Using Automated Machine Learning for Spatial Prediction—The Heshan Soil Subgroups Case Study
Abstract
1. Introduction
2. The AutoML Approach
3. Implementation of AutoML
4. Experiment Design
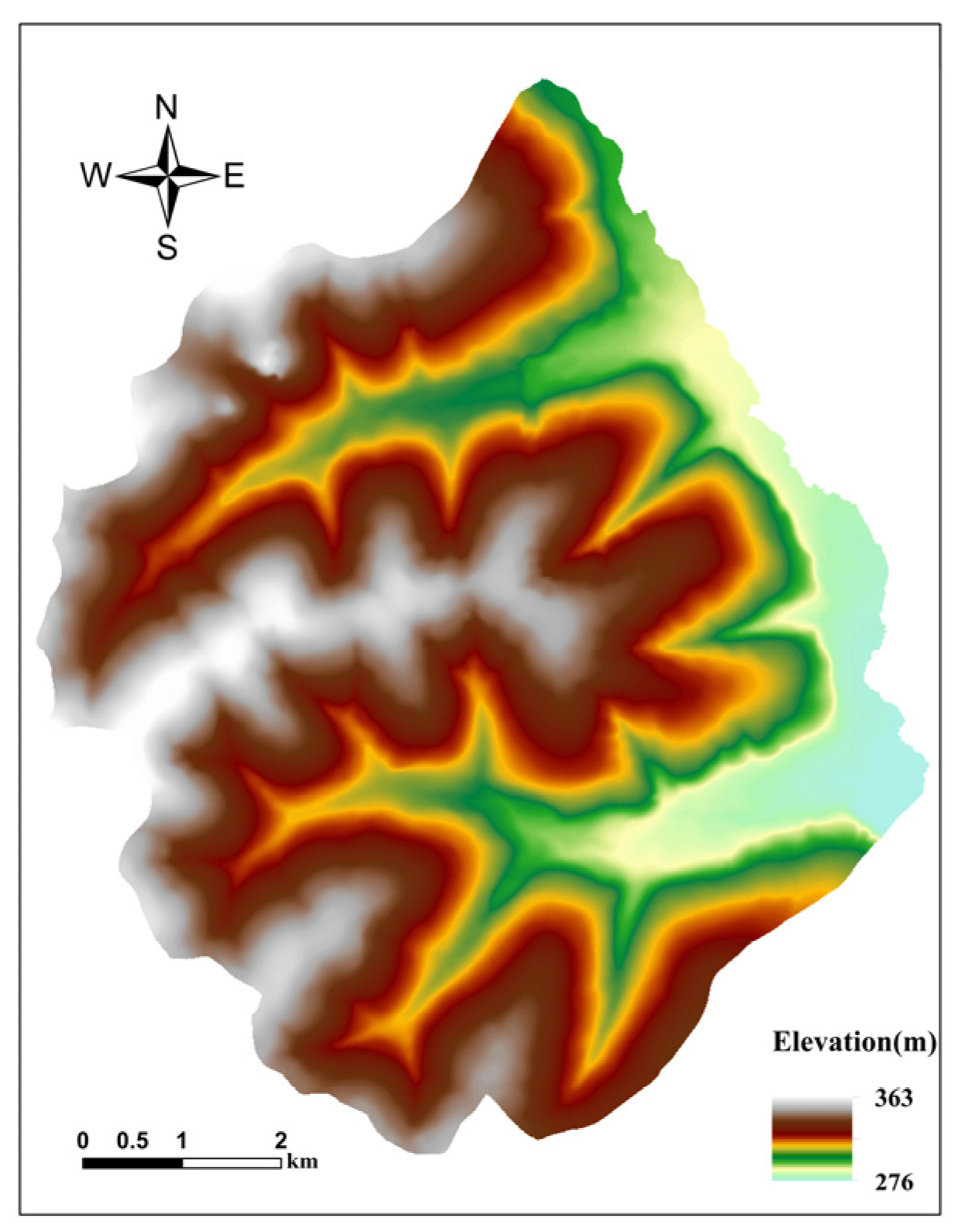
4.1. The Heshan Study Area
4.2. Dataset
4.3. Experimental Environments
4.4. Evaluation Method
4.4.1. Evaluation Aspects
4.4.2. Cross-Validation and Evaluation Index
5. Results
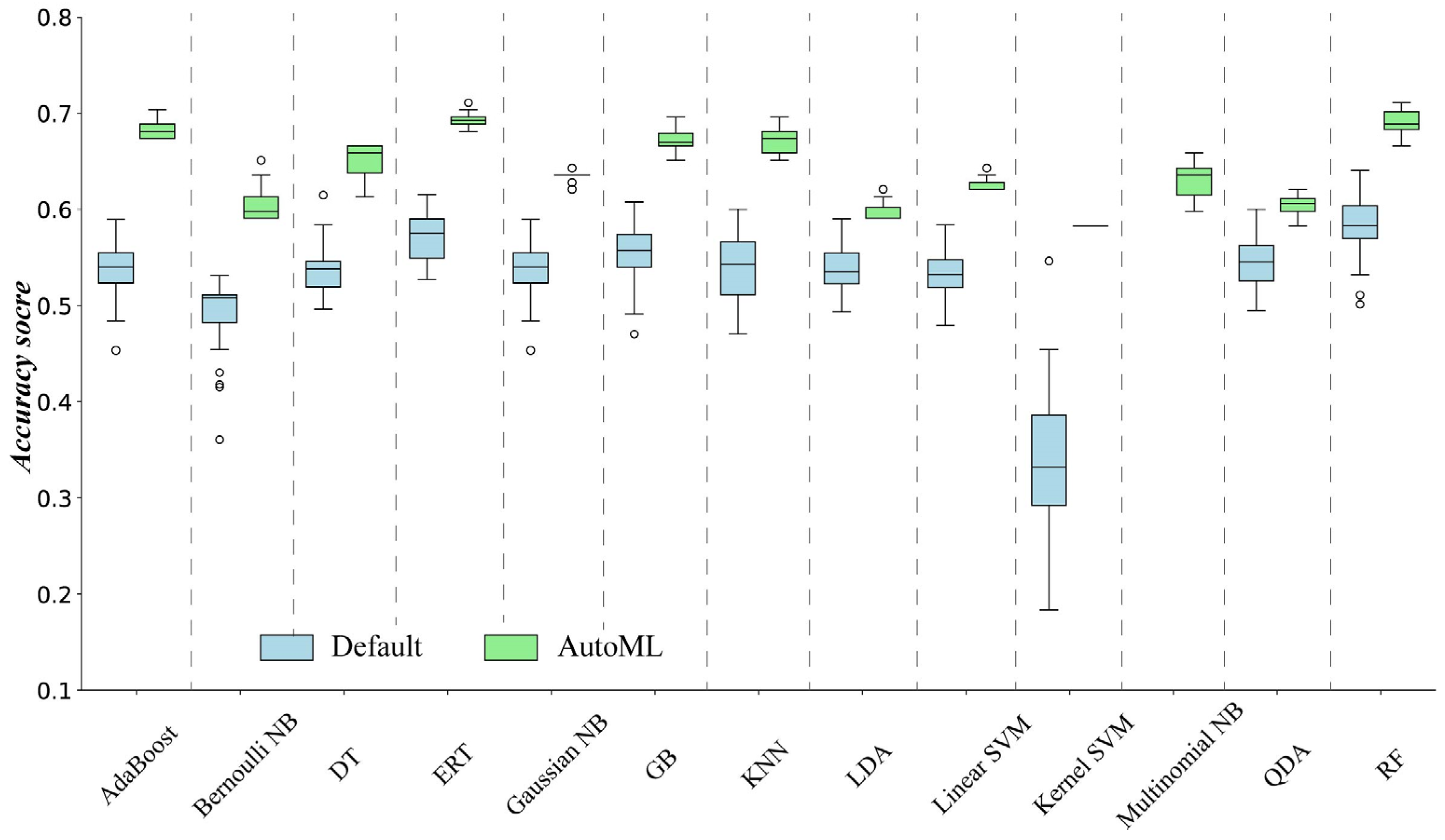
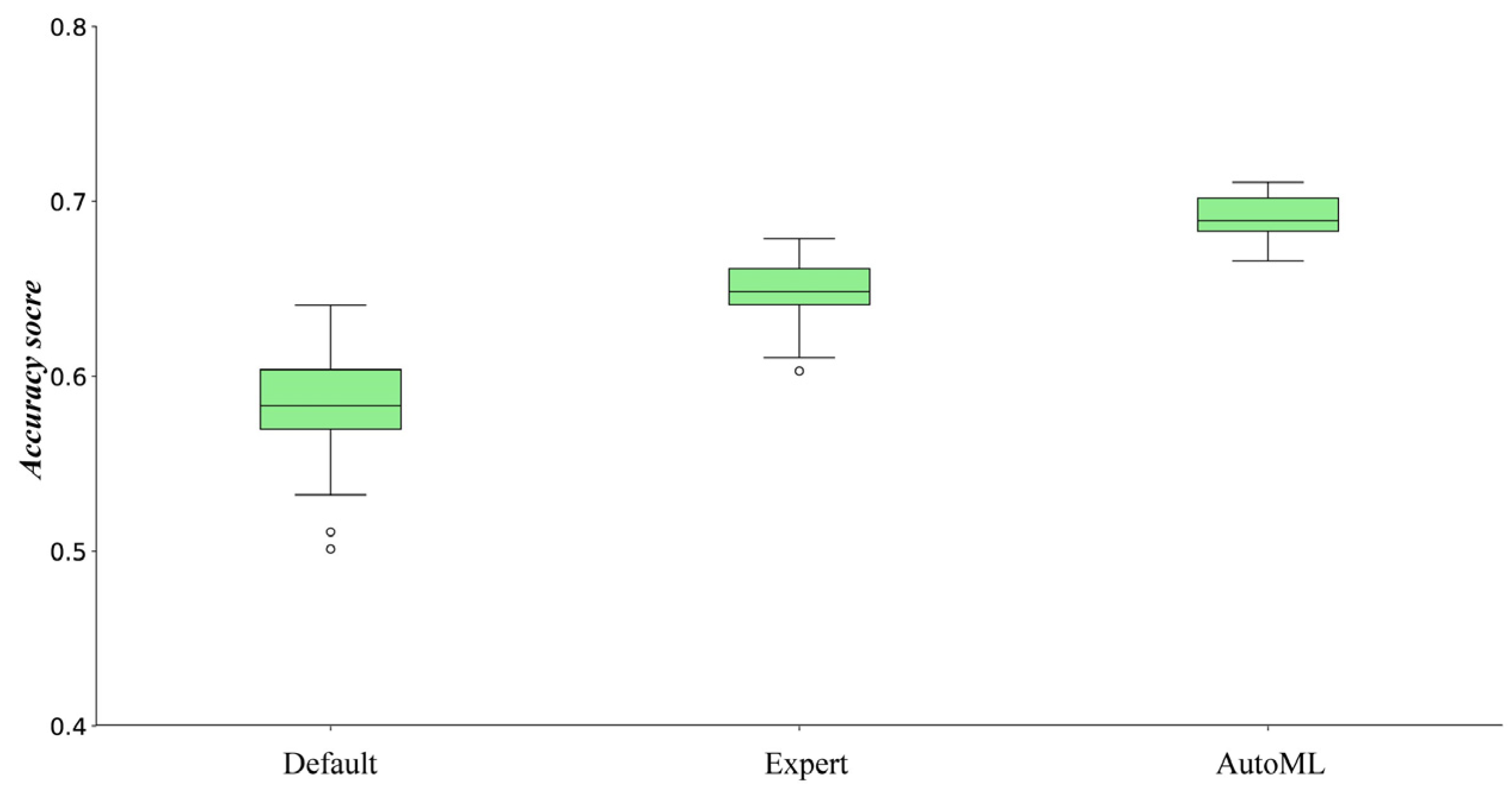
5.1. The Performance of the Determined ML Methods Using Different Ways When a Specified ML Method Exists
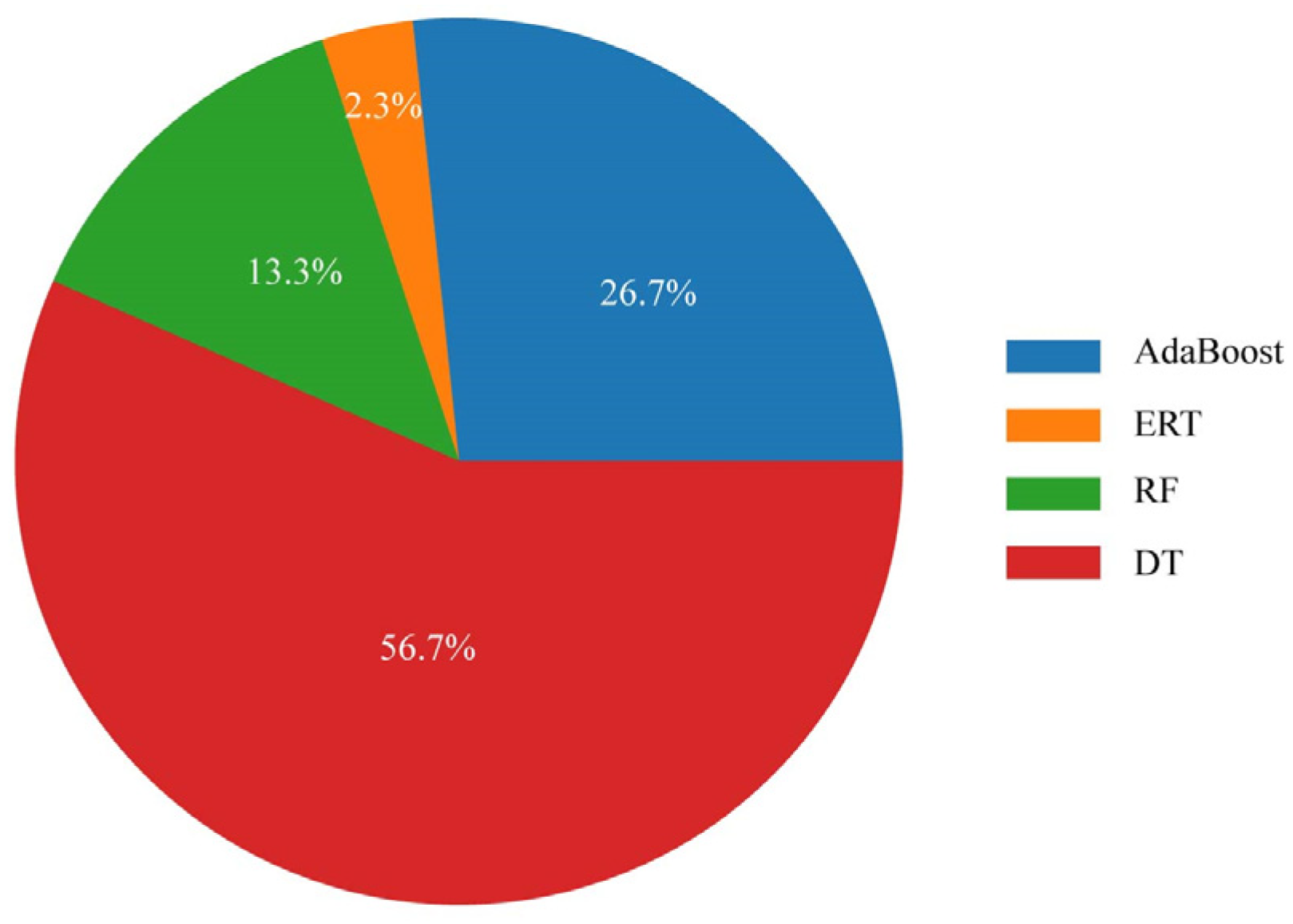
5.2. The Performance of AutoML When Multiple Potential ML Methods Exist
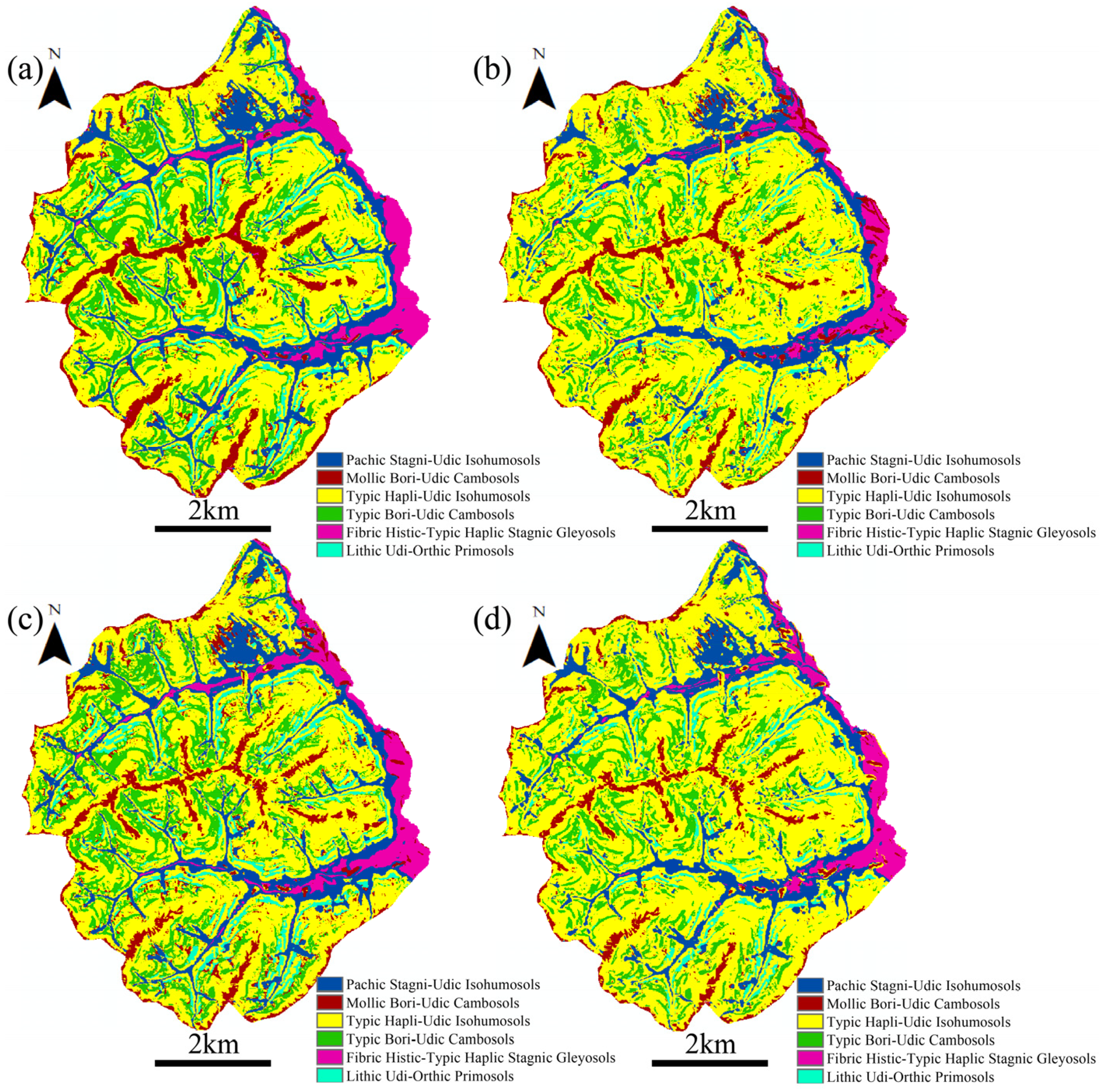
5.3. The Spatial Distribution of Soil Subgroups Predicted by AutoML
6. Discussion
6.1. The Potential Reason for the High Performance of AutoML
6.2. The Influence of the Ensemble on the Performance of AutoML
6.3. The Advantages of AutoML
6.4. The Current Limitations and Future Work of the AutoML
7. Conclusions
- (1)
- Each ML method determined by AutoML outperformed those with default parameter values or determined by expert knowledge.
- (2)
- In scenarios where numerous ML methods are available, AutoML could also identify the highest performing ML method and implement the proper data preprocessing methods.
- (3)
- The accuracy and spatial distribution of soil subgroups predicted by AutoML closely depend on the sample database.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ayalew, L.; Yamagishi, H. The application of GIS-based logistic regression for landslide susceptibility mapping in the Kakuda-Yahiko Mountains, Central Japan. Geomorphology 2005, 65, 15–31. [Google Scholar] [CrossRef]
- Reichenbach, P.; Rossi, M.; Malamud, B.D.; Mihir, M.; Guzzetti, F. A review of statistically-based landslide susceptibility models. Earth-Sci. Rev. 2018, 180, 60–91. [Google Scholar] [CrossRef]
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On Digital Soil Mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Heung, B.; Ho, H.C.; Zhang, J.; Knudby, A.; Bulmer, C.E.; Schmidt, M.G. An Overview and Comparison of Machine-Learning Techniques for Classification Purposes in Digital Soil Mapping. Geoderma 2016, 265, 62–77. [Google Scholar] [CrossRef]
- Zhu, A.X.; Lu, G.; Liu, J.; Qin, C.Z.; Zhou, C.H. Spatial prediction based on Third Law of Geography. Ann. GIS 2018, 24, 225–240. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Song, X.D.; Wang, Y.P.; Canadell, J.G.; Luo, Y.Q.; Ciais, P.; Chen, A.P.; Hong, S.B.; Wang, Y.G.; Tao, F.; et al. Size, distribution, and vulnerability of the global soil inorganic carbon. Science 2024, 384, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, W.; Ziebelin, D.; Hamilton, H. An ontology-based framework for geospatial clustering. Int. J. Geogr. Inf. Sci. 2010, 24, 1601–1630. [Google Scholar] [CrossRef]
- Li, J.; Heap, A.D. Spatial interpolation methods applied in the environmental sciences: A review. Environ. Model. Softw. 2014, 53, 173–189. [Google Scholar] [CrossRef]
- Gibert, K.; Izquierdo, J.; Sànchez-Marrè, M.; Hamilton, S.H.; Rodríguez-Roda, I.; Holmes, G. Which method to use? An assessment of data mining methods in Environmental Data Science. Environ. Model. Softw. 2018, 110, 3–27. [Google Scholar] [CrossRef]
- Hooten, M.B.; Hobbs, N.T. A guide to Bayesian model selection for ecologists. Ecol. Monogr. 2015, 85, 3–28. [Google Scholar] [CrossRef]
- Pourghasemi, H.R.; Rahmati, O. Prediction of the landslide susceptibility: Which algorithm, which precision? Catena 2018, 162, 177–192. [Google Scholar] [CrossRef]
- Daviran, M.; Maghsoudi, A.; Ghezelbash, R.; Pradhan, B. A New Strategy for Spatial Predictive Mapping of Mineral Prospectivity: Automated Hyperparameter Tuning of Random Forest Approach. Comput. Geosci. 2021, 148, 104688. [Google Scholar] [CrossRef]
- Williams, P.J.; Kendall, W.L.; Hooten, M.B. Selecting Ecological Models Using Multi-Objective Optimization. Ecol. Modell. 2019, 404, 21–26. [Google Scholar] [CrossRef]
- Clarke, B.; Fokoue, E.; Zhang, H.H. Principles and Theory for Data Mining and Machine Learning; Springer: New York, NY, USA, 2009; pp. 569–678. [Google Scholar]
- Fourcade, Y.; Besnard, A.G.; Secondi, J. Paintings predict the distribution of species, or the challenge of selecting environmental predictors and evaluation statistics. Glob. Ecol. Biogeogr. 2018, 27, 245–256. [Google Scholar] [CrossRef]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined Selection and Hyperparameter Optimization of Classification Algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 847–855. [Google Scholar]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Efficient and Robust Automated Machine Learning. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Cambridge, MA, USA, 7–12 December 2015; pp. 2755–2763. [Google Scholar]
- Samanta, B. Gear fault detection using artificial neural networks and support vector machines with genetic algorithms. Mech. Syst. Signal Process. 2004, 18, 625–644. [Google Scholar] [CrossRef]
- Bergstra, J.S.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for Hyper-Parameter Optimization. In Proceedings of the 24th International Conference on Neural Information Processing Systems, New York, NY, USA, 12–15 December 2011; pp. 2546–2554. [Google Scholar]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. In Proceedings of the 25th International Conference on Neural Information Processing Systems, New York, NY, USA, 3–6 December 2012; pp. 2951–2959. [Google Scholar]
- Solis, F.J.; Wets, R.J.B. Minimization by Random Search Techniques. Math. Oper. Res. 1981, 6, 19–30. [Google Scholar] [CrossRef]
- Zöller, M.A.; Huber, M.F. Benchmark and Survey of Automated Machine Learning Frameworks. J. Artif. Intell. Res. 2021, 70, 409–472. [Google Scholar] [CrossRef]
- Vilalta, R.; Drissi, Y. A Perspective View and Survey of Meta-Learning. Artif. Intell. Rev. 2002, 18, 77–95. [Google Scholar] [CrossRef]
- Liang, P.; Qin, C.Z.; Zhu, A.X.; Hou, Z.W.; Fan, N.Q.; Wang, Y.J. A case-based method of selecting covariates for digital soil mapping. J. Integr. Agric. 2020, 19, 2127–2136. [Google Scholar] [CrossRef]
- Guyon, I.; Saffari, A.; Dror, G.; Cawley, G. Model Selection: Beyond the Bayesian/Frequentist Divide. J. Mach. Learn. Res. 2010, 11, 61–87. [Google Scholar]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Caruana, R.; Niculescu-Mizil, A.; Crew, G.; Ksikes, A. Ensemble Selection from Libraries of Models. In Proceedings of the Twenty-First International Conference on Machine Learning, New York, NY, USA, 4–8 July 2004; p. 18. [Google Scholar]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- Mendoza, H.; Klein, A.; Feurer, M.; Springenberg, J.T.; Urban, M.; Burkart, M.; Dippel, M.; Lindauer, M.; Hutter, F. Towards Automatically-Tuned Deep Neural Networks. In Automated Machine Learning: Methods, Systems, Challenges; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; Springer: Cham, Switzerland, 2019; pp. 135–149. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Rossiter, D.G.; Zeng, R.; Zhang, G.L. Accounting for taxonomic distance in accuracy assessment of soil class predictions. Geoderma 2017, 292, 118–127. [Google Scholar] [CrossRef]
- Zeng, C.; Yang, L.; Zhu, A.X.; Rossiter, D.G.; Liu, J.; Liu, J.Z.; Qin, C.Z.; Wang, D. Mapping soil organic matter concentration at different scales using a mixed geographically weighted regression method. Geoderma 2016, 281, 69–82. [Google Scholar] [CrossRef]
- Chinese Soil Taxonomy Research Group. Keys to Chinese Soil Taxonomy, 3rd ed.; University of Science and Technology of China Press: Hefei, China, 2001. [Google Scholar]
- Qin, C.Z.; Zhu, A.X.; Shi, X.; Li, B.L.; Pei, T.; Zhou, C.H. Quantification of spatial gradation of slope positions. Geomorphology 2009, 110, 152–161. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Minasny, B.; McBratney, A.B. Machine Learning for Digital Soil Mapping: Applications, Challenges and Suggested Solutions. Earth Sci. Rev. 2020, 210, 103359. [Google Scholar] [CrossRef]
- Jeong, G.; Oeverdieck, H.; Park, S.J.; Huwe, B.; Ließ, M. Spatial soil nutrients prediction using three supervised learning methods for assessment of land potentials in complex terrain. Catena 2017, 154, 73–84. [Google Scholar] [CrossRef]
- Bouslihim, Y.; John, K.; Miftah, A.; Azmi, R.; Aboutayeb, R.; Bouasria, A.; Razouk, R.; Hssaini, L. The Effect of Covariates on Soil Organic Matter and pH Variability: A Digital Soil Mapping Approach Using Random Forest Model. Ann. GIS 2024, 1–18. [Google Scholar] [CrossRef]
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island—Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Poggio, L.; de Sousa, L.M.; Batjes, N.H.; Heuvelink, G.B.M.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing soil information for the globe with quantified spatial uncertainty. SOIL 2021, 7, 217–240. [Google Scholar] [CrossRef]
- Rossi, M.; Guzzetti, F.; Reichenbach, P.; Mondini, A.C.; Peruccacci, S. Optimal landslide susceptibility zonation based on multiple forecasts. Geomorphology 2010, 114, 129–142. [Google Scholar] [CrossRef]
- Zhu, A.X.; Wang, R.; Qiao, J.; Qin, C.Z.; Chen, Y.; Liu, J.; Du, F.; Lin, Y.; Zhu, T. An expert knowledge-based approach to landslide susceptibility mapping using GIS and fuzzy logic. Geomorphology 2014, 214, 128–138. [Google Scholar] [CrossRef]
- Liu, H.; Shi, T.; Chen, Y.; Wang, J.; Fei, T.; Wu, G. Improving Spectral Estimation of Soil Organic Carbon Content through Semi-Supervised Regression. Remote Sens. 2017, 9, 29. [Google Scholar] [CrossRef]
- Henderson, B.L.; Bui, E.N.; Moran, C.J.; Simon, D.A.P. Australia-wide predictions of soil properties using decision trees. Geoderma 2005, 124, 383–398. [Google Scholar] [CrossRef]
- Bonfatti, B.R.; Hartemink, A.E.; Giasson, E.; Tornquist, C.G.; Adhikari, K. Digital mapping of soil carbon in a viticultural region of Southern Brazil. Geoderma 2016, 261, 204–221. [Google Scholar] [CrossRef]
- Odeh, I.O.A.; McBratney, A.B.; Chittleborough, D.J. Further results on prediction of soil properties from terrain attributes: Heterotopic cokriging and regression-kriging. Geoderma 1995, 67, 215–226. [Google Scholar] [CrossRef]
- Sharma, A. Exploratory Spatial Analysis of Food Insecurity and Diabetes: An Application of Multiscale Geographically Weighted Regression. Ann. GIS 2023, 2, 485–498. [Google Scholar] [CrossRef]


| Variables | Scale/ Resolution | Mean (Range) |
|---|---|---|
| Elevation | 10 m | 322 (276–363) |
| Slope | 10 m | 2 (0–17) |
| Profile curvature (ProCur) | 10 m | 0 (−0.01–0.01) |
| Planform curvature (PlanCur) | 10 m | 0 (−1.88–1.57) |
| Topographic wetness index (TWI) | 10 m | 9.38 (4.69–22) |
| Surface Curvature Index (CS) | 10 m | 0 (−0.023–0.016) |
| Landscape Position Index (LPos) | 10 m | 0 (−0.064–0.083) |
| Relative Position Index (RPI) | 10 m | 0.47 (0–1) |
| Stream Power Index (SPI) | 10 m | 32.4 (0–8281) |
| Topographic Position Index (TPI) | 10 m | 0 (−2.57–1.81) |
| Terrain Ruggedness Index (TRI) | 10 m | 0.31 (−0.001–2.75) |
| Method Type | Methods |
|---|---|
| Data preprocessing methods | Balancing class weight, Extremely Randomized Trees, Fast Independent Component Analysis, Feature Agglomeration, Imputation of missing values, Kernel principal component analysis, Linear Support Vector Machines, No preprocessing, Normalization, Nystroem Method for Kernel Approximation, One hot encoding, Polynomial, Principal component analysis, Random Forest, Random Kitchen Sinks, Select percentile, Select rates |
| Classification methods | AdaBoost, Bernoulli Naive Bayes (Bernoulli NB), Decision tree (DT), Extremely Randomized Trees (ERT), Gaussian Naive Bayes (Gaussian NB), Gradient Boosting (GB), Kernel SVM, K-Nearest Neighbors (KNN), Linear Discriminant Analysis (LDA), Linear SVM, Multinomial Naive Bayes (Multi NB), Quadratic Discriminant Analysis (QDA), Random Forest (RF) |
| Median | Min. | Max. | Std. | |
|---|---|---|---|---|
| Workflows | 20 | 11 | 29 | 5.58 |
| The kind of ML methods | 3 | 1 | 7 | 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, P.; Qin, C.-Z.; Zhu, A.-X. Using Automated Machine Learning for Spatial Prediction—The Heshan Soil Subgroups Case Study. Land 2024, 13, 551. https://doi.org/10.3390/land13040551
Liang P, Qin C-Z, Zhu A-X. Using Automated Machine Learning for Spatial Prediction—The Heshan Soil Subgroups Case Study. Land. 2024; 13(4):551. https://doi.org/10.3390/land13040551
Chicago/Turabian StyleLiang, Peng, Cheng-Zhi Qin, and A-Xing Zhu. 2024. "Using Automated Machine Learning for Spatial Prediction—The Heshan Soil Subgroups Case Study" Land 13, no. 4: 551. https://doi.org/10.3390/land13040551
APA StyleLiang, P., Qin, C.-Z., & Zhu, A.-X. (2024). Using Automated Machine Learning for Spatial Prediction—The Heshan Soil Subgroups Case Study. Land, 13(4), 551. https://doi.org/10.3390/land13040551










