Arsenic Transformation in Swine Wastewater with Low-Arsenic Content during Anaerobic Digestion
Abstract
:1. Introduction
2. Materials and Methods
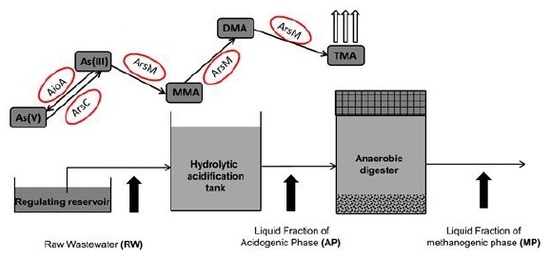
2.1. Swine Wastewater Treatment Process and Sample Collection
2.2. Determination of Physico-Chemical Properties
2.3. Total Heavy Metal and Dissolved Fractions of As Analysis
2.4. Determination of Bacterial 16S rRNA and As Metabolism Genes Copies by qPCR
3. Results and Discussion
3.1. Physico-Chemical Properties and Heavy Metals in Wastewater
3.2. Total As Changes during AD
3.3. Dissolved Fractions of As
3.4. Abundance of As Metabolism Genes
3.5. As Species Concentration vs. As Metabolism Gene Copy Numbers
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Do Amaral, A.C.; Kunz, A.; Radis Steinmetz, R.L.; Justi, K.C. Zinc and copper distribution in swine wastewater treated by anaerobic digestion. J. Environ. Manag. 2014, 141, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.J.; Yonkos, L.T.; Staver, K.W. Environmental Concerns of Roxarsone in Broiler Poultry Feed and Litter in Maryland, USA. Environ. Sci. Technol. 2015, 49, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Meharg, A.A.; Zhao, F.J. Biogeochemistry of arsenic in paddy environments. In Arsenic & Rice; Springer Science & Business Media Press: Dordrecht, The Netherlands, 2012; pp. 71–101. [Google Scholar]
- Lett, M.C.; Muller, D.; Lievremont, D.; Silver, S.; Santini, J. Unified Nomenclature for Genes Involved in Prokaryotic Aerobic Arsenite Oxidation. J. Bacteriol. 2011, 194, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Rosen, B.P.; Zhang, Y.; Wang, G.; Franke, S.; Rensing, C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.A. Functional gene surveys from ocean drilling expeditions—A review and perspective. FEMS Microbiol. Ecol. 2013, 84, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, J.; Yuan, X.; Zhu, W.; Wang, X.; Cheng, X.; Cui, Z. The effect of mixing intensity on the performance and microbial dynamics of a single vertical reactor integrating acidogenic and methanogenic phases in lignocellulosic biomass digestion. Bioresour. Technol. 2017, 238, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Marcato, C.E.; Pinelli, E.; Pouech, P.; Winterton, P.; Guiresse, M. Particle size and metal distributions in anaerobically digested pig slurry. Bioresour. Technol. 2008, 99, 2340–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbarino, J.R.; Bednar, A.J.; Rutherford, D.W.; Beyer, R.S.; Wershaw, R.L. Environmental Fate of Roxarsone in Poultry Litter. I. Degradation of Roxarsone during Composting. Environ. Sci. Technol. 2003, 37, 1509–1514. [Google Scholar] [PubMed]
- APH Association. Standard Methods for the Examination of Water and Wastewater, 20th ed.; APH Association: Washington, DC, USA, 1998; p. 1268. Available online: https://www.mwa.co.th/download/file_upload/SMWW_1000-3000.pdf (accessed on 24 October 2017).
- Zhai, W.; Wong, M.T.; Luo, F.; Hashmi, M.Z.; Liu, X.; Edwards, E.A.; Tang, X.; Xu, J. Arsenic Methylation and its Relationship to Abundance and Diversity of arsM Genes in Composting Manure. Sci. Rep. 2017, 7, 42198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, F.; Sun, G.; Su, J.; Yang, X.; Li, H.; Zhu, Y. Diversity and Abundance of Arsenic Biotransformation Genes in Paddy Soils from Southern China. Environ. Sci. Technol. 2015, 49, 4138–4146. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Liu, X.; Dai, L.; Dai, X. Changes of heavy metal speciation during high-solid anaerobic digestion of sewage sludge. Bioresour. Technol. 2013, 131, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Shin, S.G.; Cho, K.; Lee, C.; Hwang, S. Performance of methanogenic reactors in temperature phased two-stage anaerobic digestion of swine wastewater. J. Biosci. Bioeng. 2012, 114, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Parawira, W.; Murto, M.; Read, J.S.; Mattiasson, B. Profile of hydrolases and biogas production during two-stage mesophilic anaerobic digestion of solid potato waste. Process Biochem. 2005, 40, 2945–2952. [Google Scholar] [CrossRef]
- Jin, H.; Chang, Z. Distribution of Heavy Metal Contents and Chemical Fractions in Anaerobically Digested Manure Slurry. Appl. Biochem. Biotechnol. 2011, 164, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Paulo, L.M.; Stams, A.J.; Sousa, D.Z. Methanogens, sulphate and heavy metals: A complex system. Rev. Environ. Sci. Biotechnol. 2015, 14, 537–553. [Google Scholar] [CrossRef] [Green Version]
- Webster, T.M.; Reddy, R.R.; Tan, J.Y.; Van Nostr, J.D.; Zhou, J.; Hayes, K.F.; Raskin, L. Anaerobic Disposal of Arsenic-Bearing Wastes Results in Low Microbially Mediated Arsenic Volatilization. Environ. Sci. Technol. 2016, 50, 10951–10959. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, D.; Mishra, D.; Chaudhury, G.R.; Das, R.P. Removal of Arsenic from Arsenic Rich Sludge by Volatilization using Anaerobic Microorganisms Treated with Cow Dung. Soil Sediment Contam. 2008, 17, 301–311. [Google Scholar]
- Bhattacharjee, H.; Rosen, B.P. Arsenic metabolism in prokaryotic and eukaryotic microbes. In Molecular Microbiology of Heavy Metals; Springer: Berlin/Heidelberg, Germany, 2007; pp. 371–406. [Google Scholar]
- Silver, S.; Phung, L.T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 2005, 71, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Bai, Y.; Liang, J.; Qu, J. Metagenomic approach reveals variation of microbes with arsenic and antimony metabolism genes from highly contaminated soil. PLoS ONE 2014, 9, e108185. [Google Scholar] [CrossRef] [PubMed]
- Mestrot, A.; Merle, J.K.; Broglia, A.; Feldmann, J.; Krupp, E.M. Atmospheric Stability of Arsine and Methylarsines. Environ. Sci. Technol. 2011, 45, 4010–4015. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; DeMel, S.; Shi, J.; Gladysheva, T.; Gatti, D.L.; Rosen, B.P.; Edwards, B.F. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 2001, 9, 1071–1081. [Google Scholar] [CrossRef]
- Zhao, F.; Harris, E.; Yan, J.; Ma, J.; Wu, L.; Liu, W.; McGrath, S.P.; Zhou, J.; Zhu, Y. Arsenic Methylation in Soils and Its Relationship with Microbial arsM Abundance and Diversity, and As Speciation in Rice. Environ. Sci. Technol. 2013, 47, 7147–7154. [Google Scholar]
- Meyer, J.; Michalke, K.; Kouril, T.; Hensel, R. Volatilisation of metals and metalloids: An inherent feature of methanoarchaea? Syst. Appl. Microbiol. 2008, 31, 81–87. [Google Scholar] [CrossRef] [PubMed]



| Month | Treatment | pH | COD Removal Efficiency (%) | NH4+-N (mg L−1) | As (μg L−1) | Cu (μg L−1) | Zn (μg L−1) | Fe (μg L−1) | Mn (μg L−1) | Cd (μg L−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| April | RW | 7.1 ± 0.1a | - | 891.9 ± 3.1a | 294.0 ± 19.1a | 433.5 ± 101.0a | 1647.3 ± 361.9a | 2829.5 ± 1262.9a | 400.3 ± 122.6a | 0.3 ± 0.1a |
| AP | 7.1 ± 0.1a | 3.2 ± 1.5 | 927.1 ± 40.5a | 235.2 ± 102.8a | 497.6 ± 34.3a | 1100.3 ± 118.3b | 2294.1 ± 1345.8a | 374.5 ± 98.6a | 0.4 ± 0.3a | |
| MP | 7.5 ± 0.1b | 60.8 ± 8.8 | 939.3 ± 30.6a | 196.6 ± 16.9b | 211.4 ± 5.3b | 674.7 ± 19.0c | 2947.3 ± 415.7a | 187.4 ± 8.3b | 0.2 ± 0.0a | |
| June | RW | 7.2 ± 0.1a | - | 598.5 ± 5.5b | 440.1 ± 46.7a | 222.9 ± 1.7a | 718.4 ± 7.9a | 1052.3 ± 41.1a | 174.8 ± 4.1a | 0.2 ± 0.0a |
| AP | 7.1 ± 0.0a | 10.9 ± 4.9 | 572.3 ± 26.0b | 204.3 ± 10.7b | 253.3 ± 5.3a | 986.0 ± 12.6a | 1523.6 ± 25.1b | 196.3 ± 14.3a | 0.2 ± 0.0a | |
| MP | 7.4 ± 0.1b | 61.9 ± 2.2 | 785.7 ± 41.4a | 146.1 ± 32.6c | 165.9 ± 41.4b | 535.3 ± 45.9b | 597.4 ± 162.8c | 110.0 ± 29.3b | 0.2 ± 0.1a | |
| August | RW | 7.1 ± 0.0a | - | 346.0 ± 14.2c | 208.3 ± 46.7a | 268.9 ± 39.6a | 865.5 ± 123.8a | 1261.4 ± 180.3a | 219.5 ± 47.4a | 0.3 ± 0.1a |
| AP | 7.2 ± 0.0a | 13.0 ± 3.2 | 315.7 ± 22.5c | 83.2 ± 8.3b | 100.5 ± 6.9b | 306.2 ± 34.5b | 549.2 ± 28.5b | 121.9 ± 10.5b | 0.3 ± 0.0a | |
| MP | 7.3 ± 0.0a | 53.6 ± 1.8 | 365.9 ± 49.2c | 59.9 ± 5.2c | 86.2 ± 17.2c | 250.5 ± 25.1c | 401.6 ± 93.9b | 63.5 ± 19.5c | 0.2 ± 0.0a |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, W.; Qin, T.; Guo, T.; Khan, M.I.; Tang, X.; Xu, J. Arsenic Transformation in Swine Wastewater with Low-Arsenic Content during Anaerobic Digestion. Water 2017, 9, 826. https://doi.org/10.3390/w9110826
Zhai W, Qin T, Guo T, Khan MI, Tang X, Xu J. Arsenic Transformation in Swine Wastewater with Low-Arsenic Content during Anaerobic Digestion. Water. 2017; 9(11):826. https://doi.org/10.3390/w9110826
Chicago/Turabian StyleZhai, Weiwei, Tianyue Qin, Ting Guo, Muhammad Imran Khan, Xianjin Tang, and Jianming Xu. 2017. "Arsenic Transformation in Swine Wastewater with Low-Arsenic Content during Anaerobic Digestion" Water 9, no. 11: 826. https://doi.org/10.3390/w9110826
APA StyleZhai, W., Qin, T., Guo, T., Khan, M. I., Tang, X., & Xu, J. (2017). Arsenic Transformation in Swine Wastewater with Low-Arsenic Content during Anaerobic Digestion. Water, 9(11), 826. https://doi.org/10.3390/w9110826