Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Production of 10E8 CDR H3 Variants
2.3. Generation of Antigen-Binding Fragment (Fab) of 10E8v4 and 10E8VLS
2.4. Biolayer Interferometry
2.5. Expression, Purification, Crystallization and Structure Determination
2.6. Neutralization Assays
2.7. Traditional Size Exclusion Chromatography
2.8. Product-Specific Size Exclusion Chromatography
2.9. Optimized Hydrophobic Interaction Chromatography
3. Results
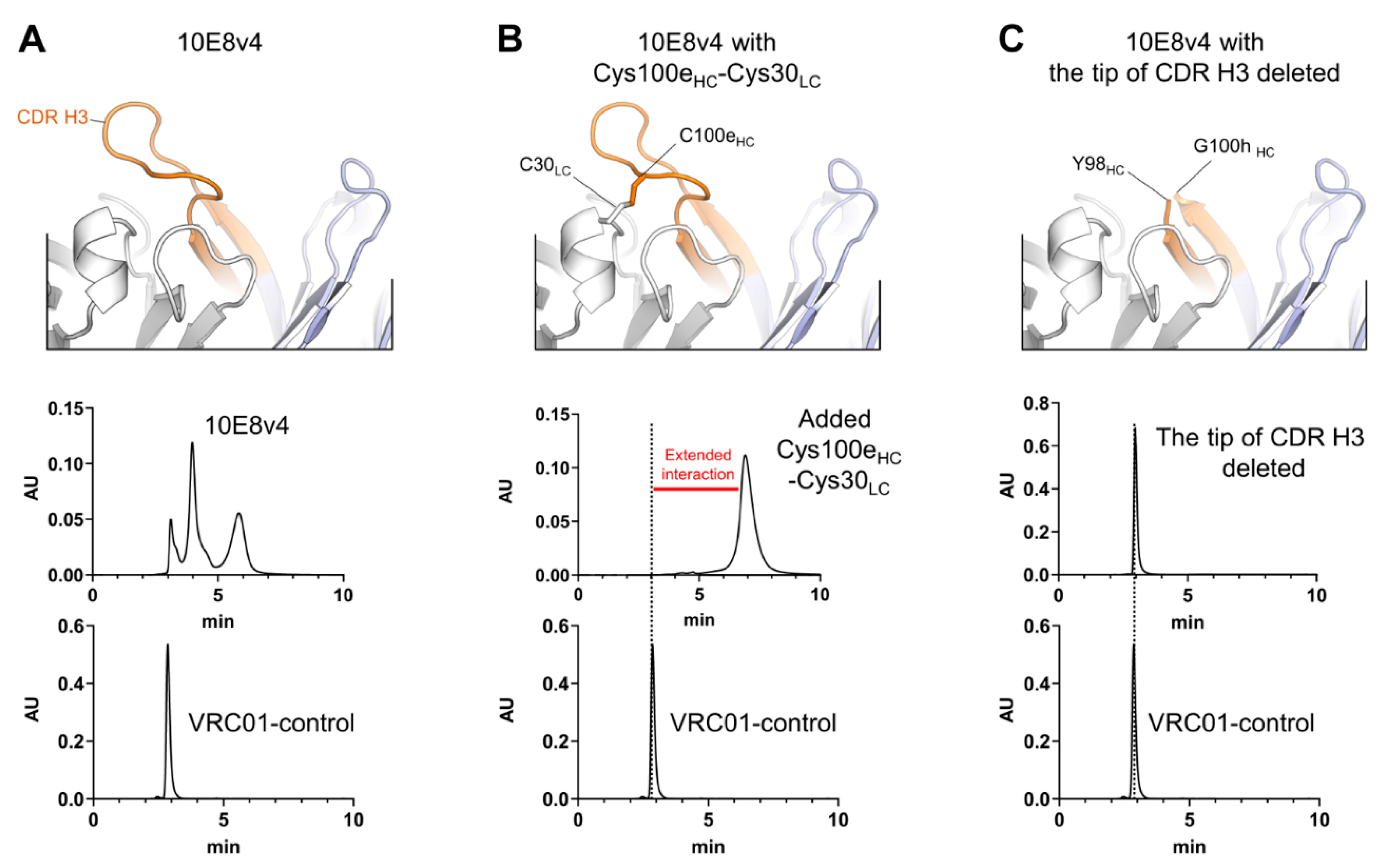
3.1. CDR H3 of 10E8 Variants Is the Cause of Both SEC Heterogeneity and Extended Column Interactions
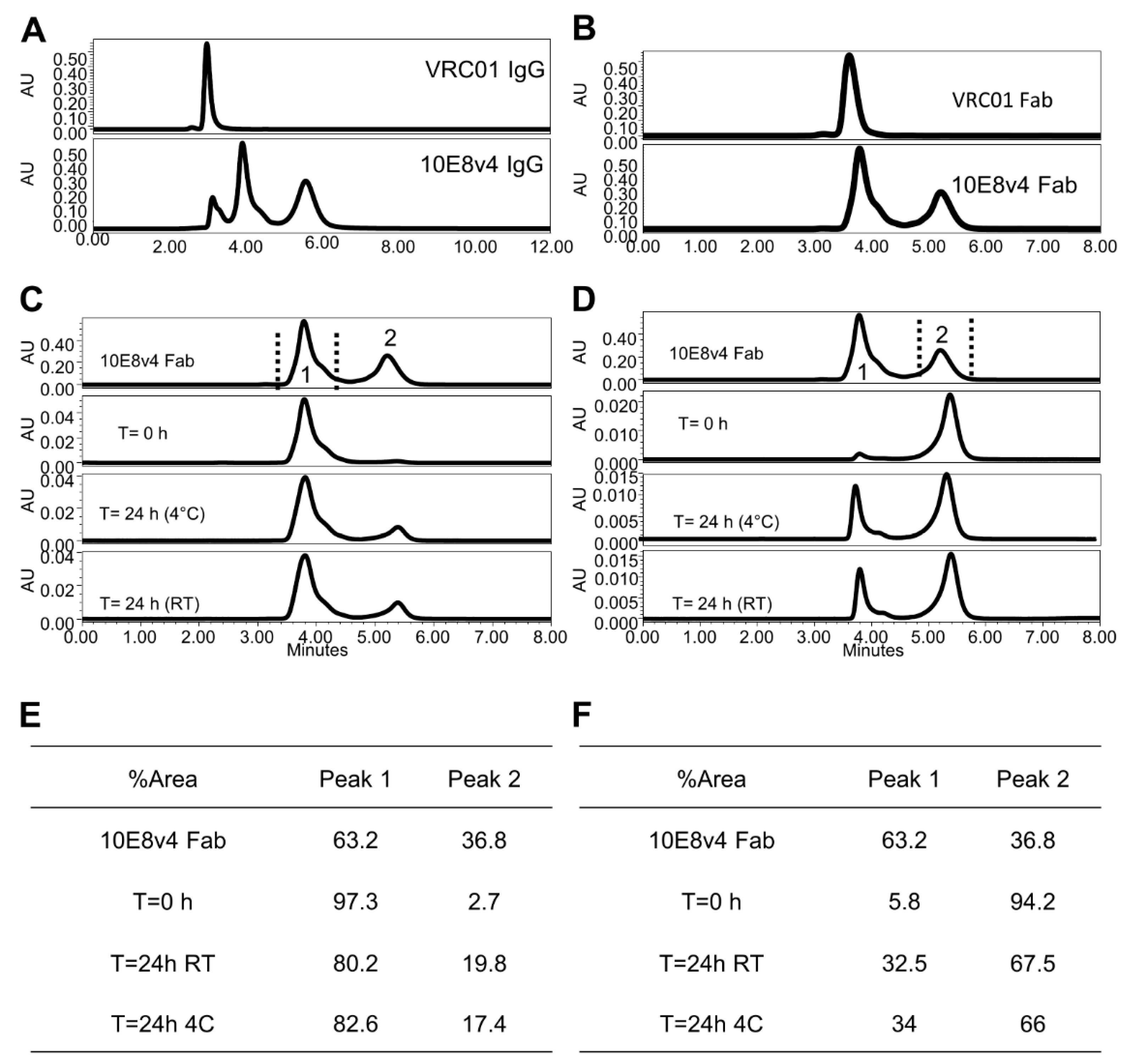
3.2. Two Predominant Slowly Interconverting Conformations of Fab Explain Three Monomeric Population Profile of 10E8 Immunoglobulin (IgG)
3.3. 10E8VLS Immunoglobulin Exists in at Least Three Equilibrium States, Each with Different Hydrophobicity
3.4. Use of Modifier Arginine with Optimized pH Coeffectively Masks Secondary Interactions between 10E8 and Column Matrices
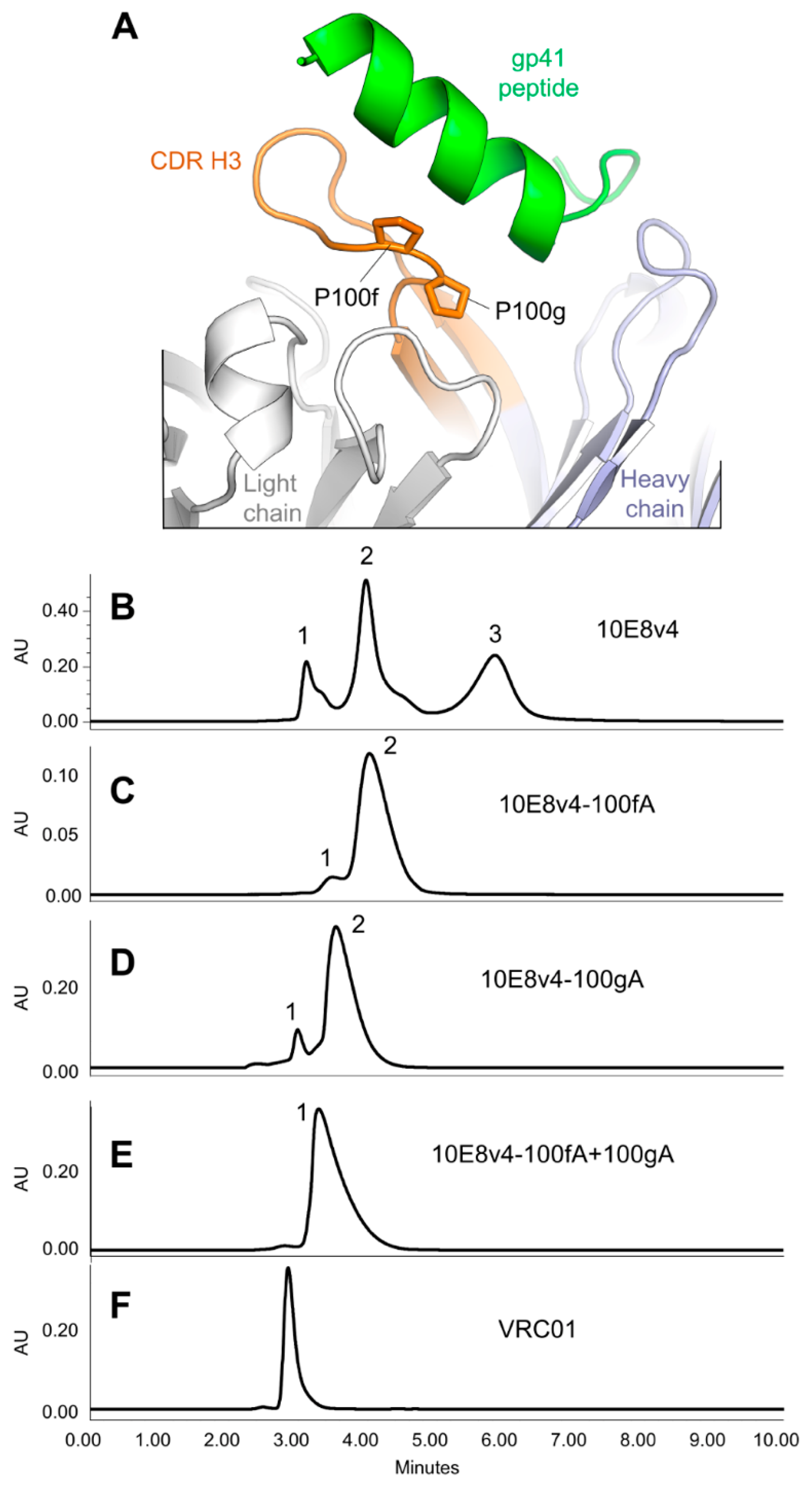
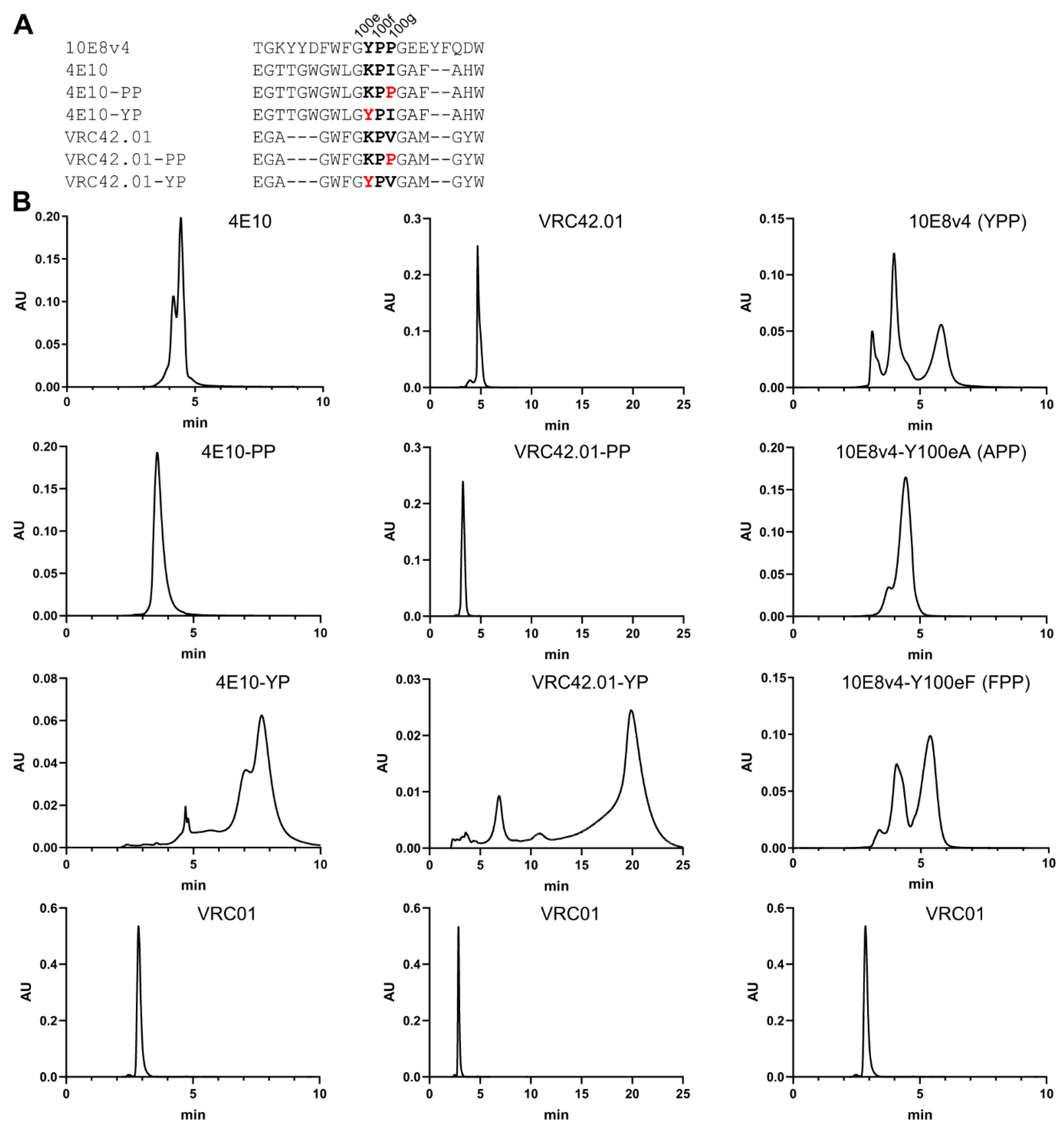
3.5. Modification of the Di-Proline Motif in the 10E8 CDR H3 Alters Both SEC and HIC Profiles
3.6. Crystal Structures of the YPP Motif Variants of 10E8 Reveal the Degree of Structural Compatibility to Trend with Binding Affinity and Neutralization Potency
3.7. Introduction of Tyrosine-Proline to CDR H3 of 4E10 and VRC42 Results in Their Recapitulating the Multi-Peak SEC Behavior of 10E8
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chawla, A.; Wang, C.; Patton, C.; Murray, M.; Punekar, Y.; De Ruiter, A.; Steinhart, C. A Review of Long-Term Toxicity of Antiretroviral Treatment Regimens and Implications for an Aging Population. Infect. Dis. Ther. 2018, 7, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Danforth, K.; Granich, R.; Wiedeman, D.; Baxi, S.; Padian, N. Global Mortality and Morbidity of HIV/AIDS, in Major Infectious Diseases; Holmes, K.K., Bertozzi, S., Bloom, B.R., Jha, P., Eds.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, USA, 2017. [Google Scholar]
- Girum, T.; Wasie, A.; Worku, A. Trend of HIV/AIDS for the last 26 years and predicting achievement of the 90–90-90 HIV prevention targets by 2020 in Ethiopia: A time series analysis. BMC Infect. Dis. 2018, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Catz, S.L.; Kelly, J.A.; Bogart, L.M.; Benotsch, E.G.; McAuliffe, T.L. Patterns, correlates, and barriers to medication adherence among persons prescribed new treatments for HIV disease. Health Psychol. 2000, 19, 124. [Google Scholar] [CrossRef]
- Günthard, H.F.; Saag, M.S.; Benson, C.A.; Del Rio, C.; Eron, J.J.; Gallant, J.E.; Hoy, J.F.; Mugavero, M.J.; Sax, P.E.; Thompson, M.A. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2016 Recommendations of the International Antiviral Society-USA Panel. Jama 2016, 316, 191–210. [Google Scholar] [CrossRef]
- Maenza, J.; Flexner, C. Combination antiretroviral therapy for HIV infection. Am. Fam. Physician 1998, 57, 2789. [Google Scholar]
- Sabin, C.A. Do people with HIV infection have a normal life expectancy in the era of combination antiretroviral therapy? BMC Med. 2013, 11, 251. [Google Scholar] [CrossRef]
- Soriano, V.; Fernandez-Montero, J.V.; Benitez-Gutierrez, L.; De Mendoza, C.; Arias, A.; Barreiro, P.; Peña, J.M.; Labarga, P. Dual antiretroviral therapy for HIV infection. Expert Opin. Drug Saf. 2017, 16, 923–932. [Google Scholar] [CrossRef]
- Huang, J.; Ofek, G.; Laub, L.; Louder, M.K.; Doria-Rose, N.A.; Longo, N.S.; Imamichi, H.; Bailer, R.T.; Chakrabarti, B.; Sharma, S.K.; et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012, 491, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.D.; Chuang, G.-Y.; Zhang, B.; Bailer, R.T.; Doria-Rose, N.A.; Gindin, T.S.; Lin, B.; Louder, M.K.; McKee, K.; O’Dell, S.; et al. Surface-Matrix Screening Identifies Semi-specific Interactions that Improve Potency of a Near Pan-reactive HIV-1-Neutralizing Antibody. Cell Rep. 2018, 22, 1798–1809. [Google Scholar] [CrossRef]
- Kwon, Y.D.; Georgiev, I.S.; Ofek, G.; Zhang, B.; Asokan, M.; Bailer, R.T.; Bao, A.; Caruso, W.; Chen, X.; Choe, M.; et al. Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design. J. Virol. 2016, 90, 5899–5914. [Google Scholar] [CrossRef]
- Irimia, A.; Serra, A.M.; Sarkar, A.; Jacak, R.; Kalyuzhniy, O.; Sok, D.; Saye-Francisco, K.L.; Schiffner, T.; Tingle, R.; Kubitz, M.; et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog. 2017, 13, e1006212. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.-L.; Guillarme, D. Theory and practice of size exclusion chromatography for the analysis of protein aggregates. J. Pharm. Biomed. Anal. 2014, 101, 161–173. [Google Scholar] [CrossRef]
- Hong, P.; Koza, S.; Bouvier, E.S.P. A Review Size-Exclusion Chromatography for the Analysis of Protein Biotherapeutics And Their Aggregates. J. Liq. Chromatogr. Relat. Technol. 2012, 35, 2923–2950. [Google Scholar] [CrossRef]
- Striegel, A.M. Modern Size-Exclusion Liquid Chromatography: Practice of Gel Permeation and Gel Filtration Chromatography, 2nd ed.; Wiley: Hoboken, NJ, USA, 2009; p. 494. [Google Scholar]
- Li, Y.; Wang, X.E.; Bender, M.F.; Yang, R.; Cozine, T.J.; Atallah, K.M.; Arnold, F.; Cooper, J.W.; Lei, Q.P. Overcoming the Multiple-Monomeric-Peak Profile of Broadly Neutralizing HIV-1 Antibody 10E8 with a Unique Size-Exclusion-Chromatography Method. Anal. Chem. 2018, 90, 12390–12394. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Padte, N.N.; Huang, Y.; Yu, J.; Rocklin, G.J.; Weitzner, B.D.; Scian, M.; Ho, D.D.; Lee, K.K. The influence of proline isomerization on potency and stability of anti-HIV antibody 10E8. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Masiero, A.; Nelly, L.; Marianne, G.; Christophe, S.; Florian, L.; Ronan, C.; Claire, B.; Cornelia, Z.; Grégoire, B.; Eric, L.; et al. The impact of proline isomerization on antigen binding and the analytical profile of a trispecific anti-HIV antibody. mAbs 2020, 12, 1698128. [Google Scholar] [CrossRef]
- Goyon, A.; Beck, A.; Colas, O.; Sandra, K.; Guillarme, D.; Fekete, S. Evaluation of size exclusion chromatography columns packed with sub-3mum particles for the analysis of biopharmaceutical proteins. J. Chromatogr. A 2017, 1498, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Roush, D.; Cramer, S. Domain contributions to antibody retention in multimodal chromatography systems. J. Chromatogr. A 2018, 1563, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Krebs, S.J.; Kwon, Y.D.; Schramm, C.A.; Law, W.H.; Donofrio, G.; Zhou, K.H.; Gift, S.; Dussupt, V.; Georgiev, I.S.; Schätzle, S.; et al. Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual. Immunity 2019, 50, 677–691.e13. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C. Phaser crystallographic software. J. Appl. Cryst. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Sarzotti-Kelsoe, M.; Bailer, R.T.; Turk, E.; Lin, C.-L.; Bilska, M.; Greene, K.M.; Gao, H.; Todd, C.A.; Ozaki, D.A.; Seaman, M.S.; et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 2014, 409, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Georgiev, I.; Wu, X.; Yang, Z.-Y.; Dai, K.; Finzi, A.; Kwon, Y.D.; Scheid, J.F.; Shi, W.; Xu, L.; et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science 2010, 329, 811–817. [Google Scholar] [CrossRef]
- Henry, C. Product Review: SEC Weighs In. Anal. Chem. 1996, 68, 431A–434A. [Google Scholar] [CrossRef]
- Hoang, N.-L.; Landolfi, A.; Kravchuk, A.; Girard, E.; Peate, J.; Hernandez, J.M.; Gaborieau, M.; Kravchuk, O.; Gilbert, R.; Guillaneuf, Y.; et al. Toward a full characterization of native starch: Separation and detection by size-exclusion chromatography. J. Chromatogr. A 2008, 1205, 60–70. [Google Scholar] [CrossRef]
- Ejima, D.; Yumioka, R.; Arakawa, T.; Tsumoto, K. Arginine as an effective additive in gel permeation chromatography. J. Chromatogr. A 2005, 1094, 49–55. [Google Scholar] [CrossRef]
- Das, U.; Hariprasad, G.; Ethayathulla, A.S.; Manral, P.; Das, T.K.; Pasha, S.; Mann, A.; Ganguli, M.; Verma, A.K.; Bhat, R.; et al. Inhibition of Protein Aggregation: Supramolecular Assemblies of Arginine Hold the Key. PLoS ONE 2007, 2, e1176. [Google Scholar] [CrossRef]
- Fitch, C.A.; Platzer, G.; Okon, M.; Garcia-Moreno, E.B.; McIntosh, L.P. Arginine: Its pKa value revisited. Protein Sci. 2015, 24, 752–761. [Google Scholar] [CrossRef]
- Harms, M.; Schlessman, J.L.; Sue, G.R. Arginine residues at internal positions in a protein are always charged. Proc. Natl. Acad. Sci. USA 2011, 108, 18954–18959. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, M.J. (Ed.) The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals; Royal Society of Chemistry: Cambridge, UK, 2013. [Google Scholar]
- Wu, T.T.; Kabat, E.A. An Analysis of the Sequences of the Variable Regions Of Bence Jones Proteins And Myeloma Light Chains And Their Implications For Antibody Complementarity. J. Exp. Med. 1970, 132, 211–250. [Google Scholar] [CrossRef] [PubMed]
- Taler-Verčič, A.; Hasanbašić, S.; Berbić, S.; Stoka, V.; Turk, D.; Žerovnik, E. Proline Residues as Switches in Conformational Changes Leading to Amyloid Fibril Formation. Int. J. Mol. Sci. 2017, 18, 549. [Google Scholar] [CrossRef]
- Fassolari, M.; Chemes, L.B.; Gallo, M.; Smal, C.; Sánchez, I.E.; de Prat-Gay, G. Minute Time Scale Prolyl Isomerization Governs Antibody Recognition of an Intrinsically Disordered Immunodominant Epitope. J. Biol. Chem. 2013, 288, 13110–13123. [Google Scholar] [CrossRef] [PubMed]
- Wedemeyer, W.J.; Welker, E.; Scheraga, H.A. Proline cis-trans isomerization and protein folding. Biochemistry 2002, 41, 14637–14644. [Google Scholar] [CrossRef]
- Xu, L.; Pegu, A.; Rao, E.; Doria-Rose, N.; Beninga, J.; McKee, K.; Lord, D.M.; Wei, R.R.; Deng, G.; Louder, M.; et al. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 2017, 358, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.M.; Zwick, M.B.; Stanfield, R.L.; Kunert, R.; Binley, J.M.; Katinger, H.; Burton, D.R.; Wilson, I.A. Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41. Immunity 2005, 22, 163–173. [Google Scholar] [CrossRef]
- Dyson, H.; Rance, M.; Houghten, R.A.; Lerner, R.A.; Wright, P. Folding of immunogenic peptide fragments of proteins in water solution: I. Sequence requirements for the formation of a reverse turn. J. Mol. Biol. 1988, 201, 161–200. [Google Scholar] [CrossRef]
- Yao, J.; Feher, V.A.; Espejo, B.F.; Reymond, M.T.; Wright, P.; Dyson, H. Stabilization of a type VI turn in a family of linear peptides in water solution. J. Mol. Biol. 1994, 243, 736–753. [Google Scholar] [CrossRef]
- Wu, W.J.; Raleigh, D.P. Local control of peptide conformation: Stabilization of cis proline peptide bonds by aromatic proline interactions. Biopolymers 1998, 45, 381–394. [Google Scholar] [CrossRef]
- Reimer, U.; Scherer, G.; Drewello, M.; Kruber, S.; Schutkowski, M.; Fischer, G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 1998, 279, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, B.; Chakrabarti, P.; Basu, G. Enhanced stability of cis Pro-Pro peptide bond in Pro-Pro-Phe sequence motif. FEBS Lett. 2007, 581, 4529–4532. [Google Scholar] [CrossRef]
- Brown, A.M.; Zondlo, N.J. A Propensity Scale for Type II Polyproline Helices (PPII): Aromatic Amino Acids in Proline-Rich Sequences Strongly Disfavor PPII Due to Proline–Aromatic Interactions. Biochemistry 2012, 51, 5041–5051. [Google Scholar] [CrossRef] [PubMed]
- Zondlo, N.J. Aromatic-proline interactions: Electronically tunable CH/pi interactions. Acc. Chem. Res. 2013, 46, 1039–1049. [Google Scholar] [CrossRef] [PubMed]


| 10E8v4-P100fA | 10E8v4-P100fA | 10E8v4-P100gA | 10E8v4-P100fA-P100gA | 10E8v4-H31FLC | |
|---|---|---|---|---|---|
| PDB accession code | 7MF9 | 7MF8 | 7MF7 | 7MFA | 7MFB |
| Data collection | |||||
| Space group | C2 | P6422 | C2 | P2 | C2 |
| Cell constants | |||||
| a, b, c (Å) | 160.9, 141.3, 137.4 | 42.0, 149.7, 160.2 | 220.2, 43.6, 226.5 | 95.8, 53.8, 328.5 | 141.8, 56.2, 69.2 |
| α, b, g (°) | 90.0, 1, 109.2, 90.0 | 90.0, 90.0, 90.0 | 90.0, 115.3, 90.0 | 90.0, 90.6, 90.0 | 90, 100.8, 90 |
| Unique reflections | 24,124 | 36,152 | 127,973 | 78,164 | 48,111 |
| Wavelength (Å) | 1 | 1 | 1 | 1 | 1 |
| Resolution (Å) | 50.0–3.70 (3.76–3.70) * | 50.0–2.20 (2.24–2.20) | 50.0–2.0 (2.04–2.0) | 50.0–2.77 (2.85–2.77) | 50–1.75 (1.78–1.75) |
| Rmerge | 18.1 (54.1) | 8.5 (7.2) | 9.5 (86.4) | 8.7(67.5) | 10.0 (59.6) |
| Rpim | 13.6 (44.6) | 3.0 (33.0) | 4.7 (49.4) | 6.4 (49.8) | 6.1 (31.6) |
| CC1/2 | 0.959 (0.646) | 0.999 (0.887) | 0.992 (0.504) | 0.990 (0.681) | 0.955 (0.826) |
| I/sI | 3.98 (1.05) | 36 (2.04) | 25.5 (1.38) | 12.7 (1.13) | 10.3 (1.7) |
| Completeness (%) | 77.2 (63.2) | 98.9 (99.8) | 97.3 (89.4) | 83.4 (85.2) | 88.2 (87.3) |
| Redundancy | 2.3 (1.7) | 8.9 (8.4) | 5.1 (3.7) | 2.3 (2.2) | 3.3 (3.4) |
| Refinement | |||||
| Resolution (Å) | 40.2–3.70 | 32.2–2.20 | 36.4–2.0 | 35.0–2.77 | 40–1.75 |
| Reflections used in refinement | 24,107 | 36,120 | 127,952 | 78,160 | 39,342 |
| Rwork/Rfree (%) | 24.1/26.9 | 20.7/23.7 | 20.3/23.1 | 23.8/28.6 | 21.5/24.5 |
| No. atoms | |||||
| Protein | 16,398 | 3259 | 13,163 | 19,637 | 3320 |
| Water | 0 | 96 | 674 | 82 | 442 |
| B-factors (Å2) | |||||
| Protein | 95.6 | 65.1 | 51.9 | 99.4 | 24.3 |
| Water | 65.3 | 52.7 | 66.2 | 38.6 | |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.02 | 0.02 | 0.006 | 0.004 | 0.012 |
| Bond angles (°) | 0.493 | 0.578 | 0.906 | 0.681 | 1.315 |
| Ramachandran | |||||
| Favored regions (%) | 97.2 | 98.6 | 97.9 | 97.9 | 98.2 |
| Allowed regions (%) | 2.8 | 1.4 | 1.9 | 2.2 | 1.8 |
| Disallowed regions (%) | 0 | 0 | 0.2 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do Kwon, Y.; Wang, X.E.; Bender, M.F.; Yang, R.; Li, Y.; McKee, K.; Rawi, R.; O’Dell, S.; Schneck, N.A.; Shaddeau, A.; et al. Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography. Antibodies 2021, 10, 23. https://doi.org/10.3390/antib10020023
Do Kwon Y, Wang XE, Bender MF, Yang R, Li Y, McKee K, Rawi R, O’Dell S, Schneck NA, Shaddeau A, et al. Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography. Antibodies. 2021; 10(2):23. https://doi.org/10.3390/antib10020023
Chicago/Turabian StyleDo Kwon, Young, Xiangchun E. Wang, Michael F. Bender, Rong Yang, Yile Li, Krisha McKee, Reda Rawi, Sijy O’Dell, Nicole A. Schneck, Andrew Shaddeau, and et al. 2021. "Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography" Antibodies 10, no. 2: 23. https://doi.org/10.3390/antib10020023
APA StyleDo Kwon, Y., Wang, X. E., Bender, M. F., Yang, R., Li, Y., McKee, K., Rawi, R., O’Dell, S., Schneck, N. A., Shaddeau, A., Zhang, B., Arnold, F. J., Connors, M., Doria-Rose, N. A., Kwong, P. D., & Lei, Q. P. (2021). Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography. Antibodies, 10(2), 23. https://doi.org/10.3390/antib10020023






