Blue Shark (Prionace glauca) Distribution in the Pacific Ocean: A Look at Continuity and Size Differences
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
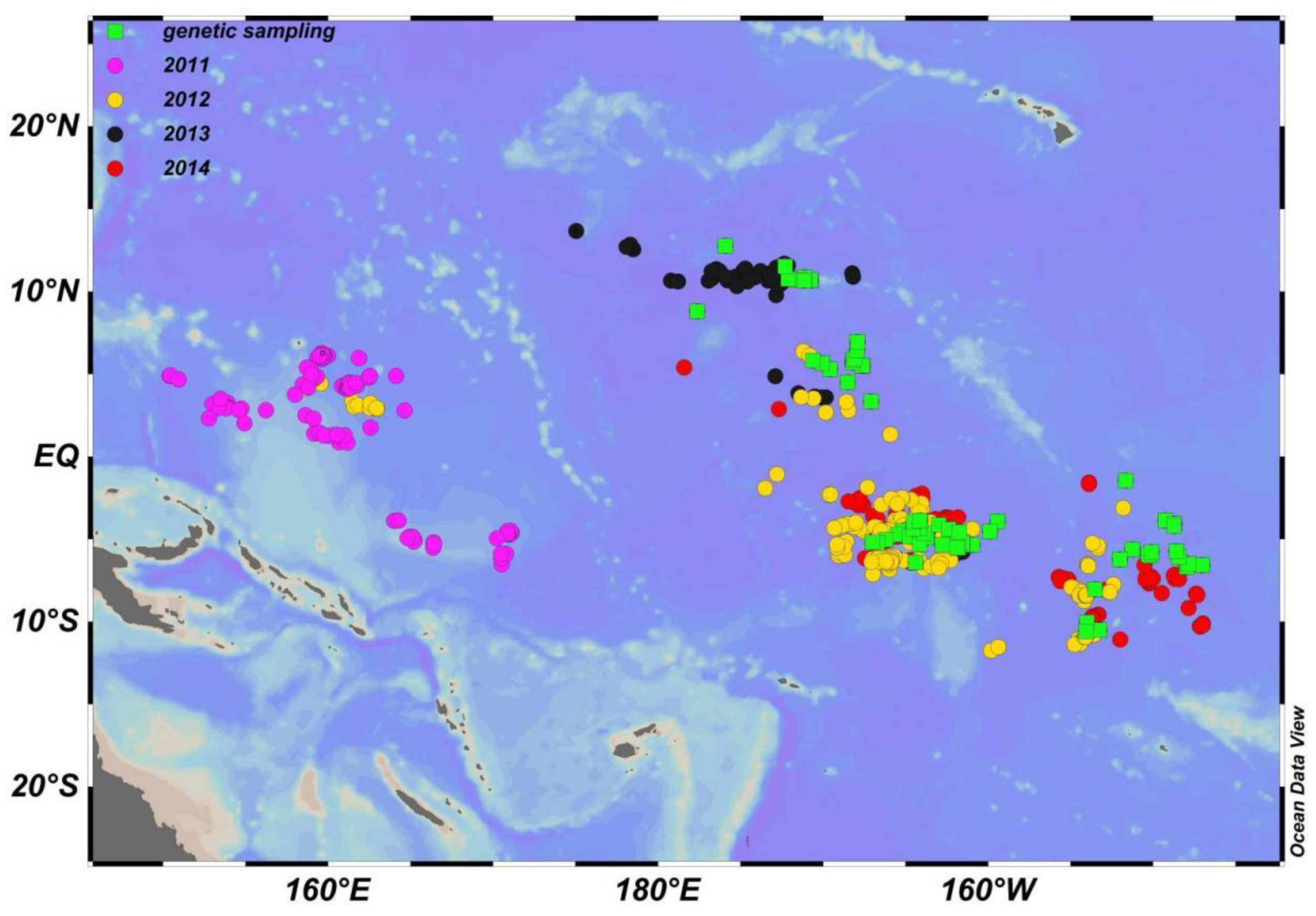
2.2. Survey Description
2.3. Biological Data Collected
2.4. Genetic Samples Collected
2.5. Genetic Marker Design and Polymerase Chain Reaction (PCR) Amplification
2.6. Genetic Data Analysis
2.7. GAM Analysis
3. Results
3.1. Population Genetics Structure and Population Expansion
3.2. Modeling Results
3.2.1. Model Selection
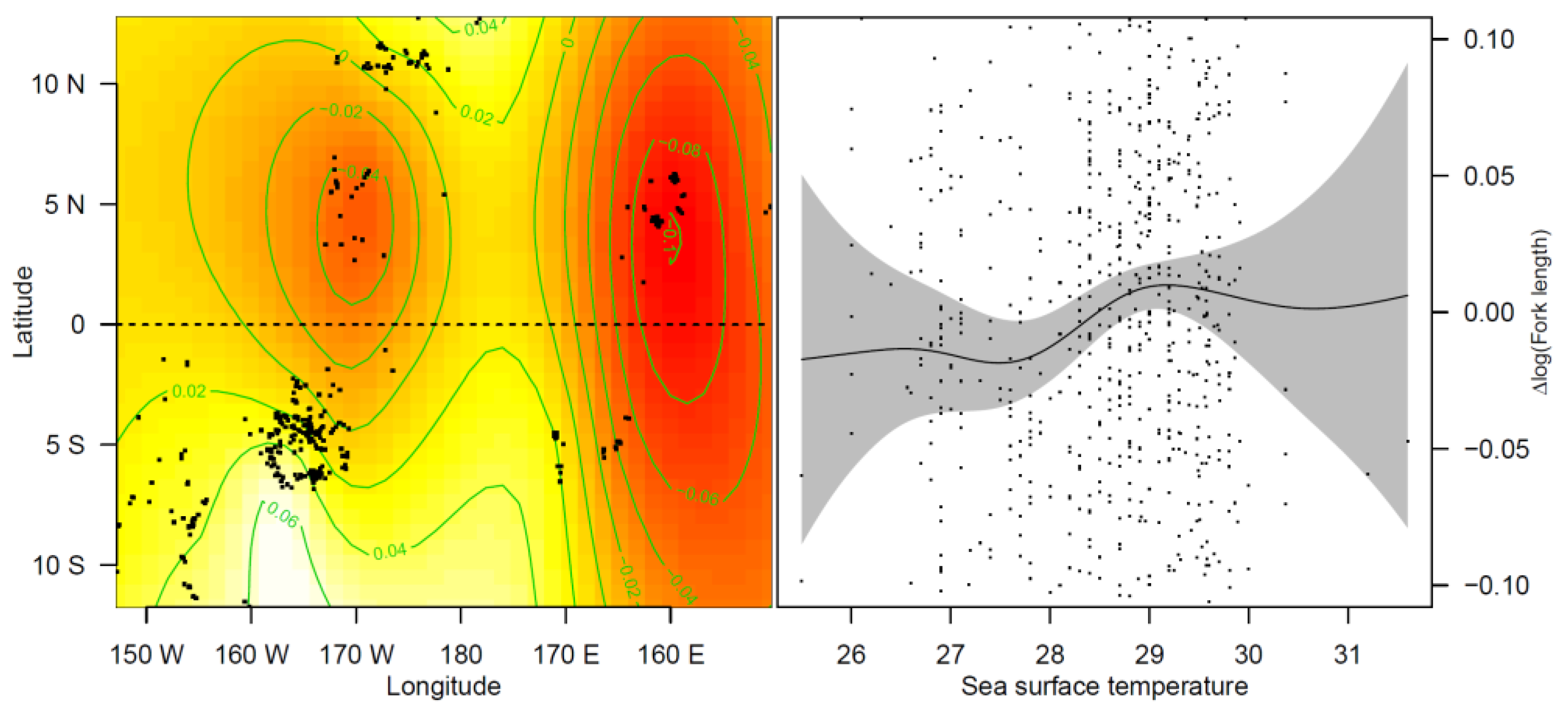
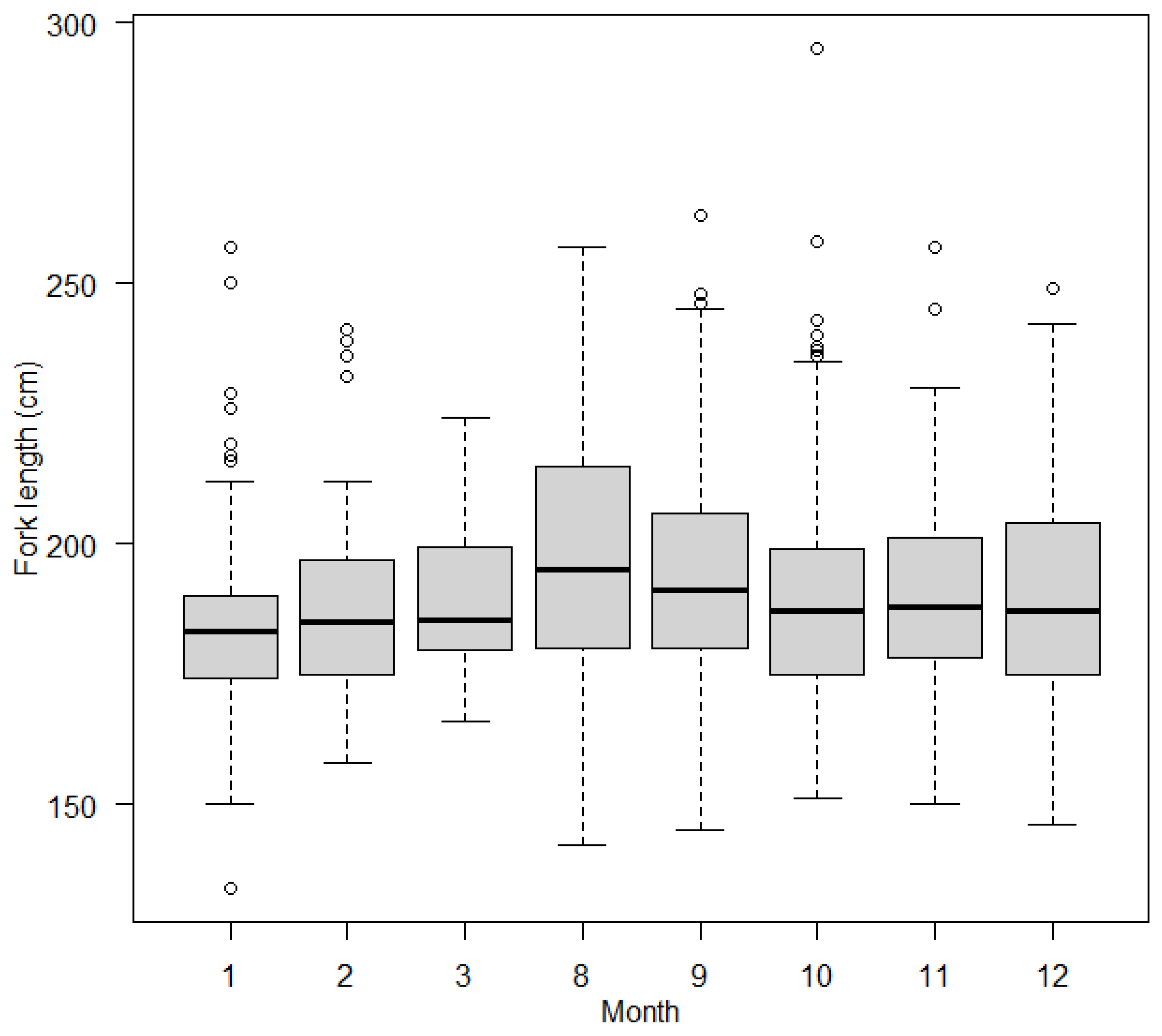
3.2.2. Effects of Covariates on FL in Blue Shark Individuals
4. Discussion
4.1. SST May Explain Body Size Differences within Sampling Areas
4.2. Age-Dependent Migration and Sex Segregation May Explain Body Size Differences within Sampling Areas
4.3. Limited Geographic Range and Sample Size for Genetic Data May Have Biased the Results
4.4. Divergence of the COI Gene May Be Too Recent to Identify Differences in Genetic Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakano, H.; Stevens, J.D. The Biology and Ecology of the Blue Shark, Prionace glauca. In Sharks of the Open Ocean: Biology, Fisheries and Conservation; Camhi, M.D., Pikitch, E.K., Babcock, E.A., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2008; pp. 140–151. [Google Scholar]
- Li, W.W.; Kindong, R.; Wu, F.; Tian, S.Q.; Dai, X.J. Catch rate and stock status of blue shark in the Pacific Ocean inferred from fishery-independent data. Indian J. Geo Mar. Sci. 2020, 49, 543–547. [Google Scholar]
- Myers, R.A.; Baum, J.K.; Shepherd, T.D.; Powers, S.P.; Peterson, C.H. Cascading effects of the loss of apex predatory sharks from a coastal ocean. Science 2007, 315, 1846–1850. [Google Scholar] [CrossRef]
- Ferretti, F.; Worm, B.; Britten, G.L.; Heithaus, M.R.; Lotze, H.K. Patterns and ecosystem consequences of shark declines in the ocean. Ecol. Lett. 2010, 13, 1055–1071. [Google Scholar] [CrossRef] [PubMed]
- Shea, B.D.; Coulter, S.K.; Dooling, K.E.; Isihara, H.L.; Roth, J.C.; Sudal, E.; Donovan, D.J.; Hoopes, L.A.; Dove, A.D.; Cooke, S.J.; et al. Recreational fishing fight times are not correlated with physiological status of blue sharks (Prionace glauca) in the northwestern Atlantic. Fish. Res. 2022, 248, 106220. [Google Scholar] [CrossRef]
- Tsai, W.P.; Sun, C.L.; Liu, K.M.; Wang, S.B.; Lo, N.C. CPUE standardization and catch estimate of blue shark by Taiwanese large-scale tuna longline fishery in the North Pacific Ocean. J. Mar. Sci. Tech. 2015, 23, 567–574. [Google Scholar]
- Sabarros, P.; Coelho, R.; Romanov, E.V.; Guillon, N.; Bach, P. Updated Standardized CPUE of Blue Shark Bycaught by the French Reunion-Based Pelagic Longline Fishery (2007-2020); IOTC-2021-WPEB17(DP)-08; Indian Ocean Tuna Commission Seychelles. 2021. Available online: https://archimer.ifremer.fr/doc/00690/80162/83227.pdf (accessed on 8 September 2021).
- Rice, J.; Harley, S.; Maunder, M.; Da-Silva, A. Stock Assessment of Blue Shark in the North Pacific Ocean Using Stock Synthesis; WCPFC-SC9-2013/SA-WP-02; Western and Central Pacific Fisheries Commission: Palikir, Micronesia, 2013. [Google Scholar]
- Taguchi, M.; King, J.R.; Wetklo, M.; Withler, R.E.; Yokawa, K. Population genetic structure and demographic history of Pacific blue sharks (Prionaceglauca) inferred from mitochondrial DNA analysis. Mar. Freshw. Res. 2015, 66, 267–275. [Google Scholar] [CrossRef]
- Sippel, T.; Wraith, J.; Kohin, S.; Taylor, V.; Holdsworth, J.; Taguchi, M.; Matsunaga, H.; Yokawa, K. A summary of blue shark (Prionaceglauca) and shortfin mako shark (Isurusoxyrinchus) tagging data available from the North and Southwest Pacific Ocean. In Proceedings of the ISC/11/SHARKWG-2/04 Working Document Submitted to the ISC Shark Working Group Workshop, La Jolla, CA, USA, 28 November–3 December 2011. [Google Scholar]
- Stevens, J.D.; Bradford, R.W.; West, G.J. Satellite tagging of blue sharks (Prionaceglauca) and other pelagic sharks off eastern Australia: Depth behavior, temperature experience and movements. Mar. Biol. 2010, 157, 575–591. [Google Scholar] [CrossRef]
- Cardeñosa, D.; Hyde, J.; Caballero, S. Genetic Diversity and Population Structure of the Pelagic Thresher Shark (Alopiaspelagicus) in the Pacific Ocean: Evidence for Two Evolutionarily Significant Units. PLoS ONE 2014, 9, e110193. [Google Scholar] [CrossRef]
- Schrey, A.W.; Heist, E.J. Microsatellite markers for the shortfin mako and cross-species amplification in lamniformes. Conserv. Genet. 2002, 3, 459–461. [Google Scholar] [CrossRef]
- Hoelzel, A.R.; Shivji, M.S.; Magnussen, J.; Francis, M.P. Low worldwide genetic diversity in the basking shark (Cetorhinusmaximus). Biol. Lett. 2006, 2, 639–642. [Google Scholar] [CrossRef]
- Schmidt, J.V.; Schmidt, C.L.; Ozer, F.; Ernst, R.E.; Feldheim, K.A.; Ashley, M.V. Low genetic differentiation across three major ocean populations of the whale shark, Rhincodon typus. PLoS ONE 2009, 4, e4988. [Google Scholar] [CrossRef] [PubMed]
- Ovenden, J.R.; Kashiwagi, T.; Broderick, D.; Giles, J.; Salini, J. The extent of population genetic subdivision differs among four co-distributed shark species in the Indo-Australian archipelago. BMC Evol. Biol. 2009, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Montealegre-Quijano, S.; Vooren, C.M. Distribution and abundance of the life stages of the blue shark Prionaceglauca in the Southwest Atlantic. Fish. Res. 2010, 101, 168–179. [Google Scholar] [CrossRef]
- Pichel, W.G.; Churnside, J.H.; Veenstra, T.S.; Foley, D.G.; Friedman, K.S.; Brainard, R.E. Marine debris collects within the North Pacific subtropical convergence zone. Mar. Pollut. Bull. 2007, 54, 1207–1211. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, S.M.; Scales, K.L.; Bograd, S.J.; Briscoe, D.K.; Dewar, H.; Hazen, E.L.; Lewison, R.L.; Welch, H.; Crowder, L.B. Seasonal spatial segregation in blue sharks (Prionace glauca) by sex and size class in the Northeast Pacific Ocean. Divers. Distrib. 2019, 25, 1304–1317. [Google Scholar] [CrossRef]
- Vandeperre, F.; Aires-da-Silva, A.; Fontes, J.; Santos, M.; Serrão Santos, R.; Afonso, P. Movements of blue sharks (Prionace glauca) across their life history. PLoS ONE 2014, 9, e103538. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, Y.; Semba, Y.; Tanaka, S. Age determination and growth of the blue shark (Prionace glauca) in the western North Pacific Ocean. Fish. Bull. 2019, 117, 107–120. [Google Scholar] [CrossRef]
- Nakano, H. Age, reproduction and migration of blue shark [Prionace glauca] in the North Pacific Ocean. Bull. Natl. Res. Inst. Far Seas Fish. 1994, 31, 141–256. [Google Scholar]
- Fei, H.; Zhang, S. Fishery Resource Science; Science and Technology Press: Beijing, China, 1990; pp. 114–285, in press. (In Chinese) [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneide, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47. [Google Scholar] [CrossRef]
- Rozas, J.; Sanchez-De, I.; Barrio, J.C.; Messeguer, X.; Rozas, R. Dna SP, DNA polymorphism analysesby the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Jakobsen, I.B.; Nei, M. Mega4: Molecular evolutionary genetics analysis (mega) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Stelbrink, B.; von Rintelen, T.; Cliff, G.; Kriwet, J. Molecular systematics and global phylogeography of angel sharks (genus Squatina). Mol. Phylogenet. Evol. 2010, 54, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.N. Stable and efficient multiple smoothing parameter estimation for generalized additive models. J. Am. Stat. Assoc. 2012, 467, 673–686. [Google Scholar] [CrossRef]
- Wood, S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 2011, 73, 3–36. [Google Scholar] [CrossRef]
- Bozdogan, H. Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika 1987, 52, 345–370. [Google Scholar] [CrossRef]
- Li, W.; Dai, X.; Zhu, J.; Tian, S.; He, S.; Wu, F. Genetic differentiation in blue shark, Prionaceglauca from the central Pacific Ocean, as inferred by Mitochondrial Cytochrome b region. Mitochondrial DNA Part A 2016, 28, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Bailleul, D.; Mackenzie, A.; Sacchi, O.; Poisson, F.; Bierne, N.; Arnaud-Haond, S. Large-scale genetic panmixia in the blue shark (Prionace glauca): A single worldwide population, or a genetic lag-time effect of the “grey zone” of differentiation? Evol. Appl. 2018, 11, 614–630. [Google Scholar] [CrossRef]
- Skomal, G.B.; Natanson, L.J. Age and growth of the blue shark (Prionace glauca) in the North Atlantic Ocean. Fish. Bull. 2003, 101, 627–639. [Google Scholar]
- Campana, S.E.; Dorey, A.; Fowler, M.; Joyce, W.; Wang, Z.; Wright, D.; Yashayaev, L. Migration pathways, behavioural thermoregulation and overwintering grounds of blue sharks in the Northwest Atlantic. PLoS ONE 2011, 6, e16854. [Google Scholar] [CrossRef] [PubMed]
- Thresher, R.E.; Koslow, J.A.; Morison, A.K.; Smith, D.C. Depth-mediated reversal of the effects of climate change on long-term growth rates of exploited marine fish. Proc. Natl. Acad. Sci. USA 2007, 104, 7461–7465. [Google Scholar] [CrossRef] [PubMed]
- Ohlberger, J. Climate warming and ectotherm body size–from individual physiology to community ecology. Funct. Ecol. 2013, 27, 991–1001. [Google Scholar] [CrossRef]
- Crawshaw, L.I.; Hammel, H.T. Behavioral temperature regulation in the California horn shark, Heterodontus francisci. Brain Behav. Evol. 1973, 7, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Nagasawa, K. Distribution of Pelagic Elasmobranchs Caught by Salmon Research Gillnets in the North Pacific. Fish. Sci. 1996, 62, 860–865. [Google Scholar] [CrossRef]
- Kohler, N.E.; Turner, P.A.; Hoey, J.; Natanson, L.J.; Briggs, R. Tag and recapture data for three pelagic shark species: Blue shark (Prionaceglauca), shortfin mako (Isurusoxyrinchus), and porbeagle (Lamnanasus) in the North Atlantic Ocean. Int. Comm. Conserv. Atl. Tunas 2002, 54, 1231–1260. [Google Scholar]
- Karnauskas, K.B.; Seager, R.; Kaplan, A.; Kushnir, Y.; Cane, M.A. Observed Strengthening of the Zonal Sea Surface Temperature Gradient across the Equatorial Pacific Ocean. J. Clim. 2009, 22, 4316–4321. [Google Scholar] [CrossRef]
- Brett, J.R. Environmental Factors and Growth. Fish Physiol. 1979, 8, 599–675. [Google Scholar]
- Nakicenovic, N.; Alcamo, J.; Davis, G.; de Vries, B.; Fenhann, J.; Gaffin, S. Special Report on Emissions Scenarios: A Special Report of Working Group III of the Intergovernmental Panel on Climate Change; Cambridge University Press: New York, NY, USA, 2000. Available online: http://www.osti.gov/scitech/servlets/purl/15009867 (accessed on 10 February 2022).
- Ganachaud, A.S.; Gupta, A.S.; Orr, J.C.; Wijffels, S.E.; Ridgway, K.R.; Hemer, M.A.; Maes, C.; Steinberg, C.R.; Tribollet, A.D.; Qiu, B.; et al. Observed and expected changes to the tropical Pacific Ocean. In Vulnerability of Tropical Pacific Fisheries and Aquaculture to Climate Change; Bell, J., Johnson, J.E., Hobday, A.J., Eds.; Secretariat of the Pacific Community: Noumea, France, 2011; pp. 115–202. [Google Scholar]
- Fernandes, J.A.; Cheung, W.W.L.; Jennings, S.; Butenschon, M.; Mora, L.; Frölicher, T.L.; Barange, M.; Grant, A. Modelling the effects of climate change on the distribution and production of marine fishes: Accounting for trophic interactions in a dynamic bioclimate envelope model. Glob. Chang. Biol. 2013, 19, 2596–2607. [Google Scholar] [CrossRef]
- Chen, Y. Fisheries management in a changing climate: Lessons from the 2012 ocean heat wave in the Northwest Atlantic. Oceanography 2013, 26, 191–195. [Google Scholar]
- Ojea, E.; Lester, S.E.; Salgueiro-Otero, D. Adaptation of fishing communities to climate-driven shifts in target species. One Earth 2020, 2, 544–556. [Google Scholar] [CrossRef]
- Szuwalski, C.S.; Punt, A.E. Fisheries management for regime-based ecosystems: A management strategy evaluation for the snow crab fishery in the eastern Bering Sea. ICES J. Mar. Sci. 2013, 70, 955–967. [Google Scholar] [CrossRef]
- Kell, L.T.; Pilling, G.M.; O’Brien, C.M. Implications of climate change for the management of North Sea cod (Gadusmorhua). ICES J. Mar. Sci. 2005, 62, 1483–1491. [Google Scholar] [CrossRef]
- Nakano, H.; Seki, M.P. Synopsis of biological data on the blue shark, Prionaceglauca Linnaeus. Bull. -Fish. Res. Agency Jpn. 2003, 6, 18–55. [Google Scholar]
- McKinnell, S.; Seki, M.P. Shark bycatch in the Japanese high seas squid driftnet fishery in the North Pacific Ocean. Fish. Res. 1998, 39, 127–138. [Google Scholar] [CrossRef]
- Waples, R.S.; Punt, A.E.; Cope, J.M. Integrating genetic data into management of marine resources: How can we do it better? Fish Fish. 2008, 9, 423–449. [Google Scholar] [CrossRef]
- Fogelqvist, J.; Niittyvuopio, A.; Ågren, J.; Savolainen, O.; Lascoux, M. Cryptic population genetic structure: The number of inferred clusters depends on sample size. Mol. Ecol. Resour. 2010, 10, 314–323. [Google Scholar] [CrossRef]
- Shen, L.; Er, M.J.; Liu, W.; Fan, Y.; Yin, Q. Population structure-learned classifier for high-dimension low-sample-size class-imbalanced problem. Eng. Appl. Artif. Intell. 2022, 111, 104828. [Google Scholar] [CrossRef]
- Ward, R.D.; Gardner, M.G. Stock Structure and Species Identification of School and Gummy Sharks in Australasian Waters; Report of FRRF 93/11 & FRDC 93/64; CSIRO Marine Research: Hobart, Australia, 1997; p. 92. [Google Scholar]
- Punt, A.E.; Pribac, F.; Walker, T.I.; Taylor, B.L.; Prince, J.D. Stock assessment of school shark, Galeorhinusgaleus, based on a spatially explicit population dynamics model. Mar. Freshw. Res. 2000, 51, 205–220. [Google Scholar] [CrossRef]
- Rogers, L.A.; Griffin, R.; Young, T.; Fuller, E.; St Martin, K.; Pinsky, M.L. Shifting habitats expose fishing communities to risk under climate change. Nat. Clim. Chang. 2019, 9, 512–516. [Google Scholar] [CrossRef]
- Hellberg, M.E.; Balch, D.P.; Roy, K. Climate-driven range expansion and morphological evolution in a marine gastropod. Science 2001, 292, 1707–1710. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.G.; Mosley-Thompson, E.; Davis, M.E. Late glacial stage and Holocene tropical ice core records from Huascaran, Peru. Science 1995, 269, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Maloney, S.K.; Fuller, A.; Mitchell, D. Climate change: Is the dark Soay sheep endangered? Biol. Lett. 2009, 5, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Sgrò, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Mildenberger, T.K.; Berg, C.W.; Kokkalis, A.; Hordyk, A.R.; Wetzel, C.; Jacobsen, N.S.; Punt, A.E.; Nielsen, J.R. Implementing the precautionary approach into fisheries management: Biomass reference points and uncertainty buffers. Fish Fish 2022, 23, 73–92. [Google Scholar] [CrossRef]
- Hilborn, R.; Maguire, J.J.; Parma, A.M.; Rosenberg, A.A. The Precautionary Approach and risk management: Can they increase the probability of successes in fishery management? Can. J. Fish. Aquat. Sci. 2001, 58, 99–107. [Google Scholar] [CrossRef]
- Cadrin, S.X. Defining spatial structure for fishery stock assessment. Fish. Res. 2020, 221, 105397. [Google Scholar] [CrossRef]






| Model Equation | Model Number | Df | AIC |
|---|---|---|---|
| FL~s(LON,LAT) + s(SST) + FeedL + Month + Sex + Gen | Model 1 | 16.000407 | −123.6367 |
| FL~s(LON,LAT) + s(SST) + FeedL + Month + Sex | Model 2 | 32.604487 | −1230.5702 |
| FL~s(LON,LAT) + s(SST) + FeedL + Sex + Gen | Model 3 | 13.000100 | −129.2055 |
| FL~s(LON,LAT) + s(SST) + Month + Sex + Gen | Model 4 | 12.000079 | −131.2627 |
| FL~s(LON,LAT) + FeedL + Month + Sex + Gen | Model 5 | 15.000729 | −124.6096 |
| FL~s(LON,LAT) + Sex + s(SST) + FeedL | Model 6 | 12.000188 | −128.8619 |
| FL~s(LON,LAT) + Sex + s(SST) + Month | Model 7 | 19.798894 | −1237.8019 |
| FL~s(LON,LAT) + Sex + s(SST) + Gen | Model 8 | 9.000123 | −136.7627 |
| FL~s(LON,LAT) + Sex + FeedL + Month | Model 9 | 26.060133 | −1235.1461 |
| FL~s(LON,LAT) + Sex + FeedL + Gen | Model 10 | 12.000188 | −128.8619 |
| FL~s(LON,LAT) + Sex + Month + Gen | Model 11 | 11.000774 | −132.1159 |
| FL~s(LON,LAT) + Sex + s(SST) | Model 12 | 24.377982 | −1579.5387 |
| FL~s(LON,LAT) + Sex + FeedL | Model 13 | 18.849146 | −1235.4664 |
| FL~s(LON,LAT) + Sex + Month | Model 14 | 26.031117 | −1580.4761 |
| FL~s(LON,LAT) + Sex + Gen | Model 15 | 8.000042 | −136.4100 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation | Fixation Index |
|---|---|---|---|---|---|
| Among population | 2 | 1.455 | 0.00935 Va | 2.16 | 0.02164 |
| Within population | 95 | 40.137 | 0.42249 Vb | 97.84 |
| CPA | CPB | CPC | |
|---|---|---|---|
| CPA | |||
| CPB | 0.0122 | ||
| CPC | 0.06737 | −0.0174 |
| Region | τ | θ0 | θ1 | FU’S | P(FU’S) |
|---|---|---|---|---|---|
| CPA | 1.15234 | 0.00000 | 99999.00000 | <0.0000 | 0.0000 |
| CPB | 0.93359 | 0.00000 | 99999.00000 | <0.0000 | 0.0000 |
| CPC | 0.76953 | 0.00000 | 99999.00000 | <0.0000 | 0.0000 |
| Total | 0.95182 | 0.00000 | 99999.00000 | <0.0000 | 0.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Dai, X.; Staples, K.W.; Chen, B.; Huang, H.; Tian, S. Blue Shark (Prionace glauca) Distribution in the Pacific Ocean: A Look at Continuity and Size Differences. Water 2023, 15, 1324. https://doi.org/10.3390/w15071324
Li W, Dai X, Staples KW, Chen B, Huang H, Tian S. Blue Shark (Prionace glauca) Distribution in the Pacific Ocean: A Look at Continuity and Size Differences. Water. 2023; 15(7):1324. https://doi.org/10.3390/w15071324
Chicago/Turabian StyleLi, Weiwen, Xiaojie Dai, Kevin W. Staples, Bin Chen, Hao Huang, and Siquan Tian. 2023. "Blue Shark (Prionace glauca) Distribution in the Pacific Ocean: A Look at Continuity and Size Differences" Water 15, no. 7: 1324. https://doi.org/10.3390/w15071324
APA StyleLi, W., Dai, X., Staples, K. W., Chen, B., Huang, H., & Tian, S. (2023). Blue Shark (Prionace glauca) Distribution in the Pacific Ocean: A Look at Continuity and Size Differences. Water, 15(7), 1324. https://doi.org/10.3390/w15071324






