Comprehensive Profiling of Klebsiella in Surface Waters from Northern Portugal: Understanding Patterns in Prevalence, Antibiotic Resistance, and Biofilm Formation
Abstract
1. Introduction
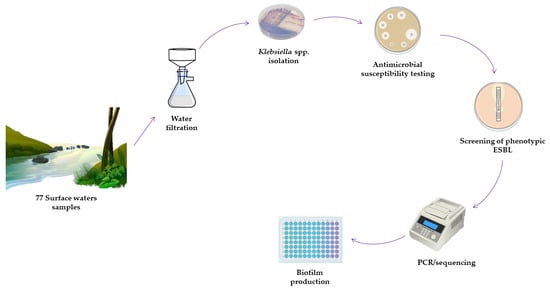
2. Materials and Methods
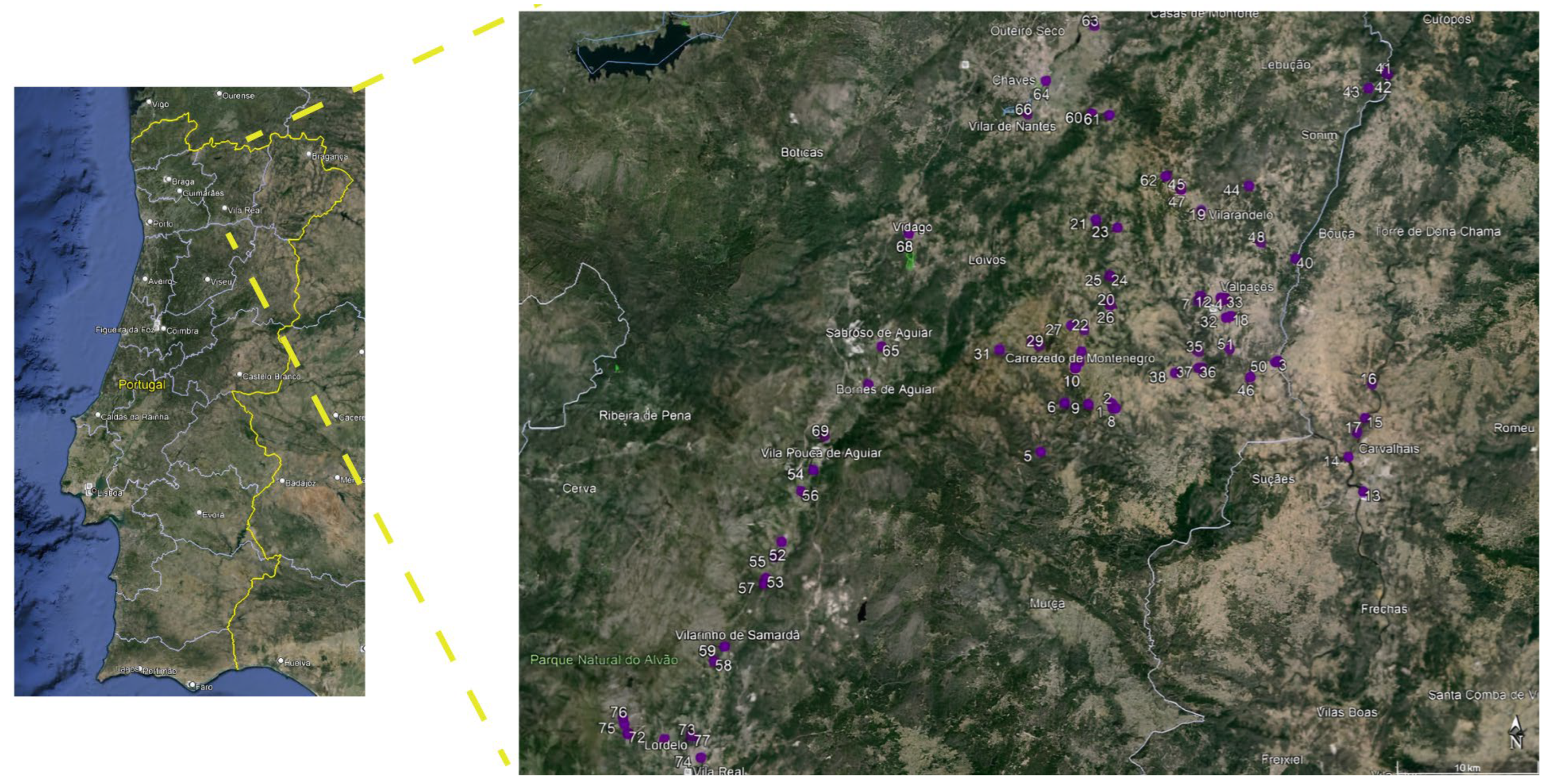
2.1. Geographical Location and Sample Collection
2.2. Bacterial Isolation
2.3. Antimicrobial Susceptibility/Resistance Assessment
2.4. Antimicrobial Resistance Genes and Virulence Factors
2.5. Biofilm Production
2.6. Statistical Analysis
3. Results
3.1. Distribution of Klebsiella in Surface Waters
3.2. Antimicrobial Resistance Phenotype in Klebsiella spp. and K. pneumoniae Strains
3.3. Characterization of Resistance Genes and Virulence Factors
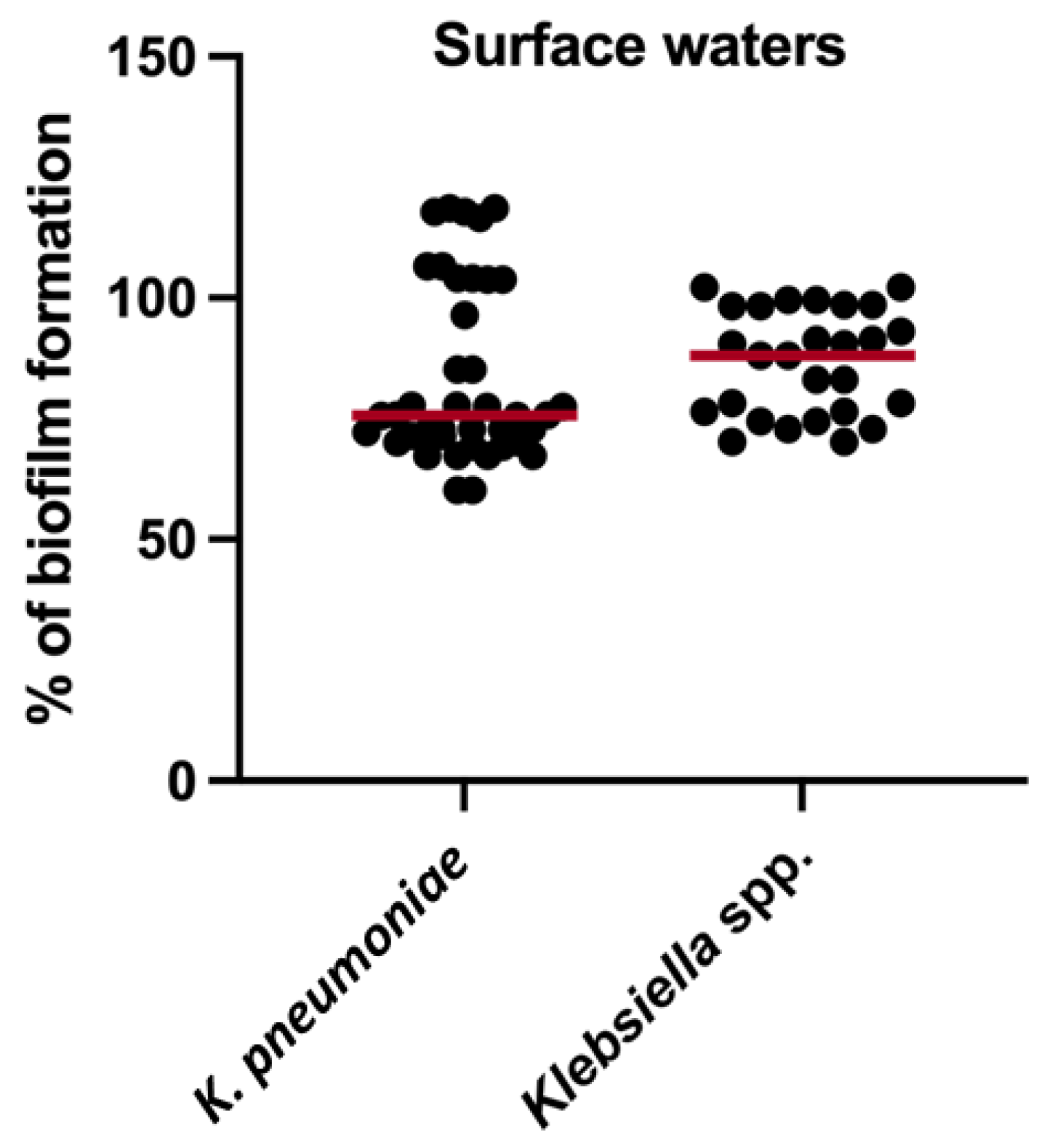
3.4. Biofilm Production
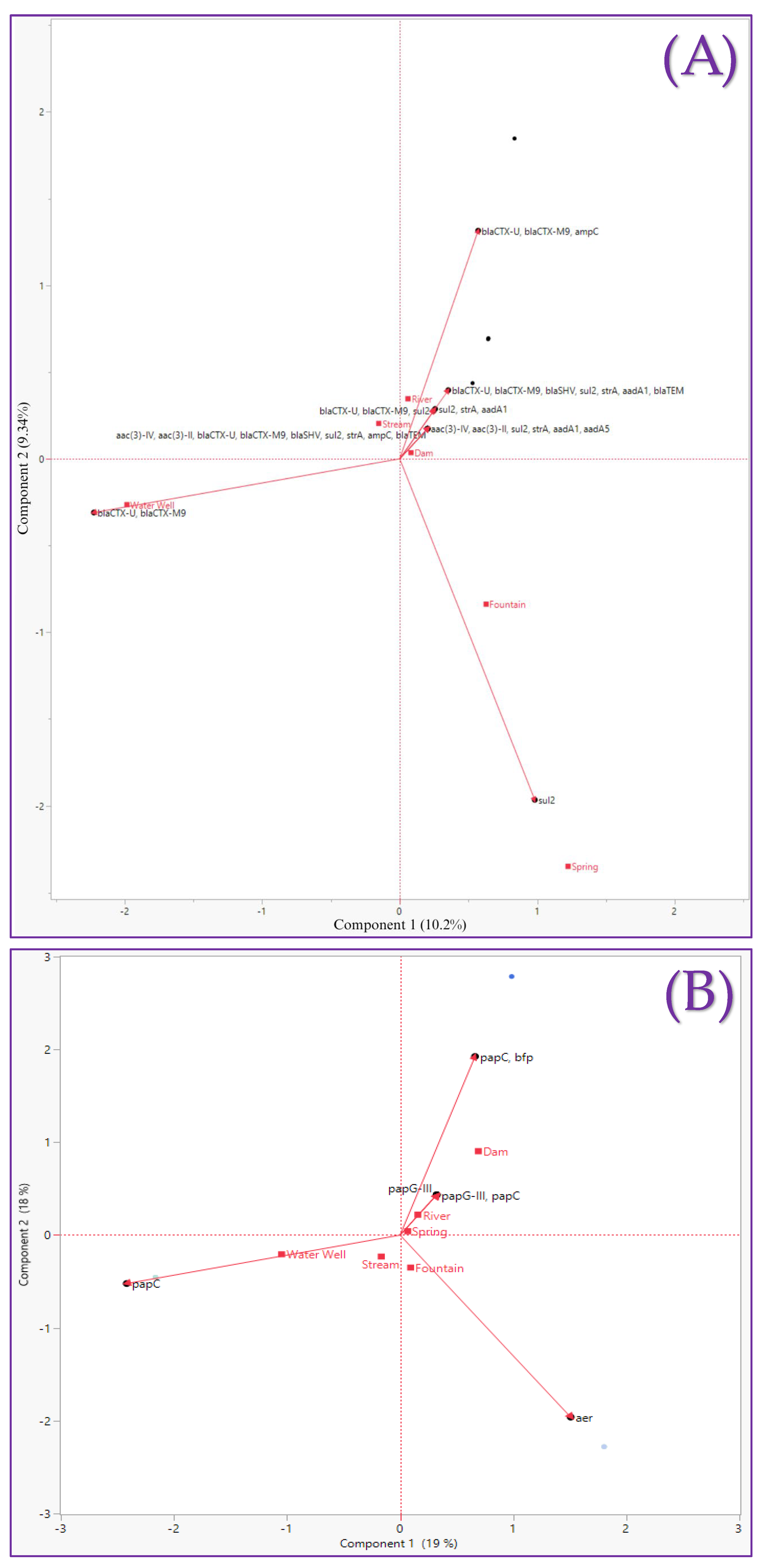
3.5. Principal Component Analysis (PCA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online: https://iiif.wellcomecollection.org/file/b28552179_AMR%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations.pdf (accessed on 19 February 2024).
- Varaldo, P.E.; Facinelli, B.; Bagnarelli, P.; Menzo, S.; Mingoia, M.; Brenciani, A.; Giacometti, A.; Barchiesi, F.; Brescini, L.; Cirioni, O.; et al. Antimicrobial Resistance: A Challenge for the Future. In The First Outstanding 50 Years of “Universita Politecnica delle Marche”: Research Achievements in Life Sciences; Springer: Cham, Switzerland, 2020; pp. 13–29. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One Health—Its Importance in Helping to Better Control Antimicrobial Resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Dagot, C.; Chesneau, O.; Bibbal, D.; Labanowski, J.; Vialette, M.; Bouchard, D.; Martin-Laurent, F.; Calsat, L.; Nazaret, S.; et al. Environmental Contamination in a High-Income Country (France) by Antibiotics, Antibiotic-Resistant Bacteria, and Antibiotic Resistance Genes: Status and Possible Causes. Environ. Int. 2022, 159, 107047. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling Antibiotic Resistance: The Environmental Framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Pal, D. Antibiotic Resistance and Wastewater: Correlation, Impact and Critical Human Health Challenges. J. Environ. Chem. Eng. 2018, 6, 52–58. [Google Scholar] [CrossRef]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and Genetic Lineages of Environmental Staphylococci: A Surface Water Overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chang, F.; Liu, Q.; Duan, L.; Li, D.; Zhang, H. Review Article Recent Advances and Perspectives on the Sources and Detection of Antibiotics in Aquatic Environments. J. Anal. Methods Chem. 2022, 2022, 5091181. [Google Scholar] [CrossRef] [PubMed]
- Barati, A.; Ghaderpour, A.; Chew, L.L.; Bong, C.W.; Thong, K.L.; Chong, V.C.; Chai, L.C. Isolation and Characterization of Aquatic-Borne Klebsiella pneumoniae from Tropical Estuaries in Malaysia. Int. J. Environ. Res. Public Health 2016, 13, 426. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Anes, J.; Devineau, S.; Fanning, S. Klebsiella pneumoniae: Prevalence, Reservoirs, Antimicrobial Resistance, Pathogenicity, and Infection: A Hitherto Unrecognized Zoonotic Bacterium. Foodborne Pathog. Dis. 2021, 18, 63–84. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, J.; Czokajło, I.; Gańko, M.; Śmiałek, M.; Koncicki, A. Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022. Animals 2022, 12, 3157. [Google Scholar] [CrossRef]
- Gómez, M.; Valverde, A.; Del Campo, R.; Rodríguez, J.M.; Maldonado-Barragán, A. Phenotypic and Molecular Characterization of Commensal, Community-Acquired and Nosocomial Klebsiella spp. Microorganisms 2021, 9, 2344. [Google Scholar] [CrossRef] [PubMed]
- Rajkumari, J.; Paikhomba Singha, L.; Pandey, P. Genomic Insights of Aromatic Hydrocarbon Degrading Klebsiella pneumoniae AWD5 with Plant Growth Promoting Attributes: A Paradigm of Soil Isolate with Elements of Biodegradation. 3 Biotech 2018, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Kimera, Z.I.; Mgaya, F.X.; Mshana, S.E.; Karimuribo, E.D.; Matee, M.I.N. Occurrence of Extended Spectrum Beta Lactamase (ESBL) Producers, Quinolone and Carbapenem Resistant Enterobacteriaceae Isolated from Environmental Samples along Msimbazi River Basin Ecosystem in Tanzania. Int. J. Environ. Res. Public Health 2021, 18, 8264. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; He, R.; Zhang, B.; Gao, C.; Liu, F. Seasonal Variations of Microbial Community Structure, Assembly Processes, and Influencing Factors in Karst River. Front. Microbiol. 2023, 14, 1133938. [Google Scholar] [CrossRef]
- Beal, C.; Matsubae, K.; Ylä-Mella, J.; Elagroudy, S.; Campanale, C.; Losacco, D.; Triozzi, M.; Massarelli, C.; Uricchio, V.F. An Overall Perspective for the Study of Emerging Contaminants in Karst Aquifers. Resources 2022, 11, 105. [Google Scholar] [CrossRef]
- Cuadrado-Quesada, G.; Holley, C.; Gupta, J. Groundwater Governance in the Anthropocene: A Close Look at Costa Rica. Water Policy 2018, 20, 475–489. [Google Scholar] [CrossRef]
- Arroita, M.; Flores, L.; Larrañaga, A.; Chauvet, E.; Elosegi, A. Hydrological Contingency: Drying History Affects Aquatic Microbial Decomposition. Aquat. Sci. 2018, 80, 31. [Google Scholar] [CrossRef]
- Giri, S.; Mishra, A.; Zhang, Z.; Lathrop, R.G.; Alnahit, A.O. Meteorological and Hydrological Drought Analysis and Its Impact on Water Quality and Stream Integrity. Sustainability 2021, 13, 8175. [Google Scholar] [CrossRef]
- He, Y.; Guo, X.; Xiang, S.; Li, J.; Li, X.; Xiang, H.; He, J.; Chen, D.; Chen, J. Comparative Analyses of Phenotypic Methods and 16S RRNA, Khe, RpoB Genes Sequencing for Identification of Clinical Isolates of Klebsiella pneumoniae. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2016, 109, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Solberg, O.D.; Ajiboye, R.M.; Riley, L.W. Origin of Class 1 and 2 Integrons and Gene Cassettes in a Population-Based Sample of Uropathogenic Escherichia coli. J. Clin. Microbiol. 2006, 44, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Silva, V.; De Lurdes, M.; Dapkevicius, E.; Azevedo, M.; Cordeiro, R.; Pereira, J.E.; Valentão, P.; Falco, V.; Igrejas, G.; et al. Unveiling Antibiotic Resistance, Clonal Diversity, and Biofilm Formation in E. Coli Isolated from Healthy Swine in Portugal. Pathogens 2024, 13, 305. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Cunha, R.; Martins, C.; Martínez-Álvarez, S.; Chenouf, N.S.; Pimenta, P.; Pereira, A.R.; Ramos, S.; Sadi, M.; Martins, Â.; et al. Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics 2021, 10, 1013. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Cerca, N.; Nicolau, A.I. Compositional Analysis of Biofilms Formed by Staphylococcus Aureus Isolated from Food Sources. Front. Microbiol. 2016, 7, 184310. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of Multiple Methods for Quantification of Microbial Biofilms Grown in Microtiter Plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Hartinger, S.M.; Medina-Pizzali, M.L.; Salmon-Mulanovich, G.; Larson, A.J.; Pinedo-Bardales, M.; Verastegui, H.; Riberos, M.; Mäusezahl, D. Antimicrobial Resistance in Humans, Animals, Water and Household Environs in Rural Andean Peru: Exploring Dissemination Pathways through the One Health Lens. Int. J. Environ. Res. Public Health 2021, 18, 4604. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.H.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Prevalence and Diversity of BlaTEM, BlaSHV and BlaCTX-M Variants among Multidrug Resistant Klebsiella spp. from an Urban Riverine Environment in India. Int. J. Environ. Health Res. 2018, 29, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Altayb, H.N.; Elbadawi, H.S.; Alzahrani, F.A.; Baothman, O.; Kazmi, I.; Nadeem, M.S.; Hosawi, S.; Chaieb, K. Co-Occurrence of β-Lactam and Aminoglycoside Resistance Determinants among Clinical and Environmental Isolates of Klebsiella pneumoniae and Escherichia coli: A Genomic Approach. Pharmaceuticals 2022, 15, 1011. [Google Scholar] [CrossRef] [PubMed]
- Hooban, B.; Fitzhenry, K.; Cahill, N.; Joyce, A.; O’ Connor, L.; Bray, J.E.; Brisse, S.; Passet, V.; Abbas Syed, R.; Cormican, M.; et al. A Point Prevalence Survey of Antibiotic Resistance in the Irish Environment, 2018–2019. Environ. Int. 2021, 152, 106466. [Google Scholar] [CrossRef] [PubMed]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and MCR-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 272364. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic Characterization of ESBL-Producing Escherichia coli and Klebsiella pneumoniae Isolated from Wastewater and River Water in Tunisia: Predominance of CTX-M-15 and High Genetic Diversity. Environ. Sci. Pollut. Res. 2020, 27, 44368–44377. [Google Scholar] [CrossRef]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm Formation and Antibiotic Resistance of Klebsiella pneumoniae Isolated from Clinical Samples in a Tertiary Care Hospital, Klaten, Indonesia. BMC Proc. 2019, 13, 20. [Google Scholar] [CrossRef]
- Teixeira, P.; Tacão, M.; Pureza, L.; Gonçalves, J.; Silva, A.; Cruz-Schneider, M.P.; Henriques, I. Occurrence of Carbapenemase-Producing Enterobacteriaceae in a Portuguese River: BlaNDM, BlaKPC and BlaGES among the Detected Genes. Environ. Pollut. 2020, 260, 113913. [Google Scholar] [CrossRef] [PubMed]
- Mahato, S.; Mahato, A.; Pokharel, E.; Tamrakar, A. Detection of Extended-Spectrum Beta-Lactamase-Producing E. coli and Klebsiella spp. in Effluents of Different Hospitals Sewage in Biratnagar, Nepal. BMC Res. Notes 2019, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Bonomo, R.A. Ceftazidime/Avibactam and Ceftolozane/Tazobactam: Second-Generation β-Lactam/β-Lactamase Inhibitor Combinations. Clin. Infect. Dis. 2016, 63, 234–241. [Google Scholar] [CrossRef]
- Paterson, D.L. Recommendation for Treatment of Severe Infections Caused by Enterobacteriaceae Producing Extended-Spectrum β-Lactamases (ESBLs). Clin. Microbiol. Infect. 2000, 6, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Hellmark, B.; Ehricht, R.; Söderquist, B.; Jass, J. Related Carbapenemase-Producing Klebsiella Isolates Detected in Both a Hospital and Associated Aquatic Environment in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2241–2251. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Prasai Joshi, T.; Nhemhaphuki, S.; Sitoula, K.; Maharjan, J.; Ranjit, R.; Shrestha, P.; Joshi, D.R. Occurrence of Antibiotic-Resistant Bacteria and Their Genes in Bagmati River, Nepal. Water Air Soil Pollut. 2023, 234, 475. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Schwengers, O.; Schmiedel, J.; Baars, C.; Lambrecht, O.; Heß, S.; Berendonk, T.U.; Falgenhauer, J.; Chakraborty, T.; Imirzalioglu, C. Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters. Front. Microbiol. 2019, 10, 482269. [Google Scholar] [CrossRef]
- Aguilar-Salazar, A.; Martínez-Vázquez, A.V.; Aguilera-Arreola, G.; de Jesus de Luna-Santillana, E.; Cruz-Hernández, M.A.; Escobedo-Bonilla, C.M.; Lara-Ramírez, E.; Sánchez-Sánchez, M.; Guerrero, A.; Rivera, G.; et al. Prevalence of ESKAPE Bacteria in Surface Water and Wastewater Sources: Multidrug Resistance and Molecular Characterization, an Updated Review. Water 2023, 15, 3200. [Google Scholar] [CrossRef]
- Fadare, F.T.; Okoh, A.I. Distribution and Molecular Characterization of ESBL, PAmpC β-Lactamases, and Non-β-Lactam Encoding Genes in Enterobacteriaceae Isolated from Hospital Wastewater in Eastern Cape Province, South Africa. PLoS ONE 2021, 16, e0254753. [Google Scholar] [CrossRef]
- Nascimento, T.; Cantamessa, R.; Melo, L.; Lincopan, N.; Fernandes, M.R.; Cerdeira, L.; Fraga, E.; Dropa, M.; Sato, M.I.Z. International High-Risk Clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in Urban Lake Waters. Sci. Total Environ. 2017, 598, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Fischer, M.A.; Neumann, B.; Kiesewetter, K.; Hoffmann, I.; Werner, G.; Pfeifer, Y.; Lübbert, C. Carbapenemase-Producing Gram-Negative Bacteria in Hospital Wastewater, Wastewater Treatment Plants and Surface Waters in a Metropolitan Area in Germany, 2020. Sci. Total. Environ. 2023, 890, 164179. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of Multi-Resistant Gram-Negative Bacteria into German Wastewater and Surface Waters. FEMS Microbiol. Ecol. 2018, 94, 5. [Google Scholar] [CrossRef] [PubMed]
- López-Banda, D.A.; Carrillo-Casas, E.M.; Leyva-Leyva, M.; Orozco-Hoyuela, G.; Manjarrez-Hernández, Á.H.; Arroyo-Escalante, S.; Moncada-Barrón, D.; Villanueva-Recillas, S.; Xicohtencatl-Cortes, J.; Hernández-Castro, R. Identification of Virulence Factors Genes in Escherichia coli Isolates from Women with Urinary Tract Infection in Mexico. Biomed. Res. Int. 2014, 2014, 959206. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Z.; Li, H.; Chen, F.; Han, K.; Hu, X.; Fang, Y.; Chen, D. Metagenomic Approaches Reveal Strain Profiling and Genotyping of Klebsiella Pneumoniae from Hospitalized Patients in China. Microbiol. Spectr. 2022, 10, e0219021. [Google Scholar] [CrossRef]
- Özad Düzgün, A.; Yüksel, G. Detection of Virulence Factor Genes, Antibiotic Resistance Genes and Biofilm Formation in Clinical Gram-Negative Bacteria and First Report from Türkiye of K. oxytoca Carrying Both blaOXA-23 and blaOXA-51 Genes. Biologia 2023, 78, 2245–2251. [Google Scholar] [CrossRef]
- Hassan, R.; El Naggar, W.; El Sawy, E.; El Mahdy, A. Characterization of Some Virulence Factors Associated with Enterbacteriaceae Isolated from Urinary Tract Infections in Mansoura Hospitals. Egypt. J. Med. Microbiol. 2011, 20, 9–18. [Google Scholar]
- Li, Y.; Ni, M. Regulation of Biofilm Formation in Klebsiella pneumoniae. Front. Microbiol. 2023, 14, 1238482. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Pereira, J.E.; Maltez, L.; Poeta, P.; Igrejas, G. Influence of Environmental Factors on Biofilm Formation of Staphylococci Isolated from Wastewater and Surface Water. Pathogens 2022, 11, 1069. [Google Scholar] [CrossRef]
- Romero, F.; Acuña, V.; Font, C.; Freixa, A.; Sabater, S. Effects of Multiple Stressors on River Biofilms Depend on the Time Scale. Sci. Rep. 2019, 9, 15810. [Google Scholar] [CrossRef]
- Türkel, İ.; Yıldırım, T.; Yazgan, B.; Bilgin, M.; Başbulut, E. Relationship between Antibiotic Resistance, Efflux Pumps, and Biofilm Formation in Extended-Spectrum β-Lactamase Producing Klebsiella pneumoniae. J. Chemother. 2018, 30, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Shadkam, S.; Goli, H.R.; Mirzaei, B.; Gholami, M.; Ahanjan, M. Correlation between Antimicrobial Resistance and Biofilm Formation Capability among Klebsiella pneumoniae Strains Isolated from Hospitalized Patients in Iran. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 13. [Google Scholar] [CrossRef]
- Sanchez, C.J.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm Formation by Clinical Isolates and the Implications in Chronic Infections. BMC Infect. Dis. 2013, 13, 47. [Google Scholar] [CrossRef]
- Hasan, M.E.; Shahriar, A.; Shams, F.; Nath, A.K.; Emran, T. Bin Correlation between Biofilm Formation and Antimicrobial Susceptibility Pattern toward Extended Spectrum β-Lactamase (ESBL)- and Non-ESBL-Producing Uropathogenic Bacteria. J. Basic Clin. Physiol. Pharmacol. 2021, 32, 20190296. [Google Scholar] [CrossRef]
- Zong, Z.; Partridge, S.R.; Thomas, L.; Iredell, J.R. Dominance of BlaCTX-M within an Australian Extended-Spectrum β-Lactamase Gene Pool. Antimicrob. Agents Chemother. 2008, 52, 4198–4202. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Heterogeneity among Virulence and Antimicrobial Resistance Gene Profiles of Extraintestinal Escherichia coli Isolates of Animal and Human Origin. J. Clin. Microbiol. 2004, 42, 5444–5452. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Chenouf, N.S.; Cunha, R.; Martins, C.; Pimenta, P.; Pereira, A.R.; Martínez-álvarez, S.; Ramos, S.; Silva, V.; Igrejas, G.; et al. Antimicrobial Resistance Genes and Diversity of Clones among ESBL- and Acquired AmpC-Producing Escherichia coli Isolated from Fecal Samples of Healthy and Sick Cats in Portugal. Antibiotics 2021, 10, 262. [Google Scholar] [CrossRef]
- Garcês, A.; Correia, S.; Amorim, F.; Pereira, J.E.; Igrejas, G.; Poeta, P. First Report on Extended-Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli from European Free-Tailed Bats (Tadarida teniotis) in Portugal: A One-Health Approach of a Hidden Contamination Problem. J. Hazard. Mater. 2019, 370, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Caroff, N.; Espaze, E.; Berard, I.; Richet, H.; Reynaud, A. Mutations in the AmpC Promoter of Escherichia coli Isolates Resistant to Oxyiminocephalosporins without Extended Spectrum β-Lactamase Production. FEMS Microbiol. Lett. 1999, 173, 459–465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vanhoof, R.; Content, J.; Van Bossuyt, E.; Dewit, L.; Hannecart-pokorni, E. Identification of the AadB Gene Coding for the Aminoglycoside-2″-O-Nucleotidyltraiisferase, ANT(2″), by Means of the Polymerase Chain Reaction. J. Antimicrob. Chemother. 1992, 29, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia coli Strains of Human, Animal, and Food Origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Jiang, X.; Yang, Z.; Chen, N.; Chen, X.; Li, G.; Lu, Y. DfrA27, a New Integron-Associated Trimethoprim Resistance Gene from Escherichia coli. J. Antimicrob. Chemother. 2009, 63, 405–406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic Resistance in the ECOR Collection: Integrons and Identification of a Novel Aad Gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Agerso, Y.; Gerner-Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of Antimicrobial Resistance Phenotypes and Resistance Genes in Enterococcus faecalis and Enterococcus faecium from Humans in the Community, Broilers, and Pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Neyestanaki, D.K.; Mirsalehian, A.; Rezagholizadeh, F.; Jabalameli, F.; Taherikalani, M.; Emaneini, M. Determination of Extended Spectrum Beta-Lactamases, Metallo-Beta-Lactamases and AmpC-Beta-Lactamases among Carbapenem Resistant Pseudomonas aeruginosa Isolated from Burn Patients. Burns 2014, 40, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Ruiz, J.; Goñi, P.; Jimenez De Anta, M.T. Detection of Mutations in ParC in Quinolone-Resistant Clinical Isolates of Escherichia coli. Antimicrob. Agents Chemother. 1996, 40, 491–493. [Google Scholar] [CrossRef]
- Rita Rebelo, A.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Andre Hammerl, J.; et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, Mcr-1, Mcr-2, Mcr-3, Mcr-4 and Mcr-5 for Surveillance Purposes. Eurosurveillance 2018, 23, 17–00672. [Google Scholar] [CrossRef] [PubMed]
- Bardiau, M.; Labrozzo, S.; Mainil, J.G. Study of Polymorphisms in Tir, Eae and TccP2 Genes in Enterohaemorrhagic and Enteropathogenic Escherichia coli of Serogroup O26. BMC Microbiol. 2011, 11, 124. [Google Scholar] [CrossRef]

| Source | Number of Samples | Klebsiella spp. | K. pneumoniae |
|---|---|---|---|
| Rivers | 18 | 7 | 8 |
| Streams | 33 | 3 | 6 |
| Irrigation ditches | 1 | - | - |
| Dams | 1 | - | 1 |
| Fountains | 11 | 2 | 3 |
| Water wells | 7 | 1 | 1 |
| Water tanks | 2 | - | - |
| Water mines | 1 | - | - |
| Springs | 2 | 1 | - |
| Total: | 77 | 14 | 19 |
| Isolate | Species | ESBL Production | Antimicrobial Resistance | Virulence Genes | |
|---|---|---|---|---|---|
| Phenotype | Genotype | ||||
| VS3296 | K. pneumoniae | N | CN, S, TOB, CIP, SXT | aac(3)-IV, aac(3)-II, sul2, strA, aadA1, aadA5 | - |
| VS3297 | K. pneumoniae | N | AUG, FOX, CAZ, CTX, MRP, CN, S, SXT | aac(3)-IV, aac(3)-II, blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, ampC, aadA1, parC, blaTEM | - |
| VS3298 | K. pneumoniae | P | AUG, CAZ, CTX, CN, S, TOB, CIP, SXT | aac(3)-IV, aac(3)-II, blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, ampC, blaTEM | - |
| VS3299 | K. pneumoniae | N | AUG, CAZ, CTX, TE, SXT | tetA, blaSHV, sul2, ampC, blaTEM | papC |
| VS3300 | K. pneumoniae | N | SXT | sul2 | papG-III, papC |
| VS3301 | K. pneumoniae | P | AUG, CTX, S, SXT | blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, ampC, aadA1 | - |
| VS3302 | K. pneumoniae | N | SXT | sul2 | - |
| VS3303 | K. pneumoniae | N | AUG, FOX | blaCTX-U, blaCTX-M9 | papC |
| VS3304 | K. pneumoniae | N | AUG | blaCTX-U, blaCTX-M9 | - |
| VS3305 | K. pneumoniae | N | CIP | - | papG-III |
| VS3306 | K. pneumoniae | P | CAZ, CTX, S, SXT | blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, aadA1, aadA5, blaTEM | - |
| VS3307 | K. pneumoniae | P | AUG, FOX, CAZ, CTX | blaCTX-U, blaCTX-M9, ampC | - |
| VS3308 | K. pneumoniae | P | FOX, CAZ, CTX | blaCTX-U, blaCTX-M9, ampC | - |
| VS3309 | K. pneumoniae | N | AUG, FOX | blaCTX-U, blaCTX-M9, ampC | - |
| VS3310 | K. pneumoniae | N | - | - | papC |
| VS3311 | K. pneumoniae | N | - | - | - |
| VS3312 | K. pneumoniae | P | CTX, S, SXT | blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, aadA1, blaTEM | - |
| VS3313 | K. pneumoniae | P | CAZ, CTX, S, SXT | blaCTX-U, blaCTX-M9, blaSHV, sul2, strA, aadA1, blaTEM | papC, bfp |
| VS3314 | K. pneumoniae | N | - | - | bfp |
| VS3315 | Klebsiella spp. | N | TE, S, SXT | sul2, strA, aadA1 | papC |
| VS3316 | Klebsiella spp. | P | CTX | blaCTX-U, blaCTX-M9 | - |
| VS3317 | Klebsiella spp. | N | CAZ | - | - |
| VS3318 | Klebsiella spp. | N | CTX, SXT | blaCTX-U, blaCTX-M9, sul2 | aer |
| VS3319 | Klebsiella spp. | N | AUG, FOX, CAZ, CTX | blaCTX-U, blaCTX-M9 | - |
| VS3320 | Klebsiella spp. | N | - | - | papC |
| VS3321 | Klebsiella spp. | N | - | - | - |
| VS3322 | Klebsiella spp. | N | AUG, FOX, CTX | blaCTX-U, blaCTX-M9 | papC, bfp |
| VS3323 | Klebsiella spp. | N | SXT | sul2 | - |
| VS3324 | Klebsiella spp. | N | AUG | - | - |
| VS3325 | Klebsiella spp. | P | - | - | - |
| VS3326 | Klebsiella spp. | N | S, CIP, SXT | sul2 | aer |
| VS3327 | Klebsiella spp. | P | AUG, CTX | blaCTX-U, blaCTX-M9 | - |
| VS3328 | Klebsiella spp. | P | CTX | blaCTX-U, blaCTX-M9 | aer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, S.; Silva, V.; de Lurdes Enes Dapkevicius, M.; Pereira, J.E.; Martins, Â.; Igrejas, G.; Poeta, P. Comprehensive Profiling of Klebsiella in Surface Waters from Northern Portugal: Understanding Patterns in Prevalence, Antibiotic Resistance, and Biofilm Formation. Water 2024, 16, 1297. https://doi.org/10.3390/w16091297
Araújo S, Silva V, de Lurdes Enes Dapkevicius M, Pereira JE, Martins Â, Igrejas G, Poeta P. Comprehensive Profiling of Klebsiella in Surface Waters from Northern Portugal: Understanding Patterns in Prevalence, Antibiotic Resistance, and Biofilm Formation. Water. 2024; 16(9):1297. https://doi.org/10.3390/w16091297
Chicago/Turabian StyleAraújo, Sara, Vanessa Silva, Maria de Lurdes Enes Dapkevicius, José Eduardo Pereira, Ângela Martins, Gilberto Igrejas, and Patricia Poeta. 2024. "Comprehensive Profiling of Klebsiella in Surface Waters from Northern Portugal: Understanding Patterns in Prevalence, Antibiotic Resistance, and Biofilm Formation" Water 16, no. 9: 1297. https://doi.org/10.3390/w16091297
APA StyleAraújo, S., Silva, V., de Lurdes Enes Dapkevicius, M., Pereira, J. E., Martins, Â., Igrejas, G., & Poeta, P. (2024). Comprehensive Profiling of Klebsiella in Surface Waters from Northern Portugal: Understanding Patterns in Prevalence, Antibiotic Resistance, and Biofilm Formation. Water, 16(9), 1297. https://doi.org/10.3390/w16091297