Distribution and Phylogenetic Position of the Antarctic Ribbon Worm Heteronemertes longifissa (Nemertea, Pilidiophora)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analysis
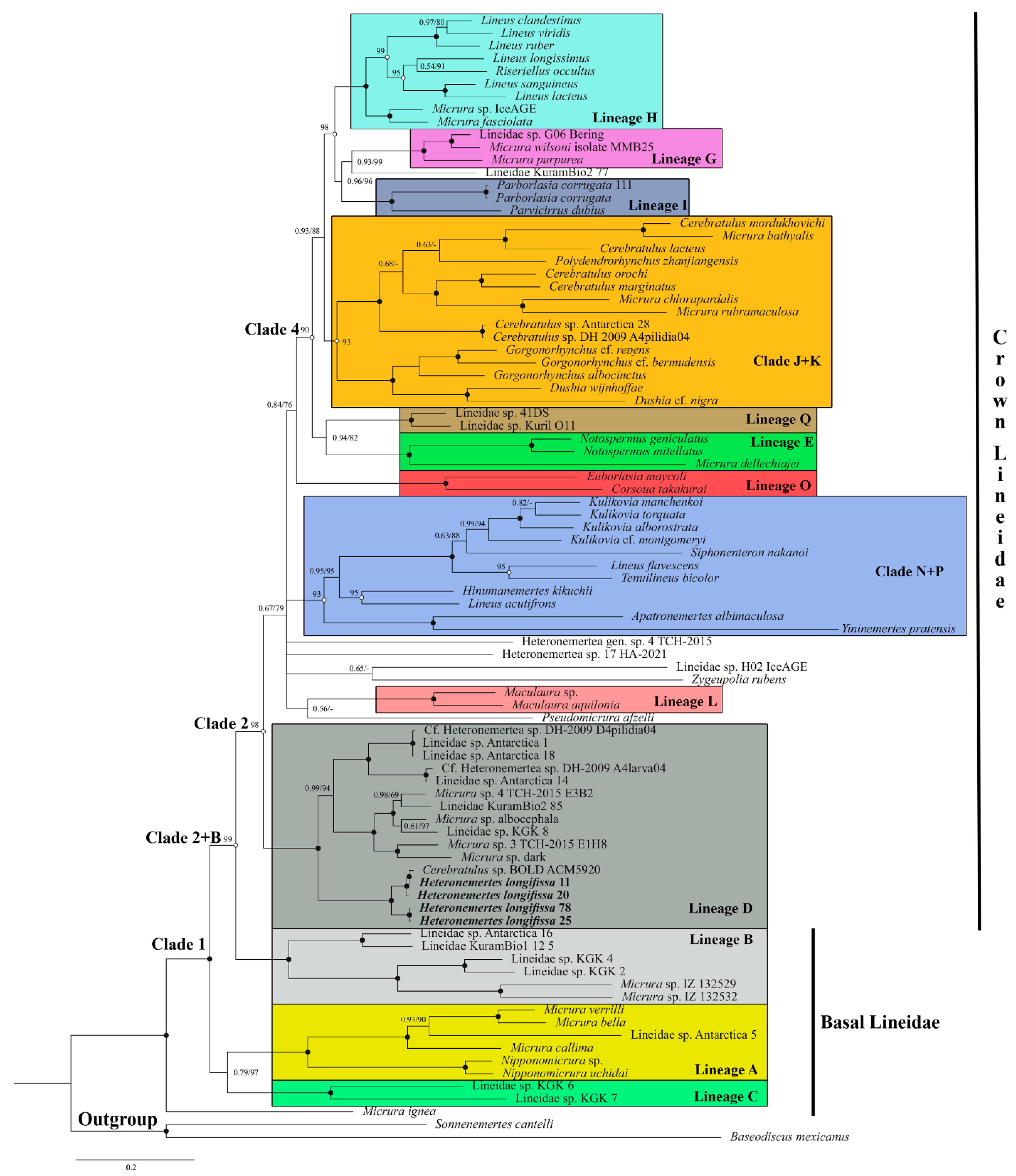
3. Results
3.1. Phylogenetic Analysis of Lineidae
3.2. Antarctic Lineids
4. Discussion
4.1. Phylogenetic Analysis of Lineids
4.2. Systematic Position of the Genus Heteronemertes
4.3. Is Heteronemertes longifissa a Bipolar Species?
4.4. Problems of Identification of Antarctic Heteronemerteans
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chernyshev, A.V. An updated classification of the phylum Nemertea. Invertebr. Zool. 2021, 18, 188–196. [Google Scholar] [CrossRef]
- McIntosh, W.C. Descriptions of some new species of Annelida from Kerguelen’s Island. Ann. Mag. Nat. Hist. 1876, 17, 318–323. [Google Scholar] [CrossRef] [Green Version]
- Bürger, O. Südgeorgische und andere exotische Nemertinen. Zool. Jahrb. Abt. Syst. Geog. Biol. Tiere 1893, 7, 207–240. [Google Scholar] [CrossRef]
- Joubin, L. Note préliminaire sur les Némertiens recueillis par l’expédition antarctique française du Dr Charcot. Bull. Mus. Natl. Hist. Nat. 1905, 11, 431–437. [Google Scholar]
- Joubin, L. Némertiens. In Expédition Antarctique Francaise 1903–1905; Vers et Brachiopodes; Masson: Paris, France, 1908; pp. 1–16. [Google Scholar]
- Joubin, L. Nemertinea. In National Antarctic Expedition 1901–1904; Royal Society: London, UK, 1910; Volume 5, pp. 1–15. [Google Scholar]
- Joubin, L. Némertiens. In Deuxième expédition antarctique francaise (1908–1910); Masson: Paris, France, 1914; Volume 4, pp. 1–33. [Google Scholar]
- Baylis, H.A. Nemertinea. In British Antarctic (“Terra Nova”) Expedition, 1910. Natural History Reports. Zoology; British Museum: London, UK, 1915; Volume 2, pp. 113–134. [Google Scholar]
- Wheeler, J.F.G. Nemerteans from the South Atlantic and southern oceans. Discov. Rep. 1934, 9, 215–294. [Google Scholar]
- Wheeler, J.F.G. Nemerteans of Kerguelen and the Southern Ocean. In Report, B.A.N.Z. Antarctic Research Expedition 1929–1931; Series B; B.A.N.Z.A.R. Expedition Committee: Adelaide, Australia, 1940; Volume 4, pp. 233–256. [Google Scholar]
- Korotkevitsch, V.S. Pelagic nemerteans of Antarctic and temperate waters of the Southern Hemisphere. Issled. Fauny Morey 1964, 2, 132–167. (In Russian) [Google Scholar]
- De Esteban, S.; De La, C.J.; Moretto, H.J.A. Heteronemertea en la bahia de Ushuaia. I. Parborlasia fueguina sp. nov. Huilkia ushuaiensis gen. et sp. nov. Physis. Buenos Aires 1968, 28, 171–181. [Google Scholar]
- Friedrich, H. Nemertinen aus Chile. Sarsia 1970, 40, 1–80. [Google Scholar] [CrossRef]
- Gibson, R. Antarctic nemerteans: Heteronemertea—descriptions of new taxa, reappraisals of the systematic status of existing species and a key to the heteronemerteans recorded south of latitude 50° S. Zool. J. Linn. Soc. 1985, 83, 95–227. [Google Scholar] [CrossRef]
- Fernandez-Álvarez, F.A.; Anadón, N. Oligodendrorhynchus hesperides gen. et sp. n. (Heteronemertea) from the Bellingshausen Sea. Pol. Polar Res. 2012, 33, 81–98. [Google Scholar] [CrossRef]
- Punnett, R.C. On the nemerteans of Norway. Bergens Mus. Årbog. 1903, 2, 1–35. [Google Scholar]
- Ushakov, P.V. To the fauna of nemerteans of the Barentz Sea. Tr. Inst. Po Izucheniju Krainego Sev. 1928, 37, 56–66. (In Russian) [Google Scholar]
- Takakura, U. A classification of nemerteans from the vicinity of Misaki. Zool. Mag. 1898, 10, 38–44, 116–120, 184–187, 331–337, 424–429. (In Japanese) [Google Scholar]
- Iwata, F. Nemertini from the coasts of Kyusyu. J. Fac. Sci. Hokkaido Univ. Ser. 6 Zool. 1952, 11, 126–148. [Google Scholar]
- Hookabe, N.; Kajihara, H. Taxonomic reappraisal of Lineus longifissus auct. (Nemertea: Pilidiophora) from Japan for the first time in 122 years. Zool. Sci. 2020, 37, 467–475. [Google Scholar] [CrossRef]
- Andrade, S.C.S.; Strand, M.; Schwartz, M.; Chen, H.-X.; Kajihara, H.; von Dohren, J.; Sun, S.; Junoy, J.; Thiel, M.; Norenburg, J.L.; et al. Disentangling ribbon worm relationships: Multi-locus analysis supports traditional classification of the phylum Nemertea. Cladistics 2012, 28, 141–159. [Google Scholar] [CrossRef]
- Thornhill, D.J.; Mahon, A.R.; Norenburg, J.L.; Halanych, K.M. Open-ocean barriers to dispersal: A test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae). Mar. Ecol. 2008, 17, 5194–5197. [Google Scholar] [CrossRef]
- Hookabe, N.; Watanabe, K.; Tsujimoto, M.; Kajihara, H. Molecular identity of the antarctic heteronemertean Parborlasia corrugata (Nemertea: Pilidiophora) from Lützow-Holm Bay. Polar Sci. 2020, 25, 100535. [Google Scholar] [CrossRef]
- Heimeier, D.; Lavery, S.; Sewell, M.A. Using DNA barcoding and phylogenetics to identify Antarctic invertebrate larvae: Lessons from a large scale study. Mar. Genom. 2010, 3, 165–177. [Google Scholar] [CrossRef]
- Mahon, A.R.; Thornhill, D.J.; Norenburg, J.L.; Halanych, K.M. DNA uncovers Antarctic nemertean biodiversity and exposes a decades-old cold case of asymmetric inventory. Polar Biol. 2010, 33, 193–202. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Lindgren, A.R.; Daily, M. The impact of length-variable data and alignment criterion on the phylogeny of Decapodiformes (Mollusca: Cephalopoda). Cladistics 2007, 23, 464–476. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [Green Version]
- Kajihara, H. Redescription of Cerebratulus marginatus auct. (Nemertea: Pilidiophora) from Hokkaido, Japan, as a new species. Zootaxa 2020, 4819, 295–315. [Google Scholar] [CrossRef]
- Chernyshev, A.V.; Polyakova, N.E. Nemerteans collected in the Bering Sea during the research cruises aboard the R/V Akademik, M.A. Lavrentyev in 2016, 2018, and 2021 with an analysis of deep-sea heteronemertean and hoplonemertean species. Deep-Sea Res. II 2022, 199, 105081. [Google Scholar] [CrossRef]
- Hookabe, N.; Schwartz, M.L.; Kajihara, H.; Norenburg, J.L. Molecular systematics of the heteronemertean genus Dushia (Nemertea, Pilidiophora), with descriptions of D. wijnhoffae sp. nov. and D. nigra species complex comb. nov. Zootaxa 2019, 4691, 333–358. [Google Scholar] [CrossRef]
- Hookabe, N.; Kajihara, H. Euborlasia Vaillant, 1890 (Nemertea: Pilidiophora) from Bocas del Toro: Description of a new species, with comments on the systematics of the genus. Mar. Biodivers. 2020, 50, 1–13. [Google Scholar] [CrossRef]
- Kajihara, H. A histology-free description of the branched proboscis ribbon worm Gorgonorhynchus albocinctus sp. nov. (Nemertea: Heteronemertea). Publ. Seto Mar. Biol. Lab. 2015, 43, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Kvist, S.; Laumer, C.E.; Junoy, J.; Giribet, G. New insights into the phylogeny, systematics and DNA barcoding of Nemertea. Invertebr. Syst. 2014, 28, 287–308. [Google Scholar] [CrossRef] [Green Version]
- Hookabe, N.; Xu, C.-M.; Tsuyuki, A.; Jimi, N.; Sun, S.-C.; Kajihara, H. A new nemertean with a branched proboscis, Gorgonorhynchus citrinus sp. nov. (Nemertea: Pilidiophora), with molecular systematics of the genus. Invertebr. Syst. 2021, 35, 350–359. [Google Scholar] [CrossRef]
- Hiebert, T.C. New nemertean diversity discovered in the Northeast Pacific, using surveys of both planktonic larvae and benthic adults. Ph.D. Thesis, University of Oregon, Eugene, OR, USA, 2016. [Google Scholar]
- Kajihara, H.; Ganaha, I.; Kohtsuka, H. Lineid heteronemerteans (Nemertea: Pilidiophora) from Sagami Bay, Japan, with some proposals for the family-level classification system. Zool. Sci. 2022, 39, 62–80. [Google Scholar] [CrossRef]
- Chernyshev, A.V.; Polyakova, N.E.; Turanov, S.V.; Kajihara, H. Taxonomy and phylogeny of Lineus torquatus and allies (Nemertea, Lineidae) with descriptions of a new genus and a new cryptic species. System. Biodivers. 2018, 16, 55–68. [Google Scholar] [CrossRef]
- Chernyshev, A.V.; Polyakova, N.E. Nemerteans from the deep-sea expedition KuramBio II with descriptions of three new hoplonemerteans from the Kuril-Kamchatka Trench. Prog. Oceanogr. 2019, 178, 102148. [Google Scholar] [CrossRef]
- Cherneva, I.A.; Chernyshev, A.V.; Ekimova, I.A.; Polyakova, N.E.; Schepetov, D.M.; Turanov, S.V.; Neretina, T.V.; Chaban, E.M.; Malakhov, V.V. Species identity and genetic structure of nemerteans of the “Lineus ruber-viridis” complex (Müller, 1774) from Arctic waters. Polar Biol. 2019, 42, 497–506. [Google Scholar] [CrossRef]
- Hiebert, T.C.; Maslakova, S. Integrative taxonomy of the Micrura alaskensis Coe, 1901 species complex (Nemertea: Heteronemertea), with descriptions of a new genus Maculaura gen. nov. and four new species from the NE Pacific. Zool. Sci. 2015, 32, 615–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, M.L. Untying a Gordian knot of worms: Systematics and taxonomy of the Pilidiophora (phylum Nemertea). Ph.D. Thesis, George Washington University, Washington, DC, USA, 2009. [Google Scholar]
- Chernyshev, A.V.; Polyakova, N.E.; Hiebert, T.C.; Maslakova, S.A. Evaluation of the taxonomic position of the genus Carinina (Nemertea: Palaeonemertea), with descriptions of two new species. Invertebr. Syst. 2021, 35, 245–260. [Google Scholar] [CrossRef]
- Thollesson, M.; Norenburg, J.L. Ribbon worm relationships: A phylogeny of the phylum Nemertea. Proc. R. Soc. Lond. B 2003, 270, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strand, M.; Sundberg, P. A DNA-based description of a new nemertean (phylum Nemertea) species. Mar. Biol. Res. 2011, 7, 63–70. [Google Scholar] [CrossRef]
- Kajihara, H.; Abukawa, S.; Chernyshev, A.V. Exploring the basal topology of the heteronemertean tree of life: Establishment of a new family, along with turbotaxonomy of Valenciniidae (Nemertea: Pilidiophora: Heteronemertea). Zool. J. Linn. Soc. 2022, 196, 503–548. [Google Scholar] [CrossRef]
- Hubrecht, A.A.W. Report on the Nemertea collected by H.M.S. Challenger during the years 1873-76. In Report on the Scientific Results of the Voyage of H.M.S. Challenger during the Years 1873-76 under the Command of Captain George S. Nares and the Late Captain Frank Tourle Thomson, R.N, Zool; Majesty: Edinburgh, UK, 1887; Volume 19, pp. 1–150. [Google Scholar]
- Chernyshev, A.V. On the higher taxa of the Nemertea, with a review of the subclass Anopla. Zool. Zh. 1995, 74, 7–18. [Google Scholar]
- Chernyshev, A.V. CLSM analysis of the phalloidin stained muscle system of the nemertean proboscis and rhynchocoel. Zool. Sci. 2015, 32, 547–560. [Google Scholar] [CrossRef]
- Friedrich, H. Nemertini. In The Zoology of Iceland; Munksgaard: Copenhagen, Denmark, 1958; Volume 2, pp. 1–24. [Google Scholar]
- Sundberg, P.; Kvist, S.; Strand, M. Evaluating the utility of single-locus DNA barcoding for the identification of ribbon worms (phylum Nemertea). PLoS ONE 2016, 11, e0155541. [Google Scholar] [CrossRef] [Green Version]
- Berg, G. Studies on Nipponnemertes Friedrich (Nemertini, Hoplonemertini). II. Taxonomy of Nipponnemertes pulcher (Johnston) and some other species. Zool. Scr. 1985, 14, 239–246. [Google Scholar] [CrossRef]
- Krämer, D.; Schmidt, C.; Podsiadlowski, L.; Beckers, P.; Horn, L.; von Döhren, J. Unravelling the Lineus ruber/viridis species complex (Nemertea, Heteronemertea). Zool. Scr. 2017, 46, 111–126. [Google Scholar] [CrossRef]



| Specimen | Station | Coordinates | Depth, m | Date |
|---|---|---|---|---|
| Heteronemertes longifissa 11 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Heteronemertes longifissa 20 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Heteronemertes longifissa 25 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Heteronemertes longifissa 103 | 7371 | 61.1977 S. 47.1032 W | 1459 | 8 February 2022 |
| Heteronemertes longifissa 107 | 7371 | 61.1977 S. 47.1032 W | 1459 | 8 February 2022 |
| Heteronemertes longifissa 75 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Heteronemertes longifissa 78 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Heteronemertes longifissa 81 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Parborlasia corrugata 111 | 7371 | 61.1977 S. 47.1032 W | 1459 | 8 February 2022 |
| Cerebratulus sp. Antarctica 28 | 6614 | 60.8862 S. 45.5282 W | 367 | 29 January2020 |
| Lineidae sp. Antarctica 1 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Lineidae sp. Antarctica 5 | 6658 | 61.0387 S. 50.6875 W | 740 | 18 February 2020 |
| Lineidae sp. Antarctica 14 | 6652. | 63.2897 S. 53.6021 W | 364 | 15 February 2020 |
| Lineidae sp. Antarctica 16 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January 2020 |
| Lineidae sp. Antarctica 18 | 6615 | 60.8879 S. 45.5342 W | 370 | 30 January2020 |
| Species | 16S | 18S | 28S | COI | H3 | Source |
|---|---|---|---|---|---|---|
| Apatronemertes albimaculosa | JF277587 | JF293030 | HQ856860 | HQ848584 | JF277733 | [21] |
| Cerebratulus lacteus | JF277575 | JF293044 | HQ856857 | HQ848576 | JF277728 | [21] |
| Cerebratulus marginatus | JF277576 | JF293042 | HQ856858 | HQ848575 | JF277729 | [21] |
| Cerebratulus orochi | LC538101 | LC538103 | LC538104 | LC538102 | LC538105 | [35] |
| Cerebratulus mordukhovichi | OM422971 | OM423090 | OM423029 | OM456681 | OM468125 | [36] |
| Cerebratulus sp. NemBar1383 | – | – | – | KP697728 | – | Strand unpubl. |
| Cerebratulus sp. Antarctica28 | OQ449306 | OQ449292 | OQ449324 | OQ450482 | OQ446609 | Present study |
| Cerebratulus sp. DH-2009 isolate A4pilidia04 | GU227009 | GU227125 | [24] | |||
| Cf. Heteronemertea sp. DH-2009_isolate_D4pilidia04 | GU227013 | – | – | GU227120 | – | [24] |
| Cf. Heteronemertea sp. DH-2009_isolate_A4larva04 | GU227014 | – | – | GU227126 | – | [24] |
| Corsoua takakurai | LC520112 | – | LC520126 | LC520106 | LC520128 | [20] |
| Dushia wijnhoffae | EF124878 | – | EF178494 | EF124967 | – | [37] |
| Dushia cf. nigra | LC389832 | LC389840 | LC389844 | LC389867 | LC389851 | [37] |
| Euborlasia maycoli | LC520114 | LC520121 | LC520125 | LC520108 | – | [38] |
| Gorgonorhynchus albocinctus | – | LC010650 | LC010651 | LC010649 | – | [39] |
| Gorgonorhynchus cf. bermudensis | KF935467 | KF935300 | KF935356 | KF935517 | KF935412 | [40] |
| Gorgonorhynchus cf. repens | LC520115 | LC520122 | LC520123 | LC520105 | LC520131 | [41] |
| Heteronemertea gen. sp. 4 TCH-2015_isolate_119 | KU197548 | – | KU365690 | KU197835 | – | [42] |
| Heteronemertea sp. 17 | LC625672 | LC625688 | LC625699 | LC625640 | LC625729 | [43] |
| Heteronemertes longifissa 11 | – | OQ449293 | OQ449325 | OQ450483 | – | Present study |
| Heteronemertes longifissa 20 | OQ449307 | OQ449294 | – | OQ450484 | OQ446610 | Present study |
| Heteronemertes longifissa 25 | OQ449308 | OQ449295 | OQ449326 | OQ450485 | OQ446611 | Present study |
| Heteronemertes longifissa 75 | – | – | – | OQ450486 | – | Present study |
| Heteronemertes longifissa 78 | OQ449309 | OQ449296 | OQ449327 | OQ450487 | OQ446612 | Present study |
| Heteronemertes longifissa 81 | – | – | – | OQ450488 | – | Present study |
| Heteronemertes longifissa 103 | – | – | – | OQ450489 | – | Present study |
| Heteronemertes longifissa 107 | – | – | – | OQ450490 | – | Present study |
| Hinumanemertes kikuchii * | OQ449310 | OQ449297 | OQ449328 | OQ450491 | OQ446613 | Present study |
| Kulikovia alborostrata | KU821503 | - | KU856679 | KU821529 | KU821552 | [44] |
| Kulikovia manchenkoi | KU821497 | KY468934 | KU856671 | KU821523 | KU821546 | [44] |
| Kulikovia cf. montgomeryi | OM422978 | OM423098 | OM423037 | OM456685 | OM456685 | [36] |
| Kulikovia torguata LtUr1 | KU821486 | KY468935 | KU856673 | KU821511 | KU821534 | [44] |
| Lineidae KuramBio1 12-5 | MN211473 | MN211375 | MN211427 | MN205497 | MN205448 | [45] |
| Lineidae KuramBio2 85 | MN211475 | MN211377 | MN211429 | MN205498 | – | [45] |
| Lineidae KuramBio2 77 | MN211481 | MN211383 | MN211434 | MN205504 | MN205454 | [45] |
| Lineidae sp. 41DS | MF512050 | MF512076 | MF512102 | – | MF512144 | [45] |
| Lineidae sp. Antarctica 1 | OM422984 | OM423104 | OM423043 | OM456687 | OM468138 | [36] |
| Lineidae_sp. Antarctica 5 | OM422988 | OM423108 | – | OM456691 | OM468142 | [36] |
| Lineidae sp. Antarctica 14 | OM422986 | OM423106 | OM423045 | OM456689 | OM468140 | [36] |
| Lineidae sp. Antarctica 16 | OM422987 | OM423107 | OM423046 | OM456690 | OM468141 | [36] |
| Lineidae sp. Antarctica 18 | OM422985 | OM423105 | OM423044 | OM456688 | OM468139 | [36] |
| Lineidae sp. G06 Bering | OM422979 | OM423099 | OM423038 | – | OM468133 | [36] |
| Lineidae sp. H02 IceAGE | OM422982 | OM423102 | OM423041 | – | OM468136 | [36] |
| Lineidae sp. KGK-2 | LC625651 | – | – | LC625624 | LC625709 | [43] |
| Lineidae sp. KGK-4 | LC625653 | LC625683 | – | LC625626 | LC625711 | [43] |
| Lineidae sp. KGK-6 | LC625656 | – | LC625690 | LC625627 | LC625714 | [43] |
| Lineidae sp. KGK-7 | LC625657 | – | LC625691 | – | – | [43] |
| Lineidae sp. KGK-8 | LC625658 | – | – | LC625628 | LC625715 | [43] |
| Lineidae sp. Kuril O11 | OM422989 | OM423109 | OM423047 | OM456692 | OM468143 | [36] |
| Lineus acutifrons | JF277573 | JF304778 | HQ856855 | GU590937 | JF277727 | [21] |
| Lineus clandestinus | MK064103 | OQ449298 | OQ449329 | MK078739 | OQ446614 | [46] present study |
| Lineus flavescens | KP682165 | – | EF178497 | KP682050 | – | [47,48] |
| Lineus lacteus | JF277584 | JF293065 | HQ856850 | HQ848583 | JF277725 | [21] |
| Lineus longissimus | MW073006 | KY468932 | MW077245 | KY561813 | KY606234 | [44,49] |
| Lineus sanquineus | KF935468 | KF935301 | KF935301 | KF935518 | KF935413 | [40] |
| Lineus ruber | MK064093 | KY468933 * | KY468929 * | MK078684 | KY606235 * | [44,46] |
| Lineus sp. Guam | KU821507 | – | KY468928 | – | KY561818 | [44] |
| Lineus viridis | MK064101 | OQ449299 | OQ449330 | MK078733 | OQ446615 | [46] present study |
| Maculaura aquilonia ** | OQ449311 | OQ449300 | OQ449331 | OQ450492 | OQ446616 | Present study |
| Maculaura sp. * | – | OQ449301 | OQ449332 | OQ450493 | OQ446617 | Present study |
| Micrura bathyalis | MN211479 | MN211381 | MN211432 | MN205502 | – | [45] |
| Micrura bella *** | OQ449312 | OQ449302 | OQ449333 | OQ450494 | OQ446618 | Present study |
| Micrura callima | MN211472 | MN211374 | MN211426 | MN205496 | MN205447 | [45] |
| Micrura chlorapardalis | KF935459 | KF935292 | KF935348 | KF935512 | KF935404 | [40] |
| Micrura dellechiajei | KF935461 | KF935294 | KF935350 | KF935514 | KF935406 | [40] |
| Micrura fasciolata | JF277585 | JF293038 | HQ856846 | HQ848578 | JF277721 | [21] |
| Micrura ignea **** | OQ449313 | OQ449303 | OQ449334 | OQ450495 | OQ446619 | Present study |
| Micrura purpurea | JF277577 | JF293036 | HQ856845 | HQ848586 | JF277726 | [21] |
| Micrura rubramaculosa | KF935460 | KF935293 | KF935349 | KF935513 | KF935405 | [40] |
| Micrura sp. albocephala | KU197574 | – | KU365712 | KU197849 | – | [42] |
| Micrura sp. dark | KU197586 | – | KU365713 | KU197858 | – | [42] |
| Micrura sp. 3 | KU197563 | – | KU365710 | KU197841 | – | [42] |
| Micrura sp. 4 | KU197581 | – | KU365711 | KU197857 | – | [42] |
| Micrura sp. IceAGE | OM422994 | OM423114 | OM423052 | OM456696 | OM468146 | [36] |
| Micrura sp. IZ 132532 | KF935457 | KF935290 | KF935346 | KF935510 | KF935402 | [42] |
| Micrura sp. IZ 132529 | KF935458 | KF935291 | KF935347 | KF935511 | KF935403 | [40] |
| Micrura verrilli | KF935455 | KF935288 | KF935344 | KF935508 | KF935400 | [40] |
| Micrura wilsoni | KU197535 | – | KU365716 | KU197827 | – | [42] |
| Notospermus geniculatus | KF935462 | KF935295 | KF935351 | – | KF935407 | [40] |
| Notospermus mitellatus | LC625660 | LC625685 | LC625693 | LC625629 | LC625717 | [43] |
| Nipponomicrura sp.* | OQ449314 | OQ449304 | OQ449335 | OQ450496 | OQ446620 | Present study |
| Nipponomicrura uchidai | KU821509 | – | KY468930 | KY561815 | KY561819 | [44] |
| Parborlasia corrugata | JF277578 | JF293037 | HQ856851 | EU194826 * | JF277732 | [21,22] |
| Parborlasia corrugata 111 | OQ449315 | OQ449305 | OQ449336 | OQ450497 | OQ446621 | Present study |
| Parvicirrus dubius | AJ436830 | – | AJ436885 | AJ436940 | – | [50] |
| Polydendrorhynchus zhanjiangensis | MT659662 | MT648831 | MT648832 | MT648511 | MT655749 | [41] |
| Pseudomicrura afzelii | GU445914 | GU445924 | GU445919 | GU392013 | – | [51] |
| Riseriellus occultus | JF277581 | JF293031 | HQ856848 | HQ848581 | JF277724 | [21] |
| Siphonenteron nakanoi | LC625678 | – | LC625706 | LC625646 | LC625737 | [43] |
| Tenuilineus bicolor | AJ436823 | – | EF124960 | AJ436933 | AJ436980 | [48,50] |
| Zygeupolia rubens | JF277574 | JF293045 | HQ856861 | HQ848585 | JF277735 | [21] |
| Yininemertes pratensis | KY274025 | KY274047 | KY274069 | KY274003 | KY274091 | [40] |
| Outgroups | ||||||
| Baseodiscus mexicanus | KF935449 | KF935281 | KF935337 | KF935503 | KF935393 | [40] |
| Sonnenemertes cantelli | MF512048 | MF512073 | MF512099 | MF512118 | MF512141 | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernyshev, A.V.; Polyakova, N.E. Distribution and Phylogenetic Position of the Antarctic Ribbon Worm Heteronemertes longifissa (Nemertea, Pilidiophora). Water 2023, 15, 809. https://doi.org/10.3390/w15040809
Chernyshev AV, Polyakova NE. Distribution and Phylogenetic Position of the Antarctic Ribbon Worm Heteronemertes longifissa (Nemertea, Pilidiophora). Water. 2023; 15(4):809. https://doi.org/10.3390/w15040809
Chicago/Turabian StyleChernyshev, Alexei V., and Neonila E. Polyakova. 2023. "Distribution and Phylogenetic Position of the Antarctic Ribbon Worm Heteronemertes longifissa (Nemertea, Pilidiophora)" Water 15, no. 4: 809. https://doi.org/10.3390/w15040809
APA StyleChernyshev, A. V., & Polyakova, N. E. (2023). Distribution and Phylogenetic Position of the Antarctic Ribbon Worm Heteronemertes longifissa (Nemertea, Pilidiophora). Water, 15(4), 809. https://doi.org/10.3390/w15040809





