S. On-site treatment of hospital wastewater in a full-scale treatment plant in Germany: SARS-CoV-2 and treatment performance. Water Sci. Technol. 2023, 87, 1747–1763. [Google Scholar] [CrossRef]
- Zhan, Q.; Babler, K.M.; Sharkey, M.E.; Amirali, A.; Beaver, C.C.; Boone, M.M.; Comerford, S.; Cooper, D.; Cortizas, E.M.; Currall, B.B.; et al. Relationships between SARS-CoV-2 in Wastewater and COVID-19 Clinical Cases and Hospitalizations, with and without Normalization against Indicators of Human Waste. ACS EST Water 2022, 2, 1992–2003. [Google Scholar] [CrossRef]










| Site-ID | County LK/Urban District SK | Sampling Point | Mean Flow Rate [m3 per d] | Size Classes (GK) | Population Served (without Industry) | Type of Sampling | Sampling Days |
|---|---|---|---|---|---|---|---|
| 1 | SK Weimar | After screen | 16,747 | 4 | 66,500 | 24 h time proportional | 31 |
| 2 | LK Gotha | After grit chamber | 3539 | 4 | 8900 | 24 h time proportional | 33 |
| 3 | LK Ilm Kreis | After screen | 6503 | 4 | 28,900 | 24 h time proportional | 33 |
| 4 | SK Gera | After grit chamber | 19,741 | 5 | 100,638 | 24 h volume proportional | 33 |
| 5 | LK Ilm Kreis | Before screen | 8317 | 5 | 72,000 | 24 h time proportional | 30 |
| 6 | LK Schmalkalden-Meiningen | After grit chamber | 9509 | 4 | 30,000 | 24 h time proportional | 32 |
| 7 | LK Saale-Holzland-Kreis | After grit chamber | 2102 | 4 | 13,768 | 24 h time proportional | 31 |
| 8 | LK Saale-Orla-Kreis | After grit chamber | 4913 | 4 | 14,020 | 24 h volume proportional | 31 |
| 9 | SK Jena | After grit chamber | 20,708 | 5 | 114,024 | 24 h time proportional | 32 |
| 10 | LK Nordhausen | After grit chamber | 9551 | 4 | 54,000 | 24 h volume proportional | 30 |
| 11 | LK Eichsfeld | After screen | 6115 | 4 | 14,358 | 24 h time proportional | 30 |
| 12 | LK Eichsfeld | After grit chamber | 2585 | 4 | 11,103 | 24 h time proportional | 30 |
| 13 | LK Eichsfeld | After screen | 4689 | 4 | 55,867 | 24 h volume proportional | 28 |
| 14 | LK Altenburger Land | After screen | 1922 | 4 | 13,550 | 24 h time proportional | 30 |
| 15 | LK Soemmerda | After screen | 3941 | 4 | 17,000 | 24 h volume proportional | 26 |
| 16 | SK Erfurt | After screen | 45,522 | 5 | 317,274 | 24 h time proportional | 28 |
| 17 | SK Suhl | After grit chamber | 17,872 | 4 | 36,000 | 24 h time proportional | 28 |
| 18 | LK Unstrut-Hainich-Kreis | Before screen | 2595 | 3 | 4569 | 24 h time proportional | 21 |
| 19 | LK Saale-Orla-Kreis | After screen | 2335 | 3 | 5400 | 24 h time proportional | 26 |
| 20 | LK Schmalkalden-Meiningen | After grit chamber | 811 | 2 | 3500 | 24 h time proportional | 24 |
| 21 | LK Saalfeld-Rudolstadt | After screen | 5154 | 4 | 28,817 | 24 h volume proportional | 22 |
| 22 | LK Saalfeld-Rudolstadt | After grit chamber | 7651 | 4 | 32,808 | 24 h volume proportional | 23 |
| 23 | LK Kyffhaeuserkreis | After screen | 952 | 3 | 7000 | 24 h time proportional | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haeusser, S.; Möller, R.; Smarsly, K.; Al-Hakim, Y.; Kreuzinger, N.; Pinnekamp, J.; Pletz, M.W.; Kluemper, C.; Beier, S. SARS-CoV-2 Wastewater Monitoring in Thuringia, Germany: Analytical Aspects and Normalization of Results. Water 2023, 15, 4290. https://doi.org/10.3390/w15244290
Haeusser S, Möller R, Smarsly K, Al-Hakim Y, Kreuzinger N, Pinnekamp J, Pletz MW, Kluemper C, Beier S. SARS-CoV-2 Wastewater Monitoring in Thuringia, Germany: Analytical Aspects and Normalization of Results. Water. 2023; 15(24):4290. https://doi.org/10.3390/w15244290
Chicago/Turabian StyleHaeusser, Sarah, Robert Möller, Kay Smarsly, Yousuf Al-Hakim, Norbert Kreuzinger, Johannes Pinnekamp, Mathias W. Pletz, Claudia Kluemper, and Silvio Beier. 2023. "SARS-CoV-2 Wastewater Monitoring in Thuringia, Germany: Analytical Aspects and Normalization of Results" Water 15, no. 24: 4290. https://doi.org/10.3390/w15244290
APA StyleHaeusser, S., Möller, R., Smarsly, K., Al-Hakim, Y., Kreuzinger, N., Pinnekamp, J., Pletz, M. W., Kluemper, C., & Beier, S. (2023). SARS-CoV-2 Wastewater Monitoring in Thuringia, Germany: Analytical Aspects and Normalization of Results. Water, 15(24), 4290. https://doi.org/10.3390/w15244290