SkipResNet: Crop and Weed Recognition Based on the Improved ResNet
Abstract
1. Introduction
- This paper constructed three different path selection algorithms for multi-path input skip-residual neural networks, which were the minimum loss value path selection algorithm, the individual optimal path selection algorithm, and the optimal path statistical selection algorithm.
- This paper proposed a multi-path input skip-residual neural network on the basis of the residual network and combined the three path selection algorithms to be verified on the plant seedling dataset and the weed–corn dataset for the efficient classification of crops and weeds.
- The proposed multi-path input and path selection algorithms were validated on the CIFAR-10 dataset, illustrating the feasibility of the method for other image classification.
2. Materials and Methods
2.1. Network Models and Algorithms
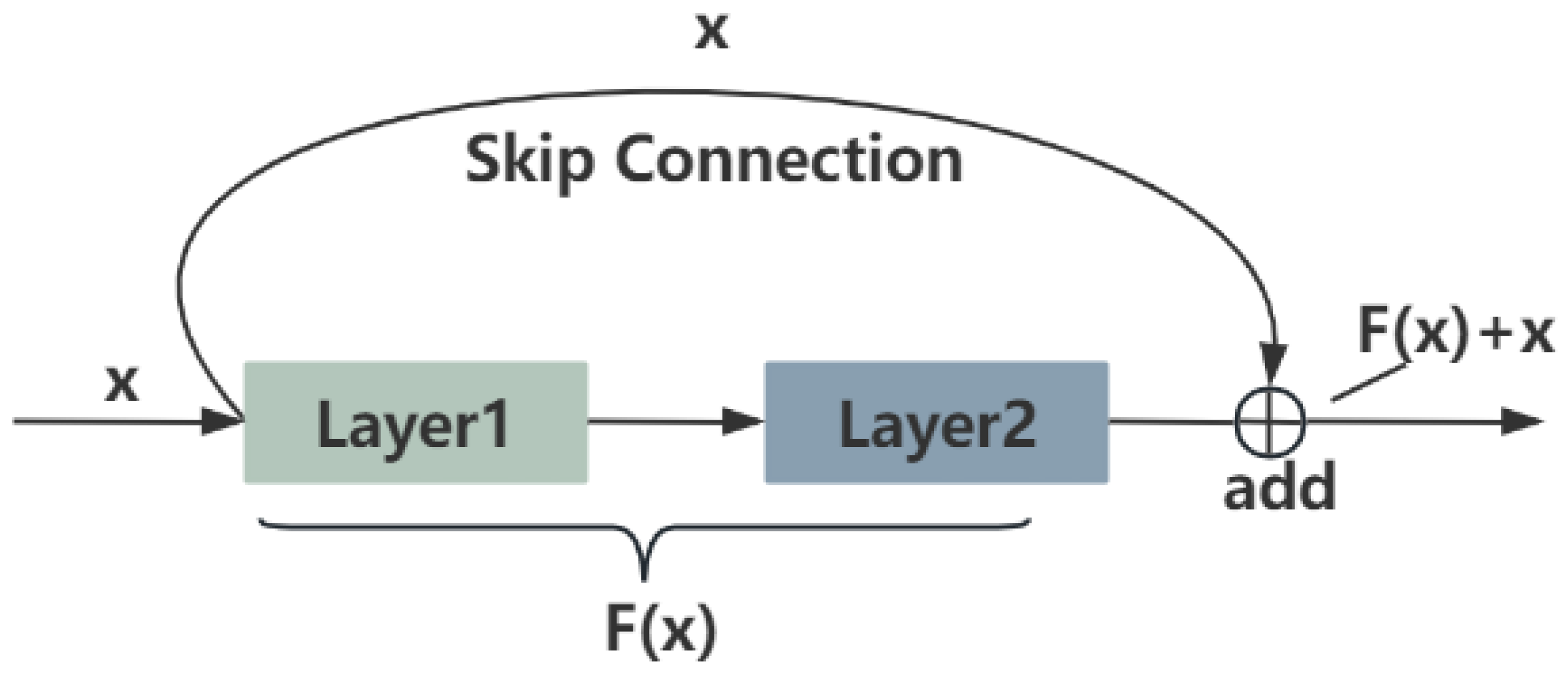
2.1.1. ResNet
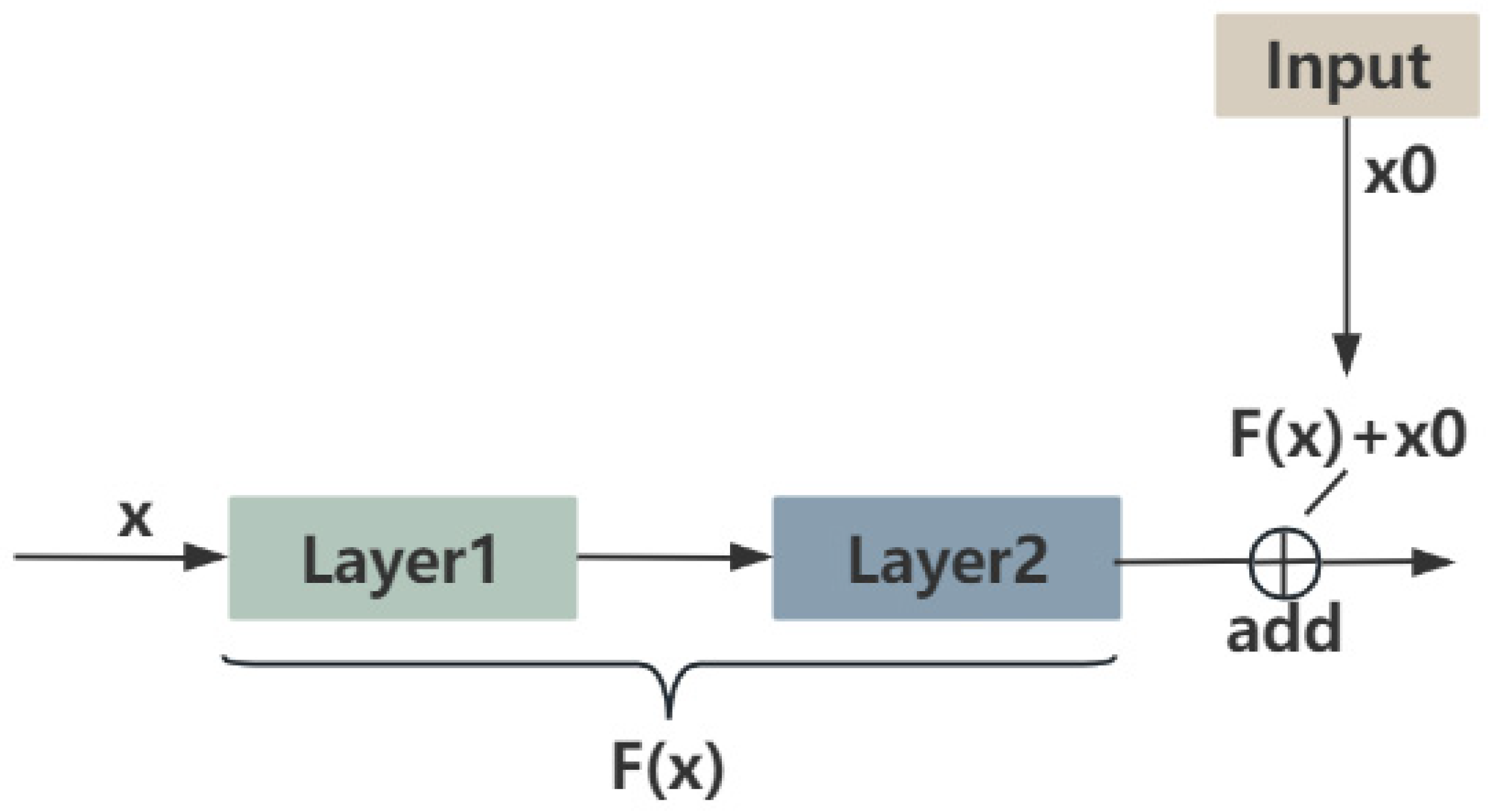
2.1.2. SkipResNet and SkipNet
2.1.3. Path Selection Algorithm
2.2. Evaluating Indicator
2.3. Dataset
2.3.1. Plant Seedling Dataset
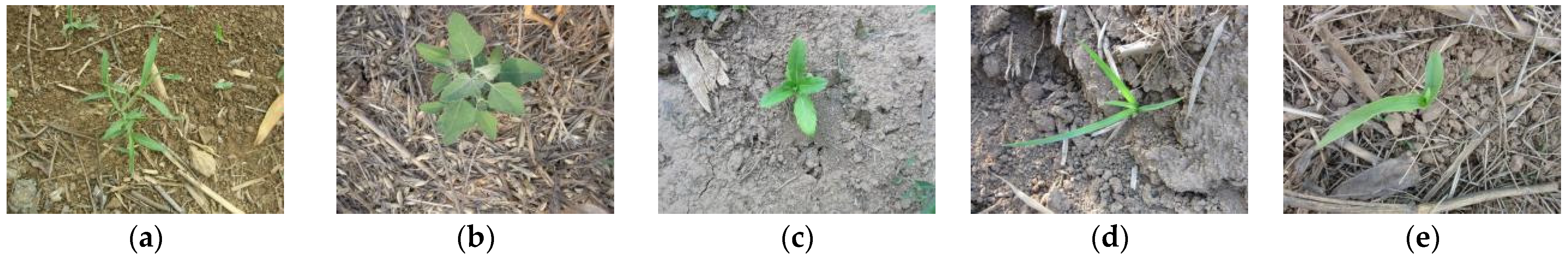
2.3.2. Weed–Corn Dataset
2.3.3. CIFAR-10 Dataset
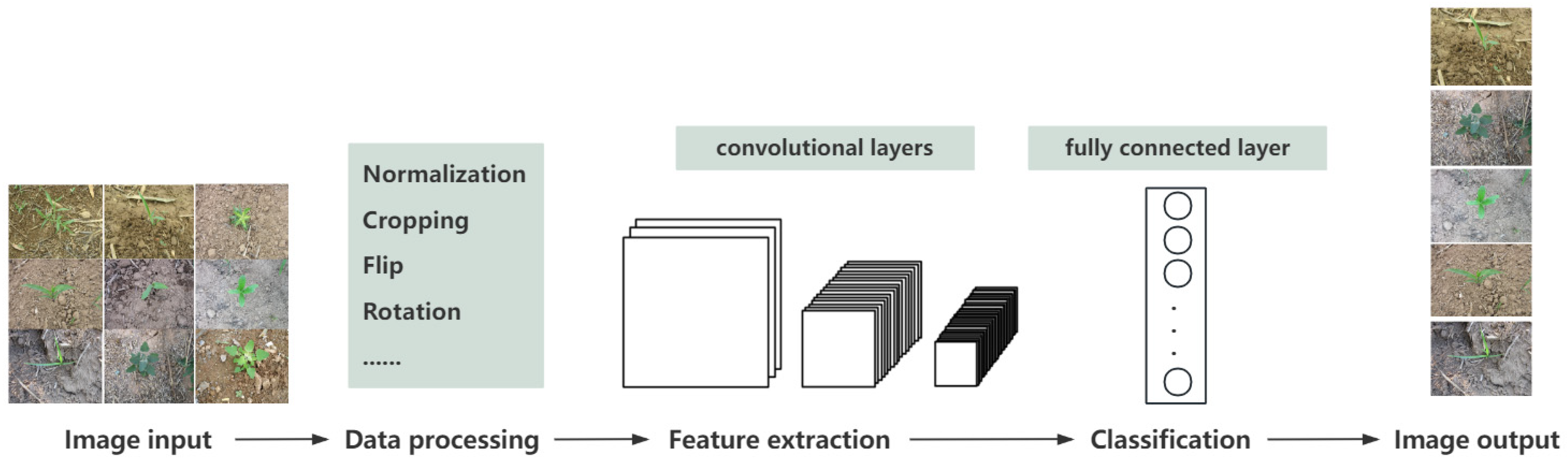
2.3.4. Data Preprocessing and Data Enhancement
3. Experiments and Results
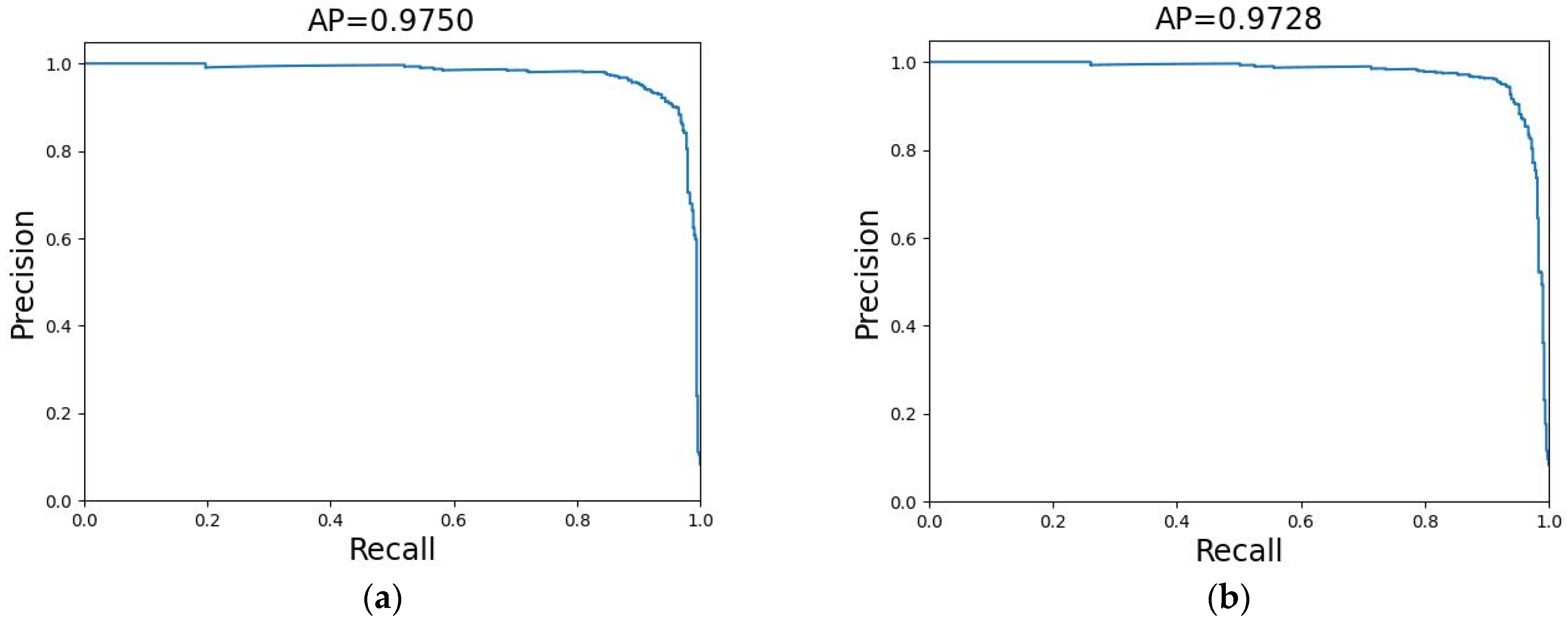
3.1. Results of SkipResNet on the Plant Seedling Dataset
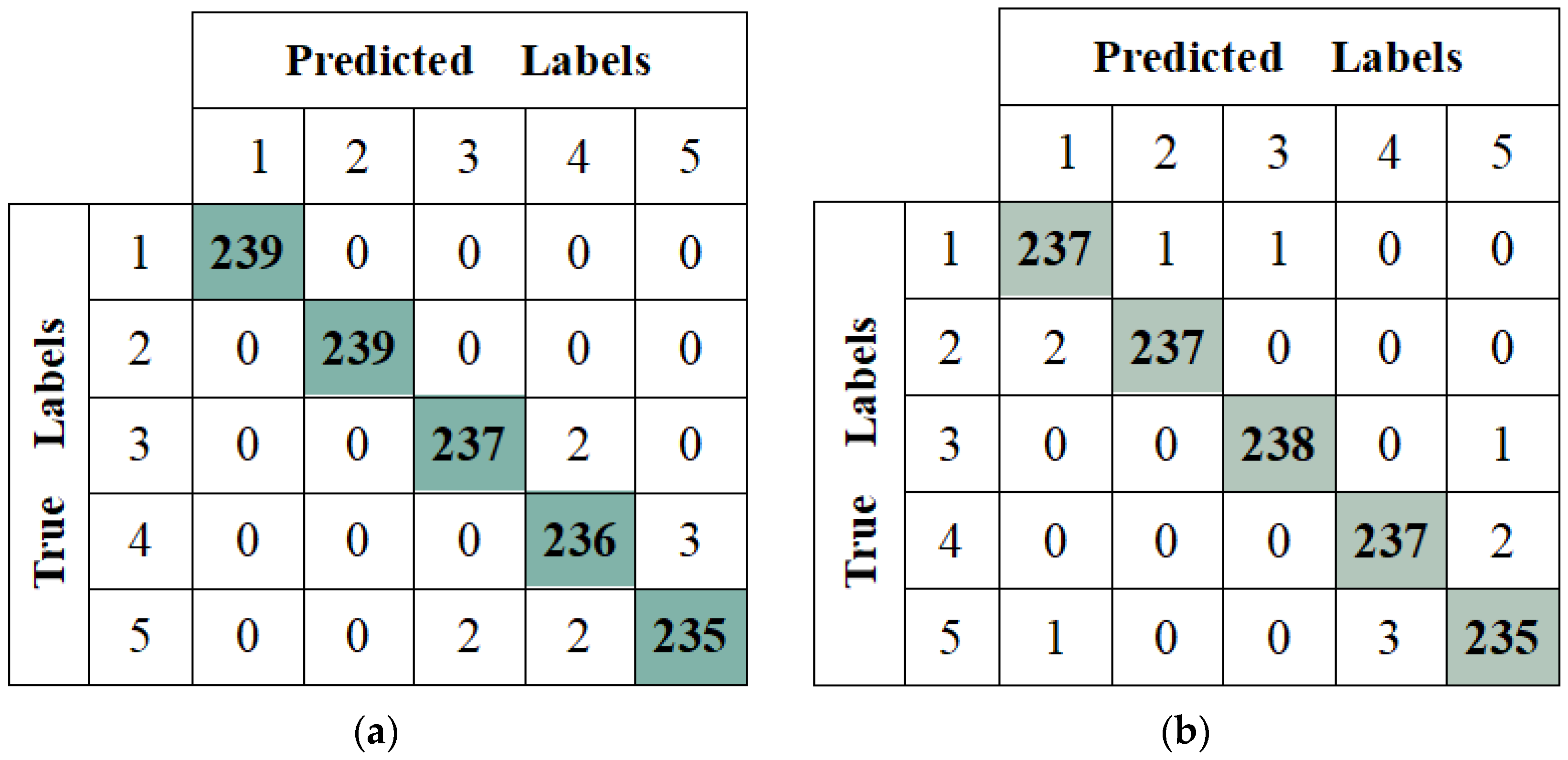
3.2. Results of SkipResNet on the Weed–Corn Dataset
| Class | SkipResNet18 | ResNet18 | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 100.0 | 100.0 | 100.0 | 100.0 | 98.75 | 98.75 | 99.16 | 98.95 |
| 2 | 100.0 | 100.0 | 100.0 | 100.0 | 99.58 | 99.58 | 99.16 | 99.37 |
| 3 | 99.16 | 99.16 | 99.16 | 99.16 | 99.58 | 99.58 | 99.58 | 99.58 |
| 4 (corn) | 98.75 | 98.33 | 98.74 | 98.53 | 98.75 | 98.75 | 99.16 | 98.95 |
| 5 | 98.32 | 98.73 | 98.32 | 98.53 | 98.73 | 98.73 | 98.32 | 98.53 |
| Average | 99.24 | 99.24 | 99.24 | 99.24 | 99.07 | 99.08 | 99.07 | 99.07 |
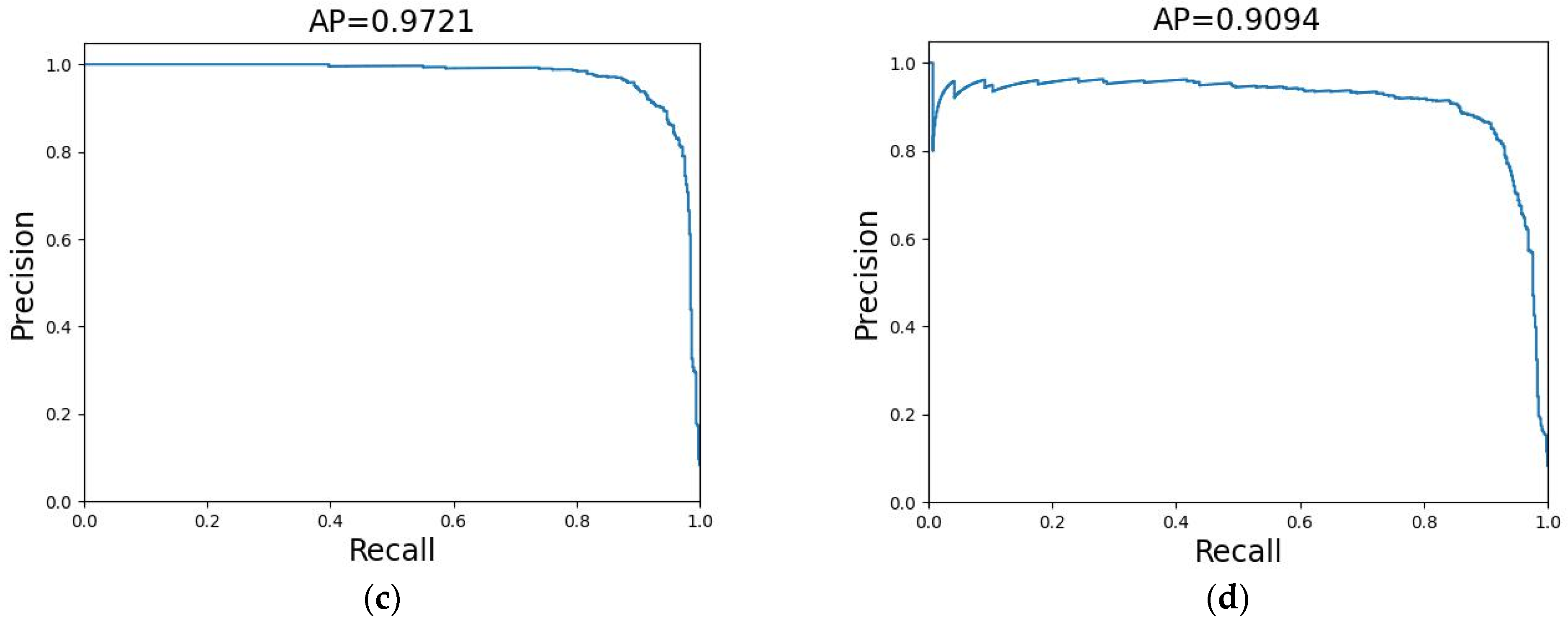
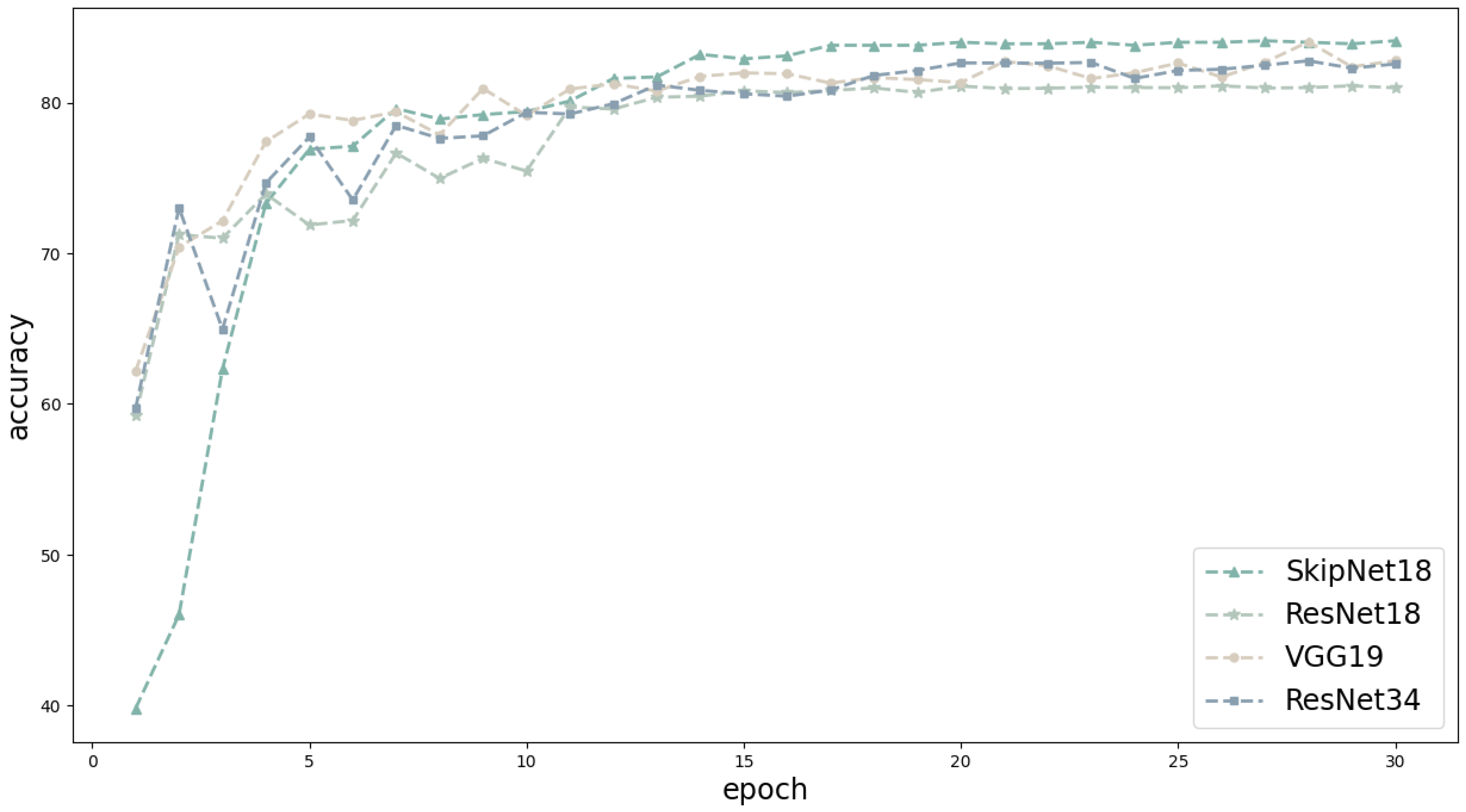
3.3. Results of SkipNet on the CIFAR-10 Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.; Liu, H.; Meng, Z.; Chen, J. Deep learning-based automatic recognition network of agricultural machinery images. Comput. Electron. Agric. 2019, 166, 104978. [Google Scholar] [CrossRef]
- Yang, K.; Liu, H.; Wang, P.; Meng, Z.; Chen, J. Convolutional neural network-based automatic image recognition for agricultural machinery. Int. J. Agric. Biol. Eng. 2018, 11, 200–206. [Google Scholar] [CrossRef]
- Adve, V.; Wedow, J.; Ainsworth, E.; Chowdhary, G.; Green-Miller, A.; Tucker, C. AIFARMS: Artificial intelligence for future agricultural resilience, management, and sustainability. AI Mag. 2024, 45, 83–88. [Google Scholar] [CrossRef]
- Sun, T.; Cui, L.; Zong, L.; Zhang, S.; Jiao, Y.; Xue, X.; Jin, Y. Weed Recognition at Soybean Seedling Stage Based on YOLOV8nGP+ NExG Algorithm. Agronomy 2024, 14, 657. [Google Scholar] [CrossRef]
- Jiang, W.; Quan, L.; Wei, G.; Chang, C.; Geng, T. A conceptual evaluation of a weed control method with post-damage application of herbicides: A composite intelligent intra-row weeding robot. Soil Tillage Res. 2023, 234, 105837. [Google Scholar] [CrossRef]
- Sheela, J.; Karthika, N.; Janet, B. SSLnDO-Based Deep Residual Network and RV-Coeącient Integrated Deep Fuzzy Clustering for Cotton Crop Classification. Int. J. Inf. Technol. Decis. Mak. 2024, 23, 381–412. [Google Scholar]
- Zheng, Y.; Zhu, Q.; Huang, M.; Guo, Y.; Qin, J. Maize and weed classification using color indices with support vector data description in outdoor fields. Comput. Electron. Agric. 2017, 141, 215–222. [Google Scholar] [CrossRef]
- Sajad, S.; Yousef, A.; Juan, I. An automatic visible-range video weed detection, segmentation and classification prototype in potato field. Heliyon 2020, 6, e03685. [Google Scholar]
- Adel, B.; Abdolabbas, J. Evaluation of support vector machine and artificial neural networks in weed detection using shape features. Comput. Electron. Agric. 2018, 145, 153–160. [Google Scholar]
- Kounalakis, T.; Triantafyllidis, G.A.; Nalpantidis, L. Weed recognition framework for robotic precision farming. In Proceedings of the 2016 IEEE International Conference on Imaging Systems and Techniques (IST), Chania, Greece, 4–6 October 2016; pp. 466–471. [Google Scholar]
- Bakhshipour, A.; Jafari, A.; Nassiri, S.M.; Zare, D. Weed segmentation using texture features extracted from wavelet sub-images. Biosyst. Eng. 2017, 157, 1–12. [Google Scholar] [CrossRef]
- Le, V.N.T.; Apopei, B.; Alameh, K. Effective plant discrimination based on the combination of local binary pattern operators and multiclass support vector machine methods. Inf. Process. Agric. 2019, 6, 116–131. [Google Scholar]
- Sun, Y.; Chen, Y.; Jin, X.; Yu, J.; Chen, Y. An artificial intelligence-based method for recognizing seedlings and weeds in Brassica napus. Fujian J. Agric. Sci. 2021, 36, 1484–1490. [Google Scholar]
- Jiang, H.; Zhang, C.; Qiao, Y.; Zhang, Z.; Zhang, W.; Song, C. CNN feature based graph convolutional network for weed and crop recognition in smart farming. Comput. Electron. Agric. 2020, 174, 105450. [Google Scholar] [CrossRef]
- Cecili, G.; De Fioravante, P.; Dichicco, P.; Congedo, L.; Marchetti, M.; Munafò, M. Land Cover Mapping with Convolutional Neural Networks Using Sentinel-2 Images: Case Study of Rome. Land 2023, 12, 879. [Google Scholar] [CrossRef]
- Tao, T.; Wei, X. A hybrid CNN–SVM classifier for weed recognition in winter rape field. Plant Methods 2022, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xue, J.; Li, D.; Zhang, W.; Chiew, T.; Xu, Z. Image recognition based on lightweight convolutional neural network: Recent advances. Image Vis. Comput. 2024, 146, 105037. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- dos Santos Ferreira, A.; Freitas, D.M.; da Silva, G.G.; Pistori, H.; Folhes, M.T. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- Zhang, L.; Jin, X.; Fu, L.; Li, S. Recognition method for weeds in rapeseed field based on Faster R-CNN deep network. Laser Optoelectron. Prog. 2020, 57, 304–312. [Google Scholar] [CrossRef]
- Garibaldi-Márquez, F.; Flores, G.; Mercado-Ravell, D.A.; Ramírez-Pedraza, A.; Valentín-Coronado, L.M. Weed Classification from Natural Corn Field-Multi-Plant Images Based on Shallow and Deep Learning. Sensors 2022, 22, 3021. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Zhao, J.; Gu, Y.; Zhang, S.; Qiao, X.; Tian, W.; Han, Y. Classification of weed seeds based on visual images and deep learning. Inf. Process. Agric. 2023, 10, 40–51. [Google Scholar] [CrossRef]
- Babu, V.S.; Ram, N.V. Deep residual CNN with contrast limited adaptive histogram equalization for weed detection in soybean crops. Trait. Signal 2022, 39, 717–722. [Google Scholar] [CrossRef]
- Manikandakumar, M.; Karthikeyan, P. Weed classification using particle swarm optimization and deep learning models. Comput. Syst. Sci. Eng. 2023, 44, 913–927. [Google Scholar] [CrossRef]
- Xu, K.; Yuen, P.; Xie, Q.; Zhu, Y.; Cao, W.; Ni, J. WeedsNet: A dual attention network with RGB-D image for weed detection in natural wheat field. Precis. Agric. 2024, 25, 460–485. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Feng, Z.; Ji, H.; Daković, M.; Cui, X.; Zhu, M.; Stanković, L. Cluster-CAM: Cluster-weighted visual interpretation of CNNs’ decision in image classification. Neural Netw. 2024, 178, 106473. [Google Scholar] [CrossRef] [PubMed]
- Krstinić, D.; Skelin, A.; Slapničar, I.; Braović, M. Multi-Label Confusion Tensor. IEEE Access 2024, 12, 9860–9870. [Google Scholar] [CrossRef]
- Giselsson, T.M.; Jørgensen, R.; Jensen, P.; Dyrmann, M.; Midtiby, H. A Public Image Database for Benchmark of Plant Seedling Classification Algorithms. arXiv 2017, arXiv:1711.05458. [Google Scholar]
- Krizhevsky, A. Learning Multiple Layers of Features from Tiny Images. Master’s Thesis, University of Toronto, Toronto, ON, Canada, 2009. [Google Scholar]
- Li, H.; Rajbahadur, G.; Lin, D.; Bezemer, C.; Jiang, Z. Keeping Deep Learning Models in Check: A History-Based Approach to Mitigate Overfitting. IEEE Access 2024, 12, 70676–70689. [Google Scholar] [CrossRef]
- Song, Y.; Zou, Y.; Li, Y.; He, Y.; Wu, W.; Niu, R.; Xu, S. Enhancing Landslide Detection with SBConv-Optimized U-Net Architecture Based on Multisource Remote Sensing Data. Land 2024, 13, 835. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chavan, T.R.; Nandedkar, A.V. AgroAVNET for crops and weeds classification: A step forward in automatic farming. Comput. Electron. Agric. 2018, 154, 361–372. [Google Scholar] [CrossRef]
- Mu, Y.; Ni, R.; Fu, L.; Luo, T.; Feng, R.; Li, J.; Li, S. DenseNet weed recognition model combining local variance preprocessing and attention mechanism. Front. Plant Sci. 2023, 13, 1041510. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Su, W. Deep Learning-Based Weed–Crop Recognition for Smart Agricultural Equipment: A Review. Agronomy 2024, 14, 363. [Google Scholar] [CrossRef]
- Søgaard, H.T.; Lund, I.; Graglia, E. Real-time application of herbicides in seed lines by computer vision and micro-spray system. In Proceedings of the 2006 ASAE Annual Meeting, Portland, Oregon, 9–12 July 2006; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2006. [Google Scholar]
- Qu, F.; Li, W.; Yang, Y.; Liu, H.; Hao, Z. Crop weed recognition based on image enhancement and attention mechanism. Comput. Eng. Des. 2023, 44, 815–821. [Google Scholar]
- Wang, X.Y.; Zhang, C.; Zhang, L. Recognition of similar rose based on convolution neural network. J. Anhui Agric. Univ. 2021, 48, 504–510. [Google Scholar]
- Chen, Y.; Xu, H.; Chang, P.; Huang, Y.; Zhong, F.; Jia, Q.; Chen, L.; Zhong, H.; Liu, S. CES-YOLOv8: Strawberry Maturity Detection Based on the Improved YOLOv8. Agronomy 2024, 14, 1353. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Wang, T.; Zhang, F.; Wang, Z. EResNet-SVM: An overfitting-relieved deep learning model for recognition of plant diseases and pests. J. Sci. Food Agric. 2024, 104, 6018–6034. [Google Scholar] [CrossRef] [PubMed]
- Deb, M.; Dhal, K.G.; Das, A.; Hussien, A.G.; Abualigah, L.; Garai, A. A CNN-based model to count the leaves of rosette plants (LC-Net). Sci. Rep. 2024, 14, 1496. [Google Scholar] [CrossRef] [PubMed]

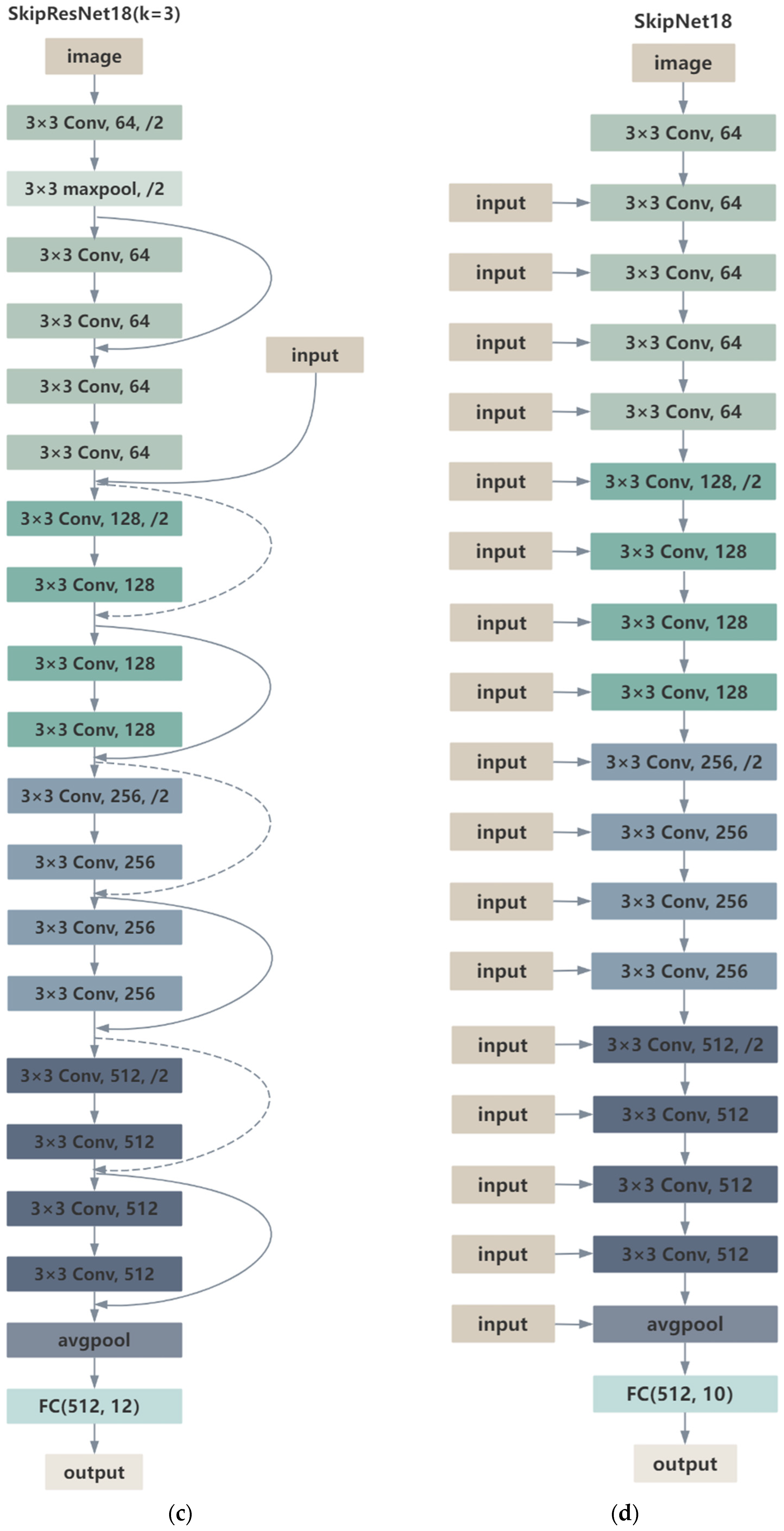




| Layer | Data Out Size | SkipResNet18 |
|---|---|---|
| conv1 | 64 × 64 | 3 × 3, 64, s = 2 |
| conv2 | 32 × 32 | 3 × 3 max pool, s = 2 |
| × 2 | ||
| conv3 | 16 × 16 | × 2 |
| conv4 | 8 × 8 | × 2 |
| conv5 | 4 × 4 | × 2 |
| 1 × 1 | average pool, 12-d fc, softmax |
| Class | Species | Training Set | Test Set | Total |
|---|---|---|---|---|
| 1 | Black-grass | 279 | 30 | 309 |
| 2 | Charlock | 407 | 45 | 452 |
| 3 | Cleavers | 302 | 33 | 335 |
| 4 | Common chickweed | 642 | 71 | 713 |
| 5 | Common wheat | 228 | 25 | 253 |
| 6 | Fat hen | 485 | 53 | 538 |
| 7 | Loose silky-bent | 686 | 76 | 762 |
| 8 | Maize | 232 | 25 | 257 |
| 9 | Scentless mayweed | 542 | 60 | 607 |
| 10 | Shepherd’s purse | 247 | 27 | 274 |
| 11 | Small-flowered cranesbill | 519 | 57 | 576 |
| 12 | Sugar beet | 417 | 46 | 463 |
| Total | 4991 | 548 | 5539 |
| Model | Average Validation Accuracy | Test Accuracy | Test Precision | Test Recall | Test F1 | Parameter |
|---|---|---|---|---|---|---|
| SkipResNet18 | 97.05 | 95.07 | 95.05 | 95.07 | 95.04 | 11,184,588 |
| ResNet18 | 95.97 | 94.34 | 94.42 | 94.34 | 94.09 | 11,174,988 |
| VGG19 | 97.07 | 94.70 | 94.74 | 94.70 | 94.70 | 70,418,892 |
| MobileNetV2 | 93.83 | 90.32 | 90.55 | 90.32 | 89.96 | 2,239,244 |
| Model | Accuracy |
|---|---|
| SkipResNet18 (our method) | 95.07 |
| AgroAVNET [35] | 93.64 |
| Improved DenseNet(without ECA) [36] | 94.34 |
| Model | Accuracy | Recall | F1 | Parameter | |
| SkipNet18 | 84.3 | 84.2 | 84.3 | 84.2 | 11,019,673 |
| ResNet18 | 81.0 | 80.7 | 81.0 | 80.8 | 11,173,962 |
| VGG19 | 82.8 | 83.0 | 82.8 | 82.8 | 38,953,418 |
| ResNet34 | 82.6 | 82.6 | 82.6 | 82.5 | 21,282,122 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Chen, T.; Lan, C.; Liu, S.; Yin, L. SkipResNet: Crop and Weed Recognition Based on the Improved ResNet. Land 2024, 13, 1585. https://doi.org/10.3390/land13101585
Hu W, Chen T, Lan C, Liu S, Yin L. SkipResNet: Crop and Weed Recognition Based on the Improved ResNet. Land. 2024; 13(10):1585. https://doi.org/10.3390/land13101585
Chicago/Turabian StyleHu, Wenyi, Tian Chen, Chunjie Lan, Shan Liu, and Lirong Yin. 2024. "SkipResNet: Crop and Weed Recognition Based on the Improved ResNet" Land 13, no. 10: 1585. https://doi.org/10.3390/land13101585
APA StyleHu, W., Chen, T., Lan, C., Liu, S., & Yin, L. (2024). SkipResNet: Crop and Weed Recognition Based on the Improved ResNet. Land, 13(10), 1585. https://doi.org/10.3390/land13101585








