Abstract
Cell-surface HLA-I molecules consisting of β2-microglobulin (β2m) associated heavy chains (HCs), referred to as Face-1, primarily present peptides to CD8+ T-cells. HCs consist of three α-domains, with selected amino acid sequences shared by all alleles of all six isoforms. The cell-surface HLA undergoes changes upon activation by pathological conditions with the expression of β2m-free HCs (Face-2) resulting in exposure of β2m-masked sequences shared by almost all alleles and the generation of HLA-polyreactive antibodies (Abs) against them. Face-2 may homodimerize or heterodimerize with the same (Face-3) or different alleles (Face-4) preventing exposure of shared epitopes. Non-allo immunized males naturally carry HLA-polyreactive Abs. The therapeutic intravenous immunoglobulin (IVIg) purified from plasma of thousands of donors contains HLA-polyreactive Abs, admixed with non-HLA Abs. Purified HLA-polyreactive monoclonal Abs (TFL-006/007) generated in mice after immunizing with Face-2 are documented to be immunoregulatory by suppressing or activating different human lymphocytes, much better than IVIg. Our objectives are (a) to elucidate the complexity of the HLA-I structural variants, and their Abs that bind to both shared and uncommon epitopes on different variants, and (b) to examine the roles of those Abs against HLA-variants in maintaining immune homeostasis. These may enable the development of personalized therapeutic strategies for various pathological conditions.
1. Introduction
Immune homeostasis is a delicately regulated balance of activation and suppression of immune and associated cells. Homeostatic imbalance may be both a cause and a consequence of pathological conditions ranging from inflammation, tissue injury, end stage organ disease, transplantation infection, malignancy to autoimmune diseases. In most immune conditions, there is no predictable pattern of recovery, and it may differ between individuals based on demographics as well as the lifestyle that include nutritional status, alcohol use, smoking, or recreational drug use. An in-depth immunological examination of the specific conditions may unravel the immune imbalance and enable developing personalized long or short duration therapeutic strategies to maintain immune homeostasis.
Antibodies play a major role in immune homeostasis, depending on the nature of their target epitopes. An antibody’s hypervariable region in F(ab)2 is specific for the physico-chemical configuration of an epitope on an antigen. Such an epitope could be specific for one antigen or shared by several other antigens. One epitope being shared by hundreds of antigens is one of the unique characteristics of the HLAs. Therefore, the immune homeostasis is examined from the perspective of HLA variants and their corresponding antibodies (Abs).
HLAs are highly polymorphic heterodimeric cell-surface molecules. Their polymorphism is exemplified by the six isoforms of HLA class-I (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G), and the three isoforms of HLA class-II (HLA-DR, HLA-DP and HLA-DQ), and by the thousands of identified alleles for each HLA-I and HLA-II gene and their respective proteins (Table 1). Several amino acid sequences of HLA polypeptide heavy chains (HCs) are highly specific for one allele or for one isoform, while some others are shared among other alleles of one isoform or all other isoforms of the HLA-class. In HLA-I, the shared epitopes are cryptic due to the presence of β2-microglobulin (β2m) and the tertiary and quaternary structures of HLA (HCs). Upon activation of immune cells under pathological conditions, the β2m-free HCs are generated on the cell-surface, which expose the cryptic shared epitopes. The exposed epitopes may elicit antibody response, in addition to binding with other ligands and Abs, depending on their antigenicity, immunogenicity, and other physicochemical characteristics.

Table 1.
The diversity of α-chain of HLA-I and α- and β-chains of HLA-II is exemplified by the number of the named alleles and proteins (Based on December 2021, IPD-IMGT/HLA Database) 1,2.
Although it is known that HLA proteins “are centrally involved in the actions of the human immune system” [1], the Abs generated against allele-specific epitopes (monospecific Abs) among individuals may differ. However, the polyreactive Abs produced against shared epitopes of different alleles may be similar and remain as allo-HLA Abs in all humans. The strength and diversity of these polyreactive allo-HLA Abs may also vary depending on the epitopes they recognize.
A prerequisite to understanding HLA-mediated immunohomeostasis is to clarify the complexity of the HLA-structural variants under normal and pathological conditions and to distinguish the shared and uncommon epitopes recognized by the HLA-Abs. With this objective, this review examines: (i) the structural variations of HLA that make the shared epitopes cryptic or exposed under different situations depending on the pathophysiological conditions; (ii) the Abs generated against the shared and uncommon epitopes; (iii) the consequences of the interaction between the epitopes and their Ab; and (iv) the diversified immunoregulatory potential of the Abs.
An understanding of the immunoregulatory roles of monospecific as well as polyreactive Abs is critical for developing personalized immunotherapies and vaccines to control various pathological conditions. The following categories are examined in this review:
- (i)
- Unique diversity of HLA polymorphism, which is reflected in the differences of the amino acid sequences in different domains, in the basic structural configurations, designated as HLA variants or Faces, and their role in immune homeostasis;
- (ii)
- Comparison of the diversity in allele specificity of HLA Abs in normal healthy individuals and in the commercial preparations of IVIg;
- (iii)
- Diversity of HLA monospecific and polyreactive monoclonal Abs (mAbs) generated by HLA molecules.
- (iv)
- Immune homeostatic role of HLA polyreactive Abs by binding
- (a)
- to intact HLA (Face-1) (e.g., mAb W6/32); and
- (b)
- to the monomer HC of HLA (Face-2) (e.g., TFL-006 & TFL-007); and
- (v)
- Immune homeostatic role of HLA-monospecific Abs.
2. The Diversity of HLA Polymorphisms
2.1. Diversity in the HLA Molecules and the Domains of Their Polypeptides
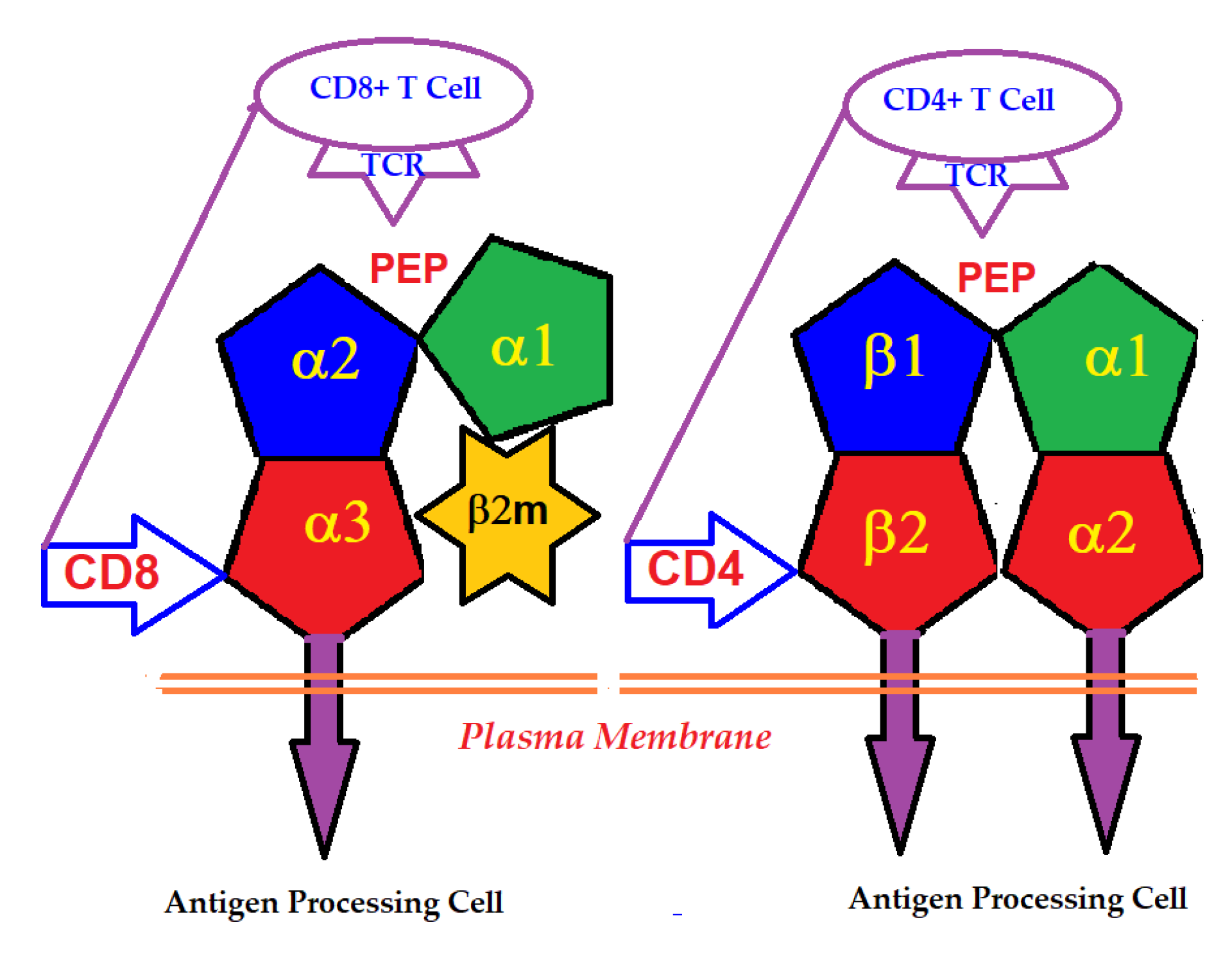
An intact HLA-I molecule consists of a polymorphic αHC and a monomorphic β chain, non-covalently linked β2m. The β2m was first identified in the urine of patients with tubular proteinurea [2,3,4]. Then its molecular size and association with human HLA-I HCs were recognized [5,6]. The HLA-I αHC genes are located on the short arm of chromosome 6, whereas that of the monomorphic β2m is located on chromosome 15. In contrast, HLA-II molecules consist of two polymorphic HCs, α and β, differing in their amino acid sequences. HLA-I α-HC contains three domains, α1, α2 and α3, whereas HLA-II α and β HCs consist of four domains, α1, α2, and β1 and β2 (Figure 1).
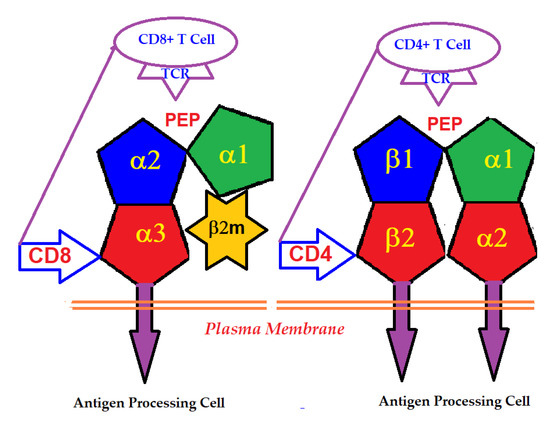
Figure 1.
Binding of T-cell co-receptors CD8 and CD4 to α3-domain of HLA-I and β2-domain of HLA-II, respectively.
2.2. The Domains Involved in the Primary Function of HLA
The primary function of the intact cell-surface HLA is antigen-presentation, and the domains serve as ligands for αβ T-lymphocyte receptors (TCRs). Most of the thymus-derived CD3+ T-cells express either the CD4 or the CD8 co-receptor molecules. CD4-expressing helper T-cells express TCRs specific for HLA class-II, whereas the CD8-expressing cytotoxic T-cells express TCRs specific for HLA class-I. The binding of the co-receptors of T-cells with HLA HCs enables the presentation of peptides to the major domain of the primary receptor of T-cells (Figure 1). The antigens presented by HLA-I are typically derived from intracellular infection caused by viruses. Presentation of these antigens to CD8+ T-cells initiates the cytolytic killing of the infected cells to prevent viral replication and spread. Since all human cells are potential targets for viral infections, the HLA-I isomers are expressed on all types of nucleated cells.
The antigens presented by HLA-II are derived from pathogens present in the extracellular spaces, which include bacteria, viruses and other pathogens. This presentation stimulates CD4+ cells to activate other immune cells, such as macrophages and B-cells. Hence, HLA-II is selectively expressed on professional antigen-presenting cells, which include macrophages, dendritic cells, and B cells. HLA-II antigen presentation activates CD4+ T-cells that stimulate the macrophages to release cytokines to further activate macrophages directly and help B cells to produce opsonizing Abs.
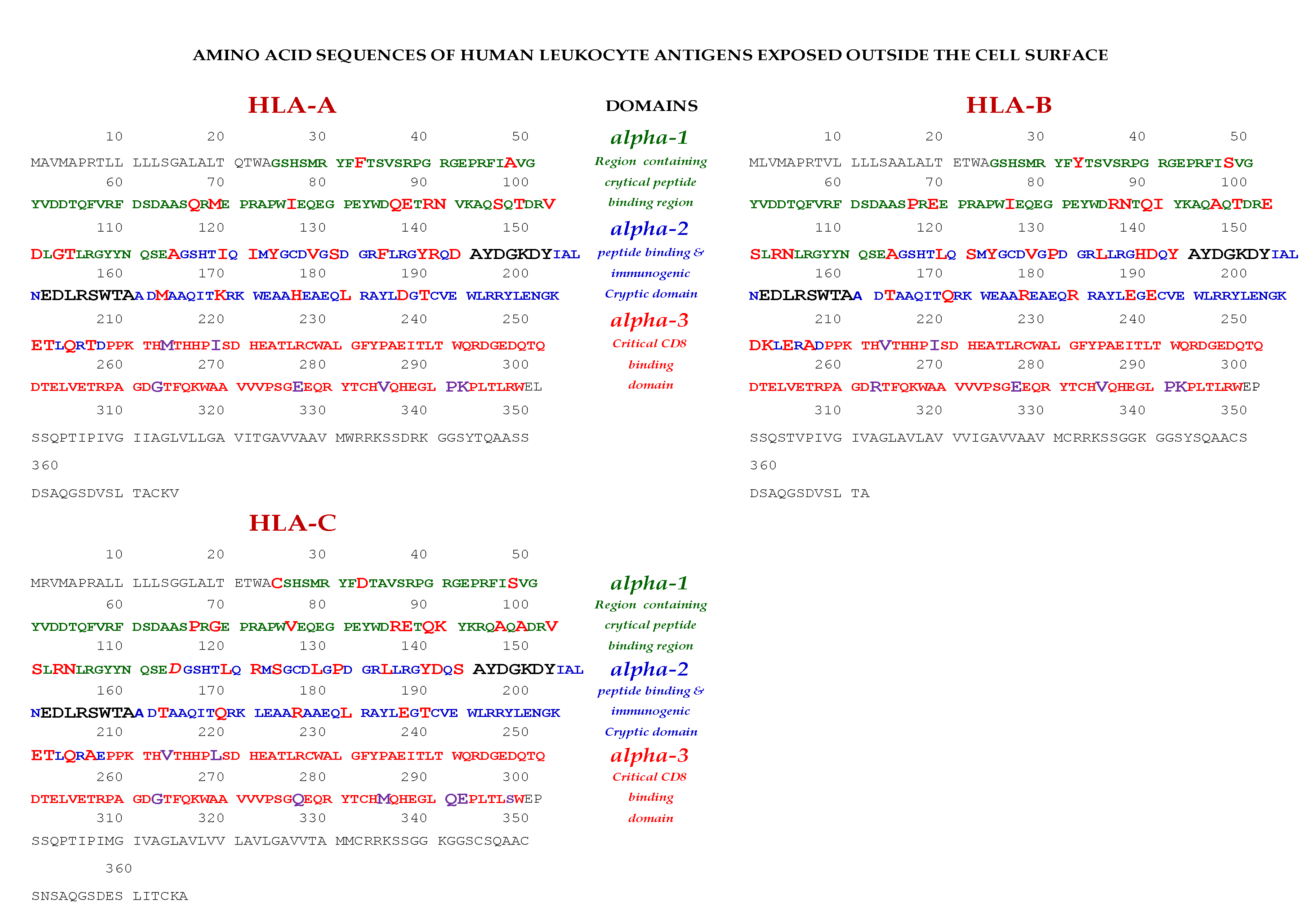
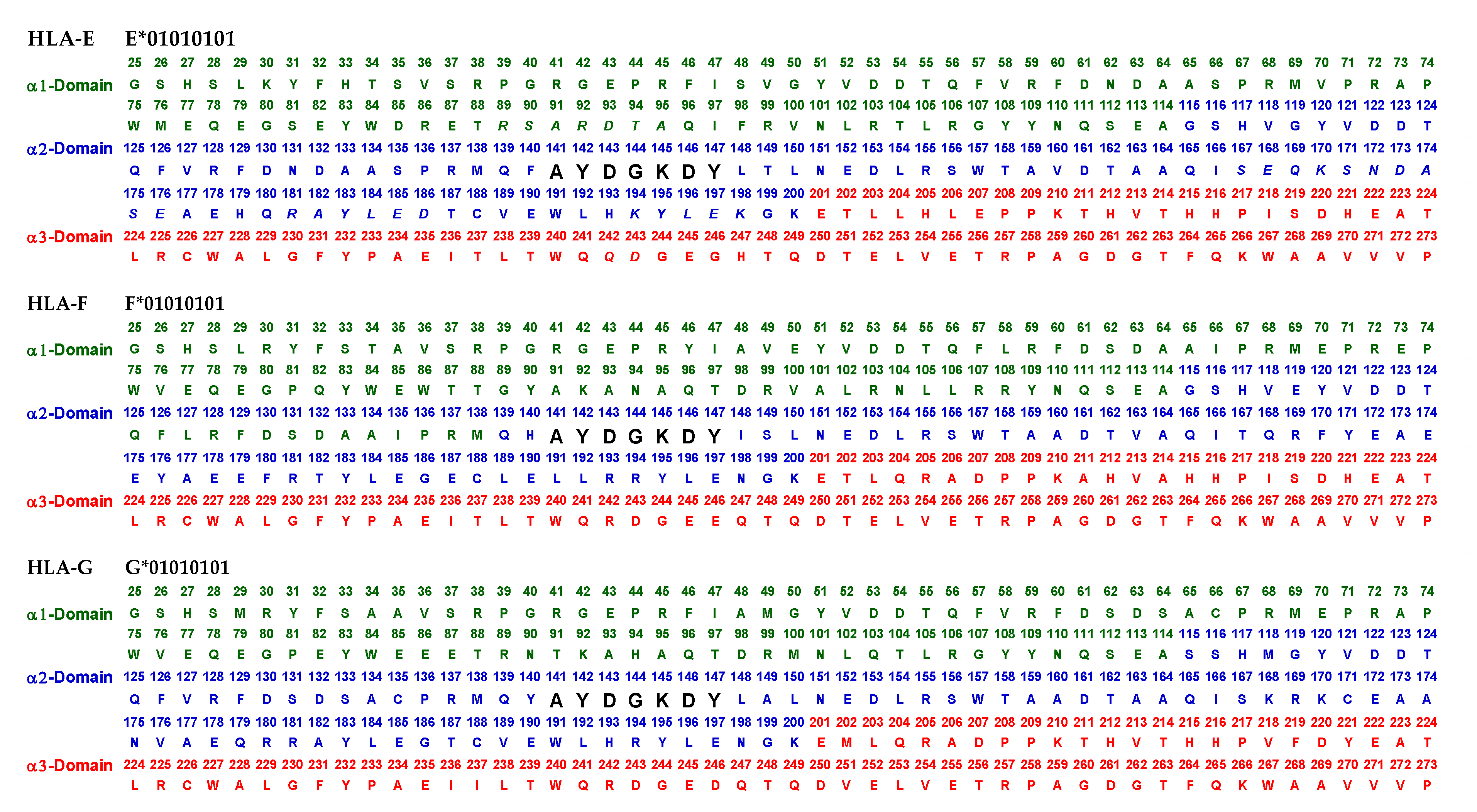
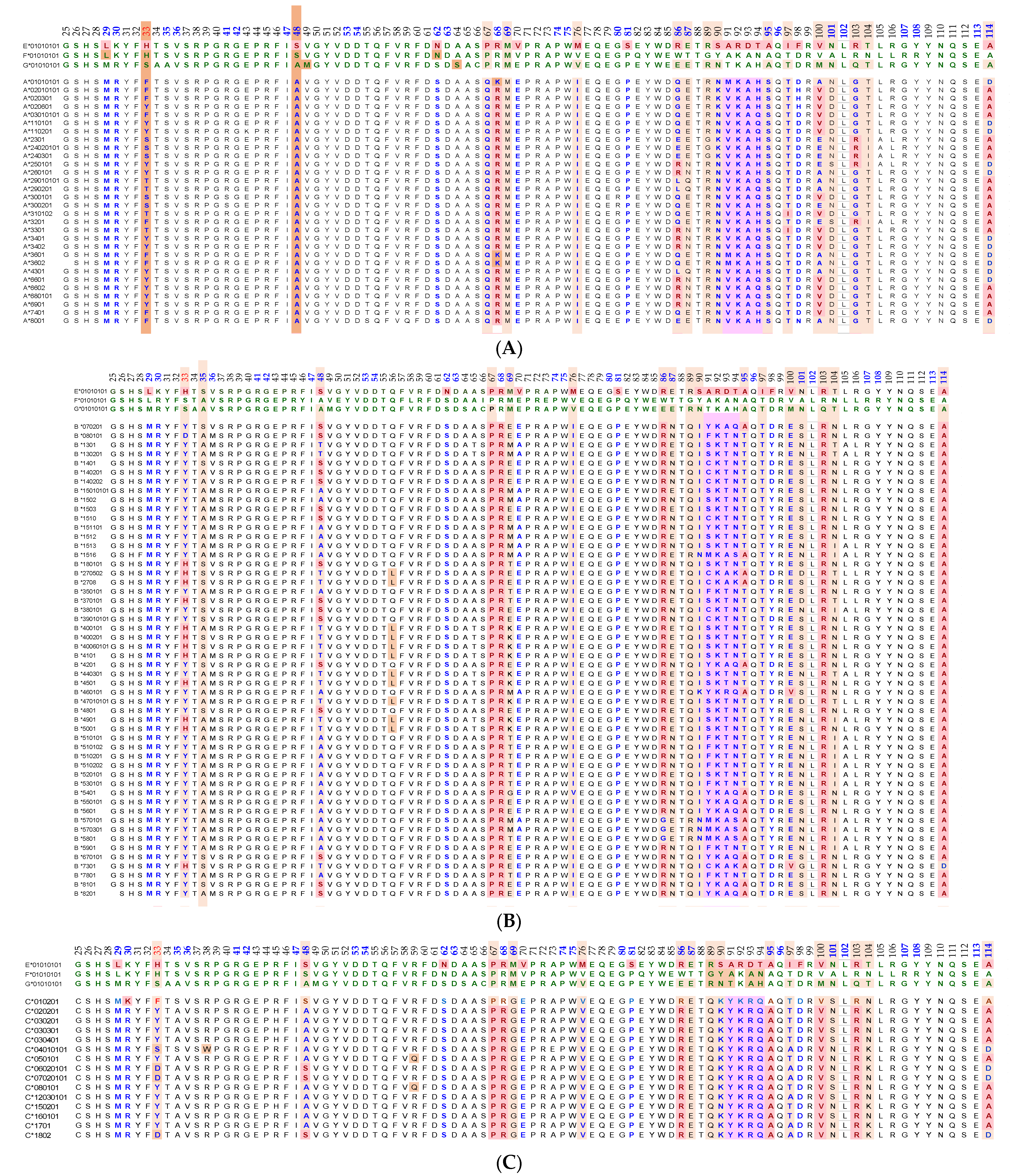
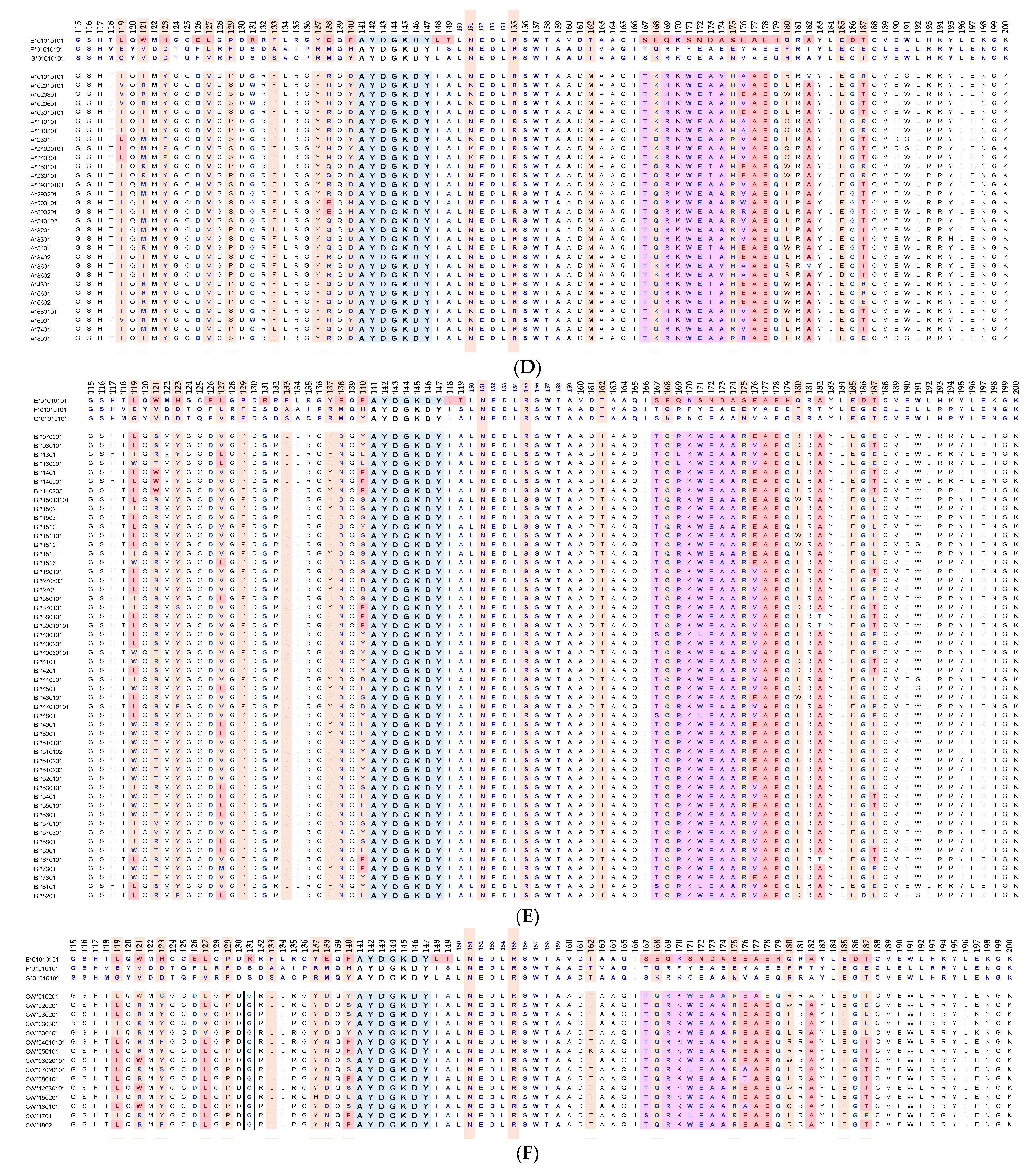
Essentially, the CD4 receptor binds to the β2 domain of the βHC. CD8-receptors of cytotoxic T-cells bind to α3 of the αHC (Figure 1). The amino acid sequences of α3 among all HLA-I HCs of isomers are almost identical with few variations. Similarly, the shared amino acid sequences of the β2-domain among all HLA-II HCs of isomers are strikingly similar. Table 2 illustrates the shared sequences in the α3-domain of HLA-I and the β2-domain of HLA-II that bind to T cell co-receptors CD8 and CD4, respectively. Figure 2 and Figure 3 show the sequences of α3 domain (in red) and also illustrate basic similarities and dissimilarities in the amino acid sequences of classical HLA-I (HLA-Ia: HLA-A, HLA-B and HLA-C, and non-classical HLA-I (HLA-Ib: HLA-E, HLA-F and HLA-G). The amino acid sequences in the α1 and α2 domains of HLA-I, and the α1 and β1 domains of HLA-II show noteworthy variability among different alleles. Such variations enable their recognition of specific peptides for antigen presentation. Amino acid sequences (shown in green) refer to the α1 domain of HLA-I, the region involved in formation of the groove and binding site of peptides. The differences seen in the specific amino acids in the sequence of the α1 domain are highly variable among different alleles of HLA-A, B, C, E, F and G (Figure 4A–C). Similarly, the sequences in the α2 domain of HLA-A, B, C, E, F and G (Figure 4D–F) are unique in that they contain a region involved in the formation of the grove for binding site of peptides and a long sequence (cryptic) domain masked by β2m (Figure 5).

Table 2.
HLA Class I and II isoforms and the shared sequences in the α3-domain in HLA-I and β2 domain HLA-II that bind to T cell co-receptors.

Figure 2.
Exposed and cryptic (in α2 domain) amino acid sequences of classical HLA-I (HLA-Ia) on the cell-surface.

Figure 3.
Exposed and cryptic amino acid sequences (in α2 domain) of non-classical HLA-I (HLA-Ib) on the cell-surface.
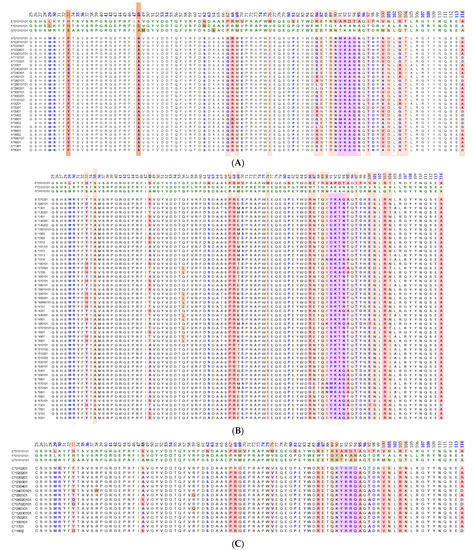
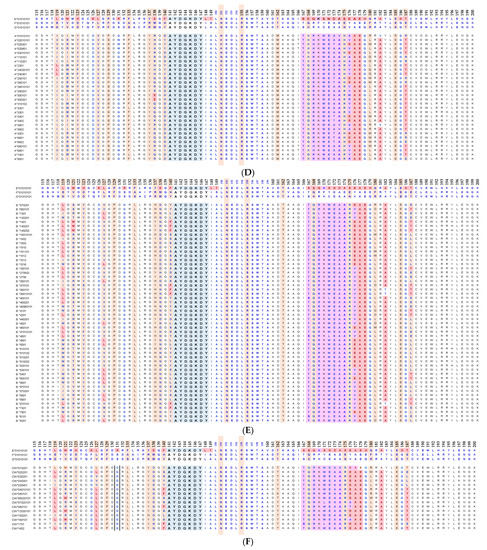
Figure 4.
An overview of the polymorphic amino acid sequences of α1 and α2 domains in the classical (HLA-Ia: HLA-A/-B/-C) and non-classical (HLA-Ib: HLA-E/-F/-G). Essentially, α1 domain of HLA isomers carry monospecific sequences characteristic of each allele. This figure illustrates a specific sequence of HLA-E, namely 90SARDTA95 in the α1-domain, which is restricted exclusively to HLA-E. This sequence is recognized by the mAb TFL-033 and the isolated sequence inhibited the mAb binding to HLA-E. Similarly, a most commonly shared amino acid sequence (141AYDGKDY147) located in α2-domain of all alleles of HLA-Ia and HLA-Ib is illustrated. Although the number of lleles of HLA-I isomers exceed several thousands as presented in Table 1, this figure is only limited to alleles of HLA-Ia and Ib commonly used to monitor antibodies using antigen coated beadsets on Luminex platform. Interestingly, further examination of several thousands of alleles revealed that the amino acid sequences of shared epitope (141AYDGKDY147) are present in all examined alleles. Sequences of all alleles or the presence of 141AYDGKDY147 in all alleles of all isoforms can be found in https://www.uniprot.org/uniprotkb/P04439/entry#sequences, accessed on 1 July 2022, or at https://ebi.a.c/uk/ipd/im/hla/, accessed on 1 July 2022. (A) HLA-A α1-domain; (B) HLA-B α1-domain; (C) HLA-C α1-domain; (D) HLA-A α2-Domain; (E) HLA-B α2-Domain; (F) HLA-C α2-domain.
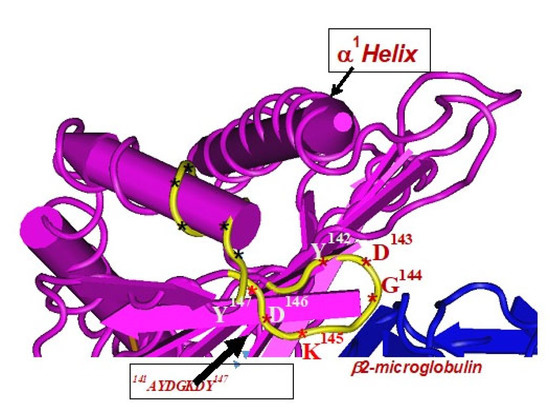
Figure 5.
The amino acid sequences of α2-domain of HLA-I shared by HLA-A, B, C, E, F and G loci, that are hidden (cryptic domain) by β2-microglobulin. These sequences are highly immunogenic. For details see Table 3.
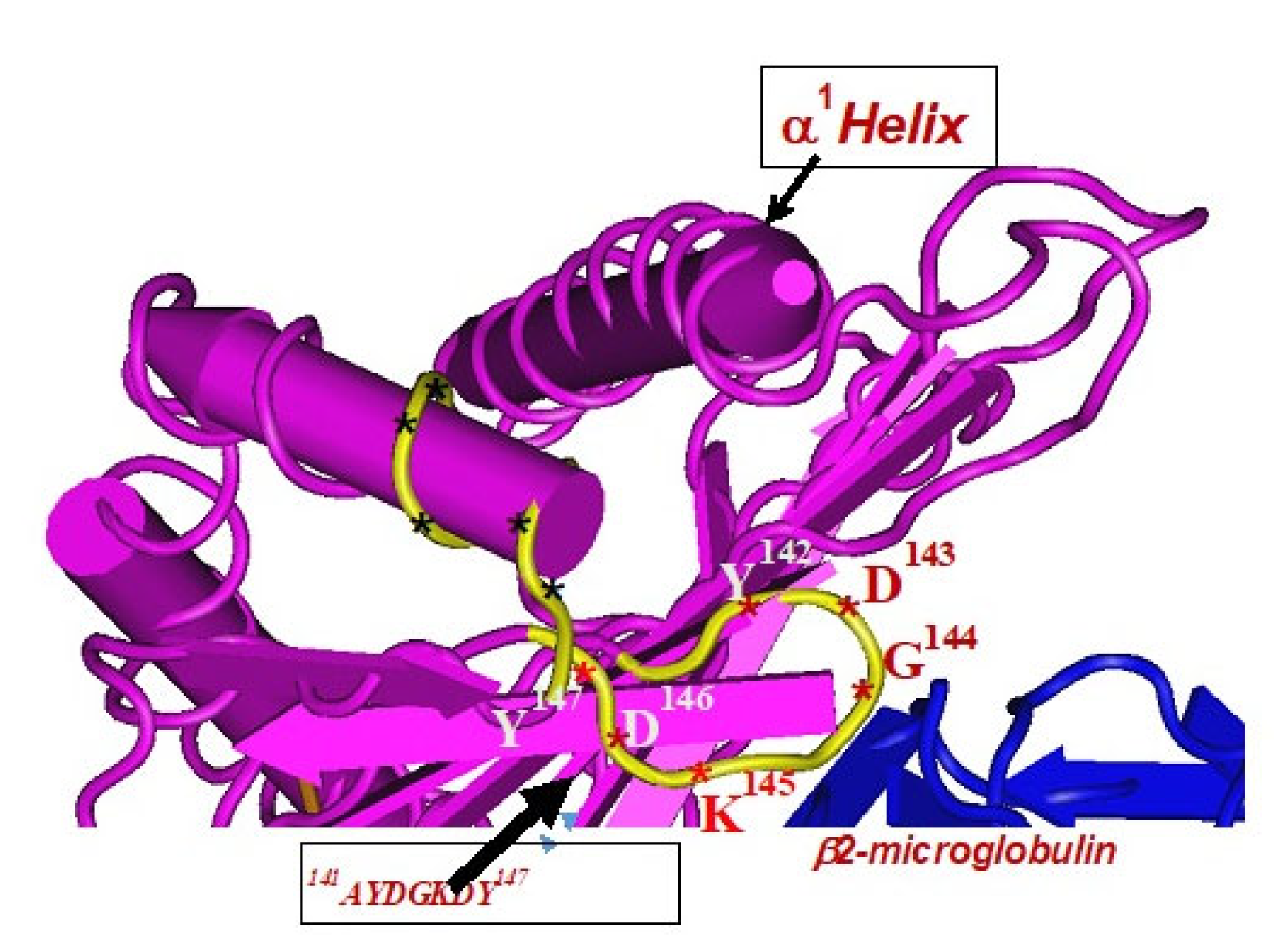
The amino acid sequences of α2-domain that are hidden by β2m (Figure 5) constitute unique cryptic epitopes, and, most importantly, they contain sequences (such as 141AYDGKDY147 and 152EDLARSWTA159) (shown in black in Figure 2) shared by almost all HLA-I isomers. Strikingly, the amino acid sequences are identical in all alleles of all six HLA-I loci (Figure 4D–F). Whereas these sequences are hidden from exposure (cryptic) by β2m in intact HLA-I (also known as Closed conformers or Face-1), they remain exposed in naturally occurring β2m-free α-HCs (also known as Open conformers or Face-2 HLA). Interestingly, the antigenicity, flexibility, hydrophobicity, and beta turn characteristics are highest for the AYDGKDY sequence compared to any other shared sequences (Table 3). The exposure of cryptic epitope in Face-2 is overexpressed in activated immune cells, resulting in immune recognition. Similarly, the sequence 161DTAAQI166 present in all alleles of HLA-B, HLA-C, HLA-E, and HLA-G, differs in position 162 in HLA-A as 161DMAAQI166 (Figure 4D) and in position 163 in HLA-F as 161DTVAQI166 (Figure 4F).

Table 3.
Most commonly shared and cryptic peptide sequences in the α2-domain of the heavy chains of all HLA-I isoforms (HLA-A/-B/-C/-E/-F and -G).
Note especially the peptide sequence AYDGKDY, which is cryptic and shared by all isoforms of HLA-I. The bioinformatics analysis was carried out by using the Immune Epitope Database (IEDB) to predict antigenicity rank of epitopes. Chou and Fasman beta turn, Kolaskar and Tongaonkar antigenicity, Karplus and Schulz flexibility and Parker hydrophilicity prediction methods in IEDB were employed. The methods predict the probability of specific sequences in HLA-E that bind to Abs being in a beta turn region, being antigenic, being flexible, and being in a hydrophilic region. Antigenicity rank is calculated by pooling the probability values.
2.3. The Four Structural Variants of HLA-I
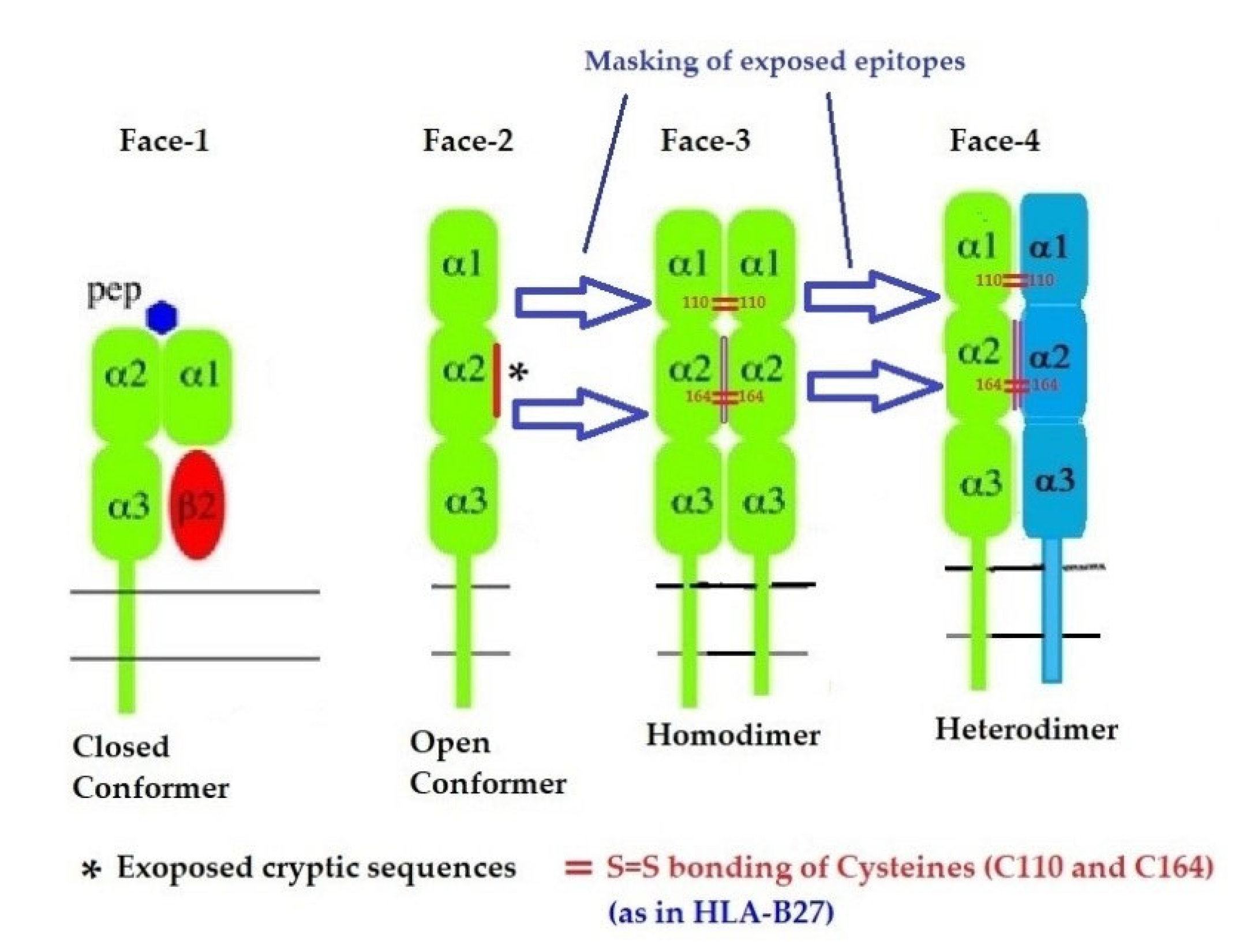
In addition to naturally occurring Face-1 and Face-2, there are two additional variants formed naturally from the Open conformers and all these variants are referred to as the four faces of HLA-I [11], as illustrated in Figure 6. Elucidation of these four faces of HLA-I variants is necessary to comprehend HLA-mediated immune homeostasis. These faces are as follows:
- Face-1: The intact HLA-I that consists of an α-HC non-covalently linked to β2m;
- Face-2: HLA-I monomeric α-HC devoid of β2m, exposing immunogenic cryptic sequences, such as 141AYDGKDY147, 152EDLARSWTA159, and 161DTAAQI166.
- Face 3: Homodimers of Face-2: Dimers of two α-HCs of the same allele formed by S-S linkage of cysteines at position 67, as observed in HLA-B27.
- Face-4: Heterodimers of Face-2: Two α-HCs of different HLA-I monomers formed by linkages of cysteines at positions 67, 101, 164, 203, or 43, as in HLA-G, or by salt linkage, H-bonding, or van der Waal forces, as observed in HLA-F and HLA-C.
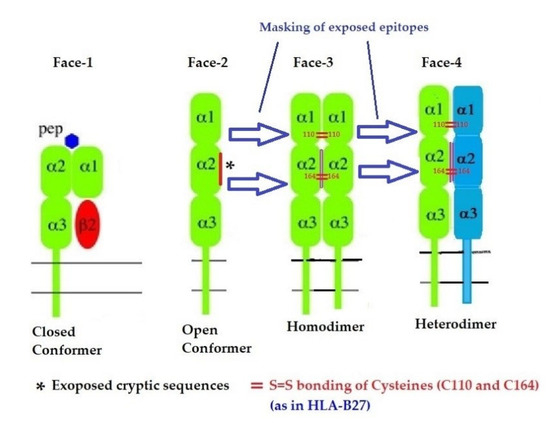
Figure 6.
Four major structural configurations or faces of HLA-I. The shared epitopes (see Table 3) remain masked by β2-m in Face-1 or intact HLA. In the absence of β2m, the HLA becomes monomeric (Face-2) on activated immune cells and on cancer cells, resulting in the exposure of cryptic sequences. It appears to be for a shorter duration since the exposed sequence is masked by homodimerization or heterodimerization. During dimerization, there is an increased propensity for pairing of cysteines and formation of disulfide bonds (as in HLA-B27, HLA-C and HLA-F), a mechanism to remask the exposed sequences.
Figure 6.
Four major structural configurations or faces of HLA-I. The shared epitopes (see Table 3) remain masked by β2-m in Face-1 or intact HLA. In the absence of β2m, the HLA becomes monomeric (Face-2) on activated immune cells and on cancer cells, resulting in the exposure of cryptic sequences. It appears to be for a shorter duration since the exposed sequence is masked by homodimerization or heterodimerization. During dimerization, there is an increased propensity for pairing of cysteines and formation of disulfide bonds (as in HLA-B27, HLA-C and HLA-F), a mechanism to remask the exposed sequences.
2.4. Transitory Expression of Face-2 on Activated Immune Cells
The presence of naturally occurring β2m-free HCs of HLA-I, Face-2, was first recognized in the human B lymphoblastoid cell line T5-1 [13]. Cell lysate was immunoprecipitated with an anti-H polyclonal serum, which does not bind to Face-1, but only to β2m-free HCs of HLA-I (Face-2). Only 1 to 2% of the HCs (Face-2) present on the surface of T5-1 cells were precipitable by anti-H. Since this investigation, cell-surface expression of Face-2 has been observed across lymphocyte subpopulations (CD3+ T-cells, CD19+ B-cells, CD56+ NK cells and CD14+ monocytes), and importantly these HCs are particularly overexpressed on activated cells, as well as on extravillous trophoblast, T-cells and monocytes [14]. Face-2 was also observed on the cell-surface of both in vitro and in vivo activated human T-cells, but not on the resting ones [12]. Immunoprecipitating the activated T-cells with both the mAb W6/32 and the mAb LA45 confirmed that W6/32 precipitated Face-1, whereas LA45 precipitated Face-2 only. Western blot analysis of the cell lysates further confirmed these results [12]. Face-2 is also expressed on human peripheral blood T-cells activated either by phorbol myristate acetate (PMA), anti-CD3 antibody, PHA, brefeldin A (PFA), or IFN-γ [15]. Furthermore, using purified and biotinylated mAb L31, HLA-C Face-2 was recognized on activated T-cells [16]. The possibility of formation of Face-3 by homodimerization of 2 identical Face-2 molecules was dependent on the unpaired cysteine at position 67 (Cys67) [17,18]. Homodimerization of Face-2 of HLA-B27 was recognized using mAbs HC-10 and ME1 [19].
The formation of Face-4 by heterodimerization of two different Face-2 molecules was reported in HLA-F [20]. Any HLA-I HC could interact with HLA-F only when the latter was in the form of Face-2. This interaction was directly observed by coimmunoprecipitation and by surface plasmon resonance and indirectly on the surface of cells through tetramers and HLA-I Face-2 colocalization. The data indicate that HLA-F is expressed as Face-2, and it has physical interaction with HCs (Face-2) of the same allele of HLA-F (Face-3) and with other alleles and isoforms of HLA-I (Face-4), based on the differential coimmunoprecipitation experiments. At least three different forms (Face-2, Face-3, Face-4) of HLA-F based on differential staining of surface HLA-F using mAbs 4A11 and 3D11 (HLA-F HC specific) and 4B4 (specific for intracellular HLA-F) over the course of lymphocyte activation were observed [20]. It is speculated that “HLA-F and MHC-I HC interactions can occur in trans between cells” (16, p. 6206). HLA-F expressed as Face-2 is not recognized by the mAb W6/32; however, the homodimer (Face-3) of HLA-F was recognized on the cell-surface by W6/32, particularly in the absence of β2m. Furthermore, it is reasonably well established that Face-2 is overexpressed in human cancers such as neuroblastoma, renal cell, colon, gastric, breast, ovarian, bladder carcinoma and melanoma, using mAbs L31, LA-45, LHC10 and M38 [21,22,23,24,25,26]. These reports led to the hypothesis that homo- and heterodimers of Face-2 exist in human cancers, and this deserves serious attention in the potential development of autologous and allogeneic tumor cell vaccines with various adjuvants.
2.5. Do the Structural Variants of HLA Have a Role in Immune Homeostasis?
Face-2 exposes cryptic epitopes and leads to immunogenicity (recognition by B cells and eliciting antibody response) and antigenicity (recognition by Abs formed against the cryptic epitopes). Both homodimerization and heterodimerization of Face-2 facilitates masking of the exposed (previously cryptic) epitopes of Face-2, such that they are blocked from immune recognition by the “polyreactive Abs”. Evidently, Face-2 seems to be a short-lived structural variant of HLA. In order to unravel the roles of the structural variants of HLA-I and the generated polyreactive antibodies in immune homeostasis, the following questions are highly pertinent:
- I.
- Is the dimerization of Face-2 a natural mechanism to prevent the exposure of cryptic epitopes otherwise masked by β2m?
- II.
- If so, what is the purpose of ephemeral exposure of these cryptic epitopes?
- III.
- Does the transient-exposure of cryptic epitopes of Face-2 generate polyreactive Abs following immune recognition by B cells? If so, what kinds of polyreactive HLA Abs are being produced?
- IV.
- Do the naturally occurring polyreactive Abs bind to the ephemeral exposure of cryptic epitopes on Face-2 to elicit salient immunoregulatory functions?
- V.
- How do the immunoregulatory effects differ between the binding of Abs directed against Face-1, such as that of mAb W6/32, and that of Abs directed against Face-2, such as that of TFL-006 and TFL-007?
Answering these questions may unravel the roles of the structural variants of HLA-I and the polyreactive Abs they generate in immune homeostasis.
3. The Diversity of HLA-I Antibodies
3.1. The Diversity of Natural Anti-HLA-I Abs in the Normal Human Sera
Avrameas et al. [27] observed Abs directed against several autoantigens such as tubulin, actin, thyroglobulin, myoglobulin, fetuin, transferrin and albumin and hypothesized that naturally occurring IgG-Abs “against a high variety of self antigens” may occur in normal healthy individuals. Indeed, Collins et al. [28] first reported a naturally occurring anti-HLA-A8 Ab. Tongio et al. [29] confirmed the presence of natural IgM Abs in the sera of 21 normal healthy individuals reacting with HLA-I alleles. Subsequently, more reports [30,31] emanated on naturally occurring Abs to HLA-I and HLA-II. Since these Abs in the sera of normal and healthy individuals recognize several HLA alleles not found in oneself, they were considered as allo-HLA Abs. Morales-Buenrostro et al. [32] examined in detail these anti-HLA-I Abs in the sera of 424 nonalloimmunized healthy males using a single antigen bead assay (SAB assay) with Luminex beadsets coated with more than 90 different purified recombinant HLA molecules. They found allo-HLA Abs to class-I in 42%, class-II in 11%, and both in 12 of the individuals examined. Interestingly, it was reported that the serum Abs also bound to beadsets treated with a mild acid to convert Face-1 to Face-2, suggesting that the normal non-alloimmunized male sera contain Abs reacting to both Face-1 and Face-2. This is the first report documenting the occurrence of Abs against both Face-1 and Face-2.
We have examined the sera of normal healthy males (non-alloimmunized) and females for the presence of IgM and IgG reacting to HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G using the SAB assay on the Luminex platform on single antigen beads (SABs) provided by different vendors [33,34]. Our focus was to distinguish between serum HLA Abs reacting to Face-1 and Face-2. The beadsets differed significantly in the composition of the antigens present on their surface. LIFECODES SABs manufactured by Immucor (Norcross, GA, USA) contain only intact HLA (Face-1) [33], whereas the LABScreen SABs (Thermofisher-One Lambda Inc., Canoga Park, CA, USA) contain both Face-1 and Face-2 [34].
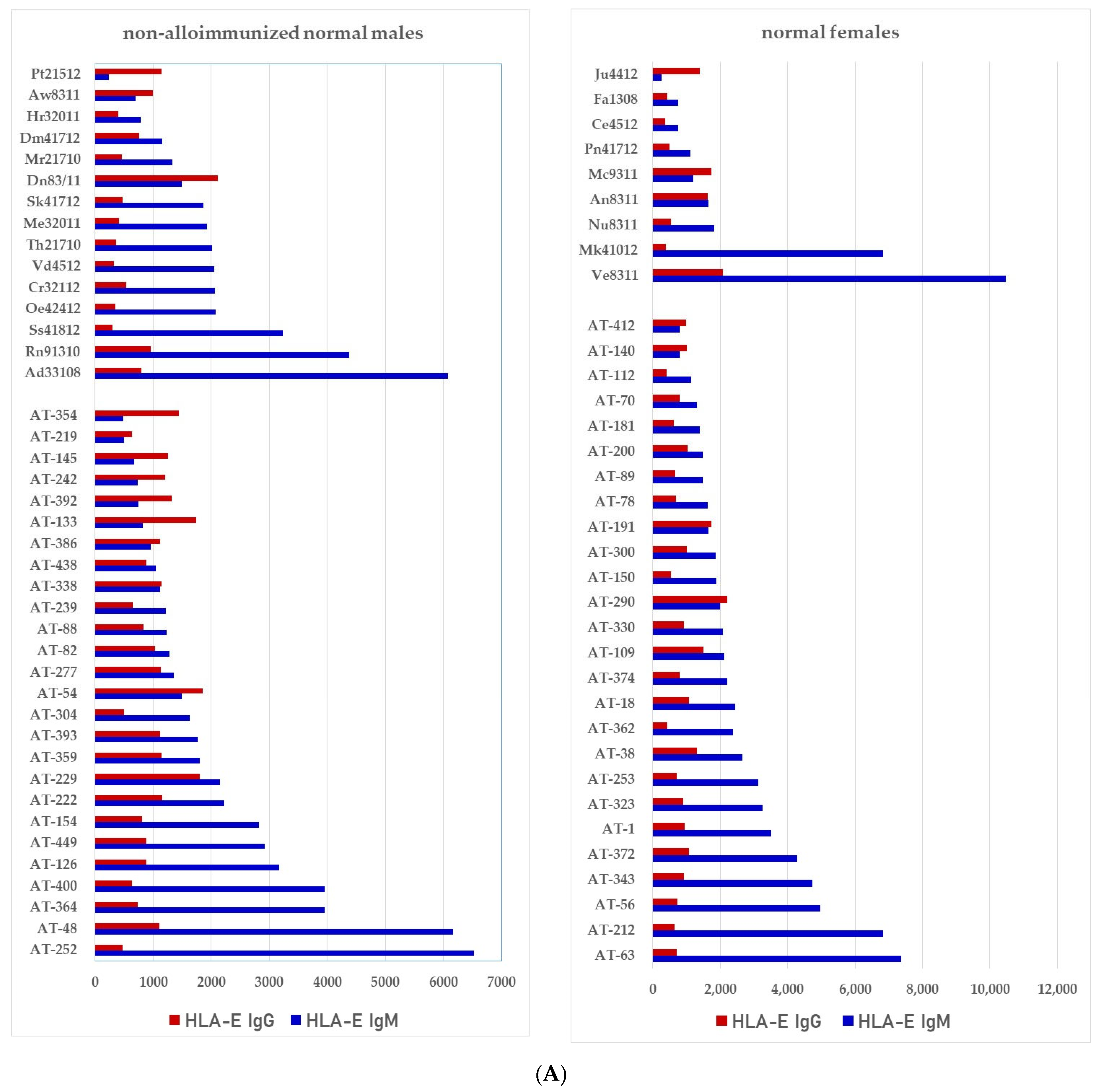
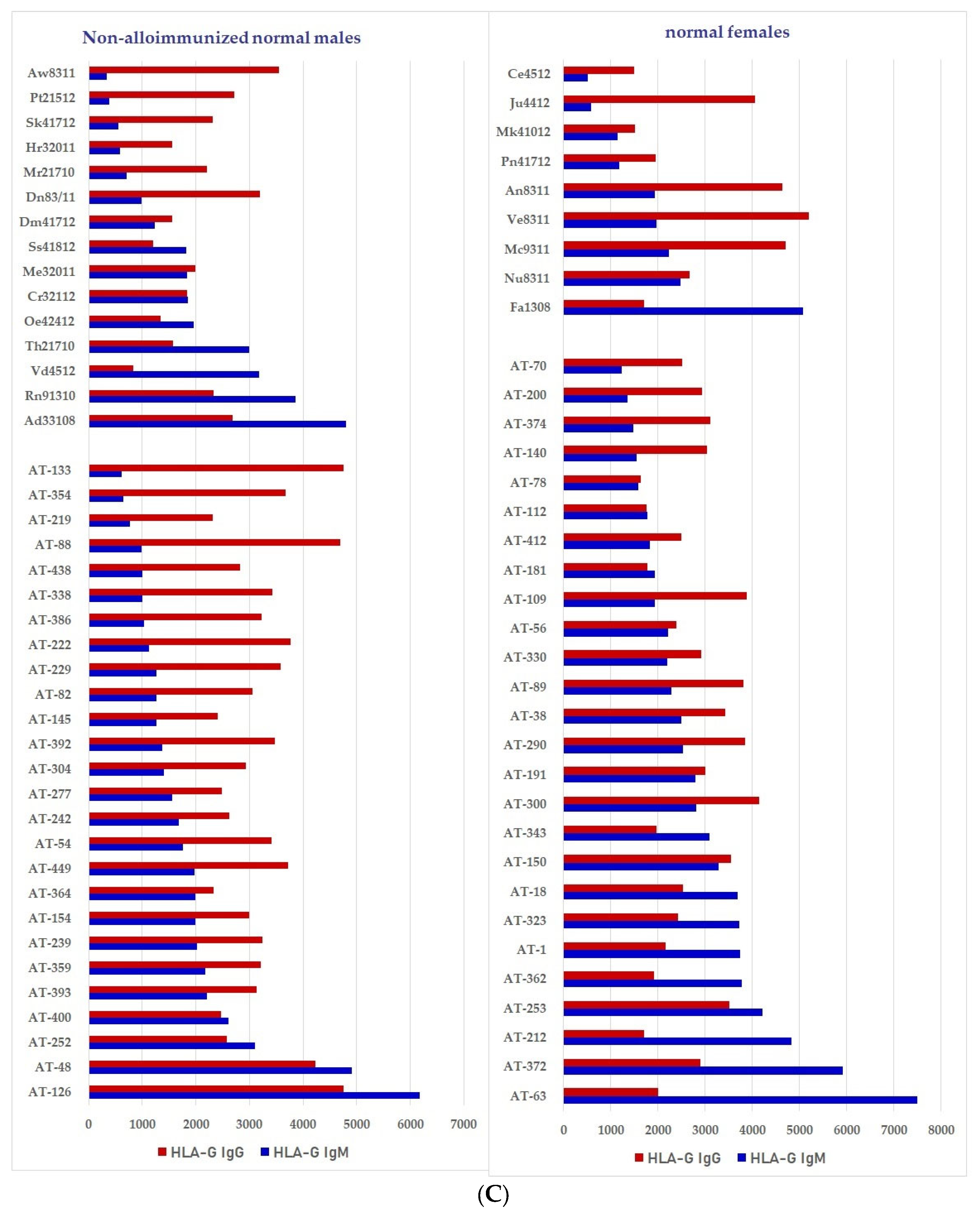
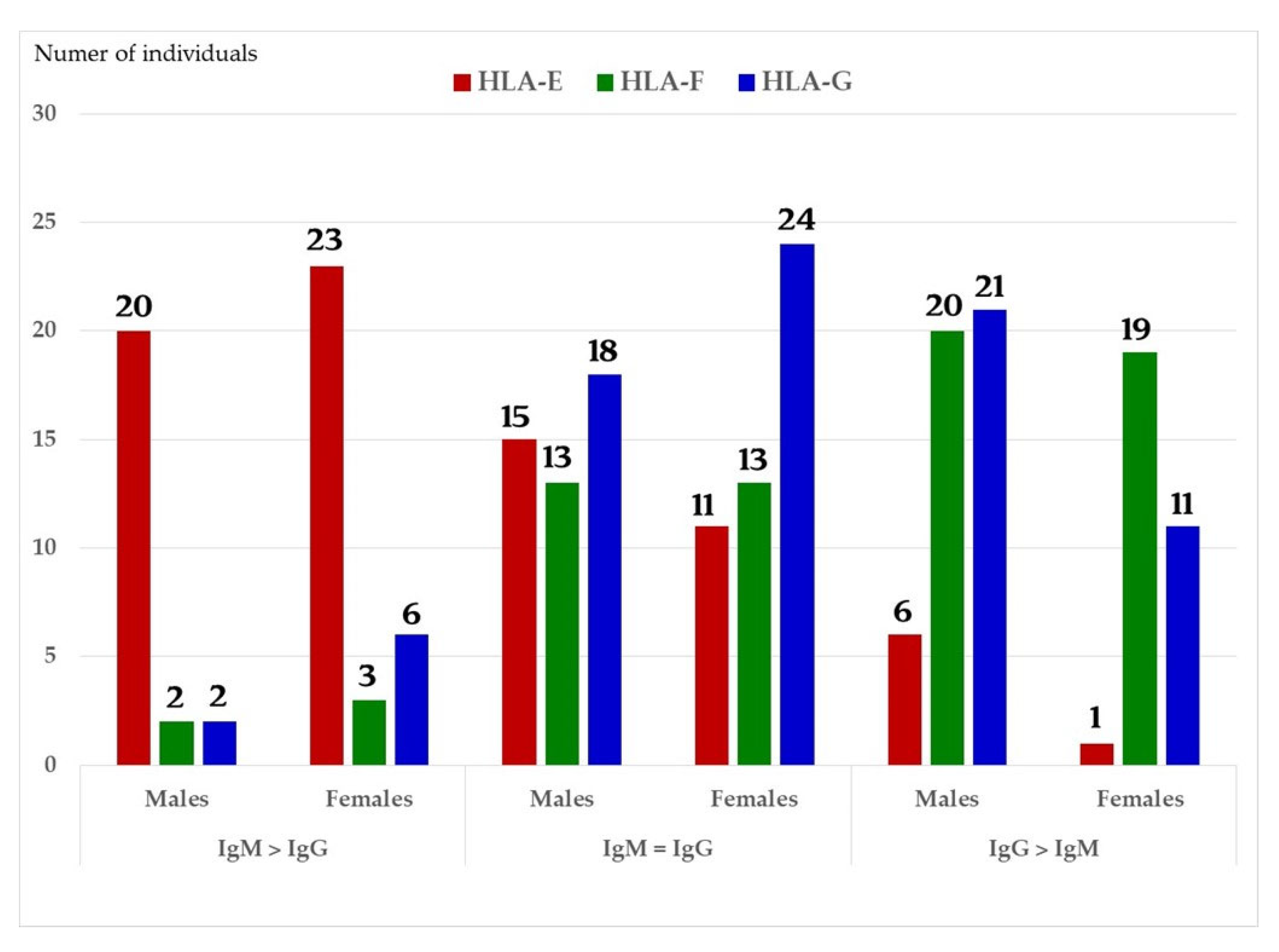
We have examined both IgM and IgG in the sera of normal healthy males (non-alloimmunized) and females. Table 4 documents the profile and strength (expressed as mean fluorescence intensity or MFI) of HLA-A, HLA-B and HLA-C reactivities of the normal non-alloimmunized male sera. Since IgG in human sera can occur as immune complexes in conjunction with anti-idiotypic IgG or IgM Abs and other proteins, we have also examined the normal sera after purifying the Abs with a Protein-G affinity column. Table 5 documents the profile of serum anti-HLA reactivity after removing the immune complexes and other factors bound to IgG. It is interesting to note that the IgG Abs in these sera bound to almost all alleles of HLA-A, HLA-B and HLA-C. We have also examined the sera of several normal healthy male and female volunteers for IgM and IgG reactivity to HLA-E, HLA-F and HLA-G. Figure 7A–C illustrate the relative proportions (MFIs) of IgM and IgG reacting to HLA-E, HLA-F, and HLA-G in the unpurified sera of the normal individuals. The relative ratios of anti-HLA-Ib IgM and IgG in the sera of normal males (non-alloimmunized) and females are depicted in Figure 8. Note the high prevalence of IgM-Abs against HLA-E in a number of males and females, and the high prevalence of IgG reacting to HLA-G.

Table 4.
Documentation of antibodies against HLA class Ia molecule in the sera of non-alloimmunized male volunteers. The profiles of antibodies against HLA-A, HLA-B and HLA-C differ among different individuals. Antibodies against different alleles of each HLA isomers differ as follows: HLA-C > HLA-A > HLA-B. One male (Ho) has antibodies against almost all alleles of HLA-Ia tested suggesting that the individual may possibly carry naturally occurring antibodies formed against most commonly shared epitopes such as that of 141AYDGKDY147 illustrated in Figure 4. No or low MFI in several individuals does not imply the absence of antibodies, as antibodies against almost all alleles can be seen after purification of IgG using the Protein-G affinity purification column as illustrated in Table 5. It implies that the naturally occurring HLA antibodies may be masked by other factors, such as anti-idiotype antibodies and immune complexes.

Table 5.
HLA-Ia Ab profiles in the sera of normal non-alloimmunized male volunteers after purification of IgG using Protein-G affinity purification columns, elaborate the profile of the naturally occurring anti-HLA-I antibodies, suggesting that under natural conditions these antibodies may be masked by other factors, such as anti-idiotypic antibodies and immune complexes.
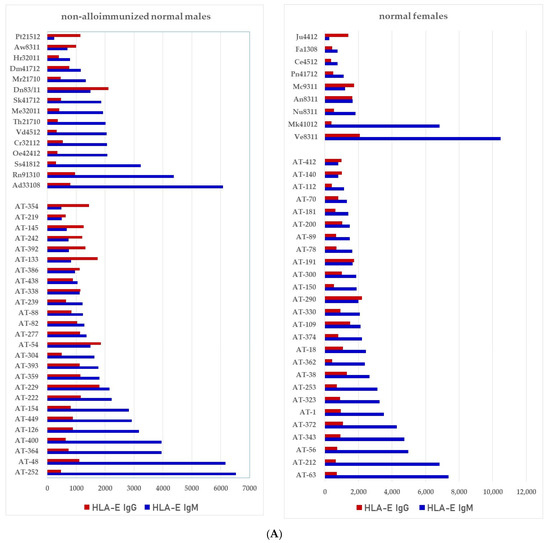
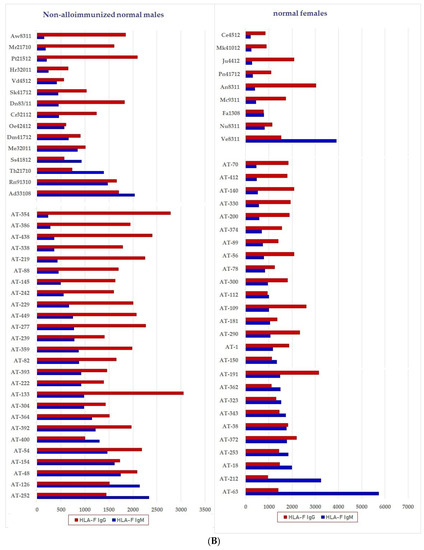
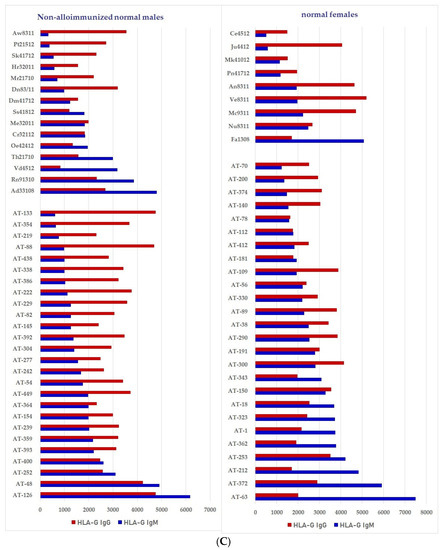
Figure 7.
Documentation of IgM and IgG antibodies against HLA class Ib molecules in the sera of male (non-alloimmunized) and female (volunteers). Interestingly, naturally occurring IgM antibodies are more prevalent than IgG for HLA-E (A). In contrast, naturally occurring IgG antibodies are more prevalent than IgM for HLA-F and HLA-G (B,C). Simultaneous prevalence of higher IgM than IgG in HLA-E, HLA-F and HLA-G, suggests that these individuals may be experiencing induction of IgM due to various pathological conditions such as inflammation, injury and other disorders. (A) Anti-HLA-E IgM and IgG; (B) PosiAnti-HLA-F IgM and IgG; (C) Anti-HLA-G IgM and IgG.
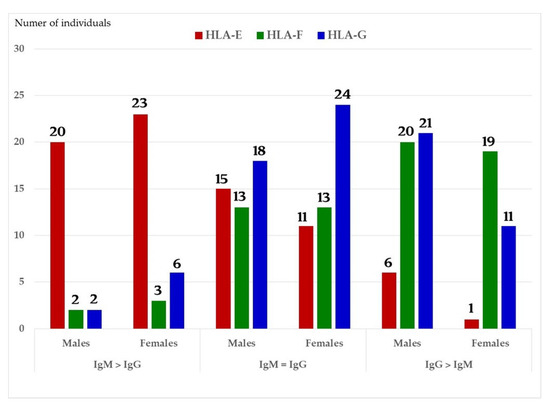
Figure 8.
The relative ratios of IgM and IgG in the sera of normal males (non-alloimmunized) and females. Note the high prevalence of IgM Abs against HLA-E in a number of males and females, and high prevalence of IgG Abs against HLA-G.
3.2. The Diversity of Abs in Intravenous Immunoglobulin
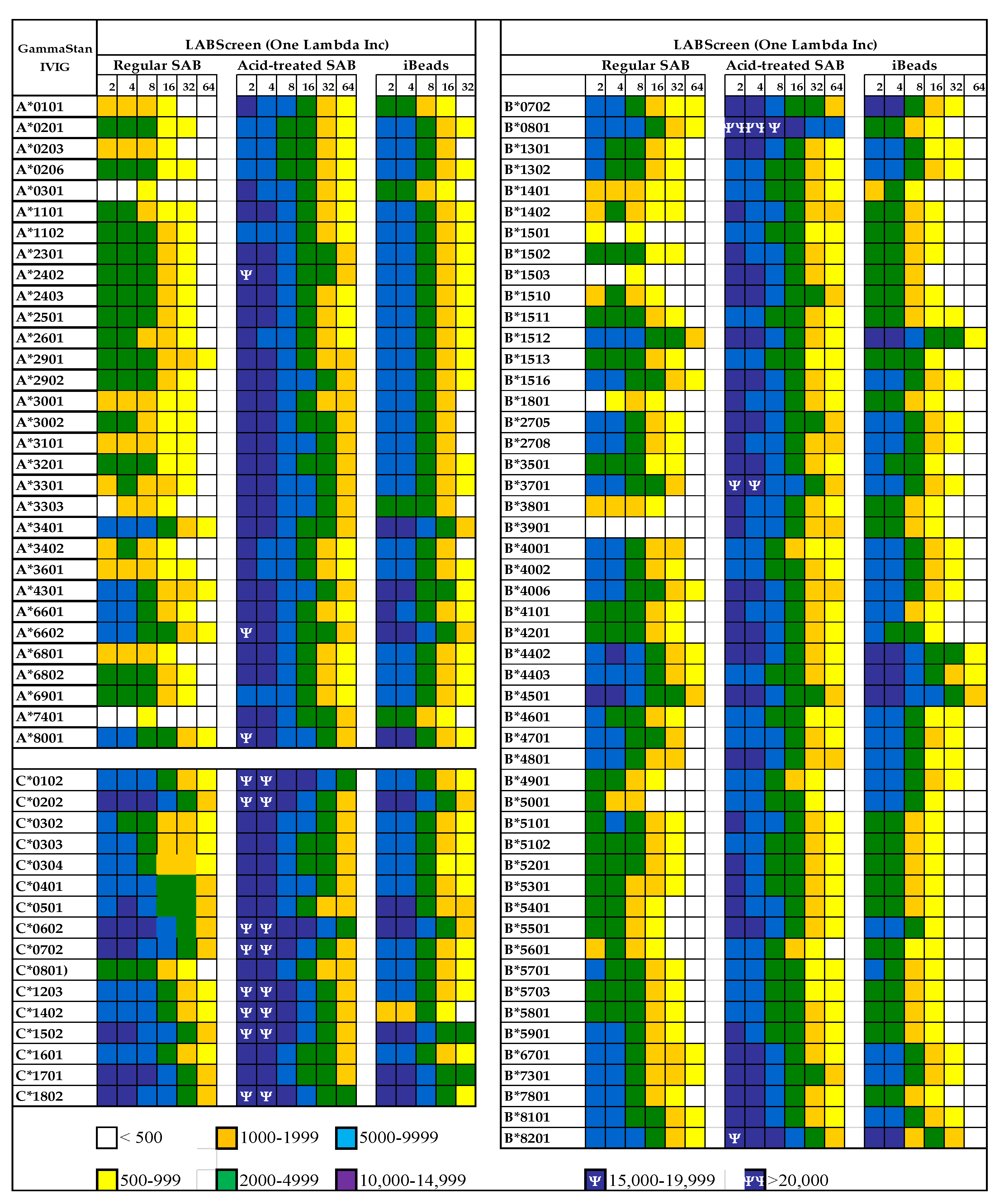
The presence of natural allo-HLA Abs in human plasma implies that intravenous immunoglobulin (IVIg), the IgG purified and pooled from the plasma of thousands of normal donors, contains such natural HLA antibodies. IVIg has been extensively used to prevent microbial and fungal infections and to reduce inflammation after transplantation [35,36,37,38,39] and in other inflammatory and autoimmune diseases [40,41,42,43]. IVIg is often administered to lower the level of anti-HLA Abs in end-stage organ disease patients and transplant recipients [40,41,42,43,44,45,46,47,48,49,50,51,52,53,54]. These findings lead Kaveri and his team [55] to propose a role for IVIg in immune homeostasis. The different vendors’ commercial preparations of IVIg are known to suppress activated CD4+ T-cells and their production of proinflammatory cytokines when the cells are activated by various stimuli in vitro [56,57,58,59,60,61,62,63,64,65,66]. Interestingly, Kaveri et al. [67] reported that IVIg has Abs, to a conserved region of HLA-I, that are capable of modulating CD8+ T cell mediated function. We have examined the presence of Abs against different alleles of HLA-I in different therapeutic preparations of IVIg [68] and tested IVIg HLA-I reactivity on a Luminex platform using three different beadsets: (1) Regular LABScreen SABs, which are coated with Face-1 admixed with Face-2; (2) the mild acid-treated Regular LABScreen SABs exposing the cryptic epitopes and only carry Face-2; and (3) iBeads, which are mildly trypsinized regular LABScreen SABs for removal of Face-2 so that they carry only Face-1. The results (Figure 9) confirm that IVIg reacts not only with Face-1 but also to Face-2. However, all preparations of IVIg showed greatly reduced reactivity for iBeads, suggesting that they predominantly recognize the Face-1 of HLA-I. At this stage, it is still uncertain whether the immunoregulatory properties of IVIg are due to the Abs binding to Face-1 or Face-2 of HLA.
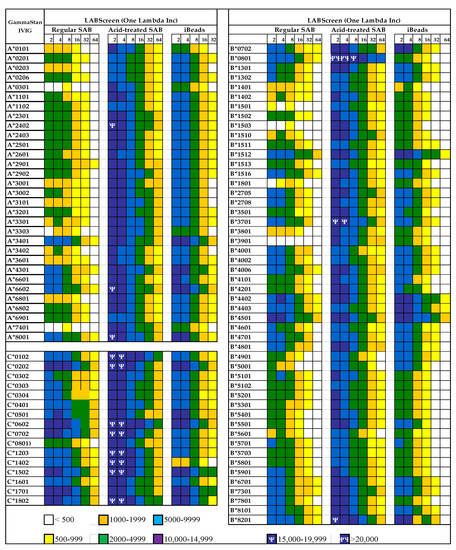
Figure 9.
Documentation of antibodies against HLA class Ia molecules (HLA-A, HLA-B and HLA-C) in a commercial preparation (GamaStan) of Intravenous Immunoglobulin (IVIg). * Suggesting that the individuals may possibly carry naturally occurring antibodies formed against most commonly shared epitopes. The profiles of IgG antibodies were obtained on three different kinds of beadsets (LABScreen, One Lambda Inc, Canoga Park, CA). The different kinds of beads are (1) Regular LABScreen single antigen beadsets (SABs) which are coated with Face-1 admixed with Face-2 or open conformers, (2) the same SABs after mild acid treatment resulting in the exposure of cryptic epitopes, resulting in molecules similar to naturally occurring Face-2 on activated immune cells, (3) iBeads, the same regular LABScreen SABs which are gently trypsinized to remove Face-2, to leave only Face-1 on the solid matrix. The difference in the profile between regular beads and iBeads also illustrates the admixture of Face-1 and Face-2 in regular LABScreen SAB commonly used in the clinical laboratories. If the objective of the clinicians is to monitor antibodies against Face-1, then only iBeads or Immucor Beadsets will serve the specific purpose. Mean Fluorescent Intensities (MFI) are indicated in different colors and symbols.
It was necessary to delineate the functions of HLA Abs by generating HLA polyreactive monoclonal Abs mimicking the Face-2 polyreactive IVIg. As was reported for IVIg, there were infrequent reports indicating that anti-HLA mAbs suppressed T cell proliferation [69,70,71,72], T cell activation [69], interleukin (IL)-2 and IL-2R synthesis [70], and apoptosis induction [73,74]. These reports, except for the mAb W6/32, did not identify the specific epitopes or amino acid sequences recognized by the anti-HLA-I mAbs.
Not only were we able to develop mAbs mimicking HLA-I reactivities of IVIg, but also documented that these IVIg-mimetics perform the unique immunoregulatory roles of IVIg similar to or better than the current formulation of IVIg while minimizing the adverse effects of IVIg. Possibly, they may minimize the adverse effects of IVIg by eliminating other non-HLA Abs.
4. The Pattern of Diversity of HLA Monoclonal Abs (mAbs)
4.1. Is the mAb W6/32 Specific for Closed Conformers (Face-1) of HLA-I?
Many investigators studying the functional role of HLA-I Abs [75,76,77,78,79,80,81,82,83,84,85,86,87,88,89] have used a well-known murine monoclonal antibody W6/32 on both naïve and activated cells that express Face-1. The mAb W6/32 has been used for quality control by the manufacturers of HLA beadsets (One Lambda Inc, Thermo Fisher, and Immucor) to prove that Face-1 are coated on the beadsets. The mAb W6/32 (IgG2a) has been developed to monitor HLA-I on the human cell-surface, since the mAb reacted with a wide range of cells but not with Daudi Burkitt lymphoma cells, which are devoid of β2m-associated HCs [75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93]. The same team established that W6/32 immunoprecipitated both a 43kDA HLA HC and a 12-kDa β2m. Interestingly, it formed immune complexes with β2m-associated HCs of HLA but not with β2m-free HCs. The mAb bound to both peptide-carrying β2m-associated HCs and peptide-free β2m-associated HCs [94,95]. However, W6/32 failed to bind to HC-associated β2m in the MHC (Major Histocompatibility Complex) class-I of mice, rats, rabbits, and guinea pigs [96,97,98,99,100]. Interestingly, when these non-human MHC HCs were reconstituted with β2m of cattle or humans, they regained the binding affinity. A comparative study of the amino acid sequences of the HCs and β2m of all animal MHC antigens with human HLAs identified the critical epitope as amino acid residue 121 in the HCs of HLA-I, the glutamic acid and glutamine residues in β2m at positions 44 and 89, respectively, [101] and the amino acid residue at position 3 [102].
Most interestingly, Tran et al. [103] reported that the mAb W6/32 “actually recognizes an epitope present on isolated non-reduced α-chains of most HLA-B allelic forms.” The specificity of W6/32 for Face-2 of HLA-B raises concern about the use of mAb W6/32 to validate the expression of intact HLA-B coated on the LABScreen and Immucor beadsets used on the Luminex platform.
4.2. Are There True HLA-I Polyreactive mAbs That Recognize Face-2 of All HLA-I Isoforms and Their Alleles?
To develop mAbs for HLA-E for immunodiagnosis, we have used recombinant β2m-free HCs (Face-2) of HLA-ER107and HLA-EG107 (Immune Monitoring Laboratory, Fred Hutchinson Cancer Research Center) [104]. The mAbs generated were analyzed using multiplex bead assays on a Luminex platform for HLA-I reactivity. Beads coated with an array of HLA heterodimers admixed with HCs (LABScreen) were used to examine the binding of IgG to different HLA-Ia (31 HLA-A, 50 HLA-B, and 16 HLA-C) and HLA-Ib (2 HLA-E, one of each HLA-F and HLA-G) alleles, as detailed elsewhere. A striking diversity in the HLA-Ia and/or HLA-Ib reactivity of mAbs was observed. The hybridomas and their mAbs obtained are shown in Table 6.

Table 6.
HLA-I isoform reactivity of the monoclonal Abs generated against two alleles of recombinant β2m-free HLA-E heavy chains (Face-2).
We have reported a detailed analysis of HLA-Ia and -Ib reactivities of a monospecific mAb (TFL-033) [104,105,106,107,108,109] and the polyreactive mAbs (TFL-006 and TFL-007) [109,110,111,112,113]. The binding affinity of the monospecific mAb TFL-033 was tested using the HLA-E specific amino acid sequence (SARDATA) as well as the binding affinity of the mAb TFL-006 with the most shared sequence 141AYDGKDY147, by inhibiting the binding of mAbs to the beadsets (LABScreen) used on Luminex Platform. The affinity of the polyreactive mAb TFL-006 to shared sequences which are cryptic in Face-1 but exposed in Face-2 was tested. For this purpose we have compared the mAb-binding to LABScreen (LS) beadsets which contain Face-1 admixed with Face-2, to acid treated LS SABs which contain only Face-2, and to iBeads [33] or to LIFECODES (LC) [34] SABs, which contain only Face-1. The results are recapitulated in Table 7. The mAb TFL-006, which binds to exposed cryptic epitopes found in Face-2, did not recognize theFace-1 HLA-I molecules coated on the iBeads or LC SABs. Interestingly, the mAb TFL-006 recognized regular LS SAB lots, confirming the presence of Face-2 on the regular LS SABs. The results revealed explicitly that the mAb TFL-006 only recognize the shared peptides when exposed on Face-2 HLA, but not when cryptic on the Face-1.

Table 7.
Recognition pattern of the TFL-006 that binds to exposed cryptic epitopes found in β2m-free HLA HCs (Face-2).
5. The Immunoregulatory Potential of Anti-HLA-I mAbs unravels the Roles of the HLA-I Face-1 and Face-2 and Their Polyreactive Abs in Immune Homeostasis
5.1. The Immunopotential of Anti-HLA Face-1 mAb W6/32
Several mAbs were used on the assumption that they are directed against intact HLA-I (Face-1). Most of them are poorly defined and used by few groups, without characterizing their epitope specificity. The most common mAb used to localize intact HLA-I (Face-2) on the cell-surface is mAb W6/32, which binds to both the HCs and β2m of all HLA-I isomers. Interpreting the affinity of the mAb requires caution in view of two findings:
- (i)
- Reports published from 1989 [12,15,16,17,18,19,20,24,25,26,114,115,116,117,118,119,120,121] document the prevalence of Face-2 on activated immune cells. Publications on W6/32 have ignored these findings until 2001.
- (ii)
- Tran et al. [103] reported that “although it (W6/32) does recognize in a monomorphic fashion all HLA-A, B, and C molecules when present in their native state (Face-1) on the cell-surface, under partially denaturalized conditions, it recognizes an epitope preserved in free nonreduced α chains of most HLA-B antigens but not in other HLA class-I α chains. Essentially identical results were also obtained with another pan-HLA class-I mAb MEM-147 which cross-blocks W6/32” (p. 443, column 1, para 3). Therefore, one should be cautious while interpreting the results stemming from using W6/32 on activated cells, which express Face-2.
Two categories of studies shed light on the immunoregulatory role of the mAb. The first one investigates the impact of W6/32 on endothelial cells (ECs), smooth muscle cells (SMCs), and epithelial cells. The second one investigates the effects of the mAb made on immune cells, particularly CD3+/CD4+ and CD3+/CD8+ T lymphocytes and CD3+/CD19/20+ B lymphocytes.
5.1.1. Impact of the mAb W6/32 on Endothelial, Smooth Muscle and Epithelial Cells
Augmentation of serum anti-HLA IgG during Transplant Atherosclerosis (TA) results as a consequence of the intimal proliferation of smooth muscle cells, endothelial cells, and fibroblasts, leading to vessel obstruction, fibrosis, and graft loss. Reed and her team [76,77,78,79,80,81,82,83,84,85,86,87,88,89] investigated the role of the anti-HLA-I Abs in TA. On the assumption that the mAb W6/32, represents a typical anti-HLA-I IgG Ab, they studied its impact on the activation and proliferation of endothelial cells (ECs) and smooth muscle cells (SMCs). Indeed, the binding of W6/32 to ECs increased tyrosine phosphorylation of intracellular proteins, inositol phosphate generation, and cell proliferation [77,78]. In addition, W6/32 added to ECs and smooth muscle cells (SMCs) in vitro increased mRNA expression of high affinity fibroblast growth factor receptor (FGFR-1), and enhanced basic fibroblast growth factor ligand binding, culminating in augmented cell proliferation [76,78]. The FGFR-1 is a member of the superfamily of tyrosine kinase growth factor receptors and is capable of transducing proliferative signals. These findings suggest that the binding of W6/32 to cell-surface HLA-I is able to transduce signals and generate intracellular messengers. Furthermore, the treatment of ECs with mAb W6/32 stimulated a higher level of FGFR expression than treatment with serum Abs to individual A locus or B locus molecules, suggesting that the intensity of the signal transduction is related to the aggregation of HLA-I molecules on the cell-surface [76,78]. Evidently, the mAb W6/32 promotes cell proliferation and upregulates cell survival genes. The intracellular events initiated by W6/32 binding are concentration-dependent. At high concentration, the mAb upregulated cell proliferation, whereas at low levels, the mAb activated the PI3K/Akt pathway and promoted expression of cell survival proteins including Bcl-2 and Bcl-xL. These findings were further substantiated by others [122].
Similarly, Mohanakumar’s group [123,124] reported that the incubation of airway epithelial cells (AECs) with the W6/32 induced a significant cell proliferation within 24 h comparable with the proliferation observed in the presence of LHC-9 growth medium. However, the proliferation of the AECs induced by the W6/32 was only observed within the first 24 h. AECs treated with W6/32 exhibited significantly greater proliferation and tyrosine phosphorylation of proteins compared to AECs treated with normal human serum or mouse IgG, respectively.
5.1.2. Impact of the mAb W6/32 on Immune Cells
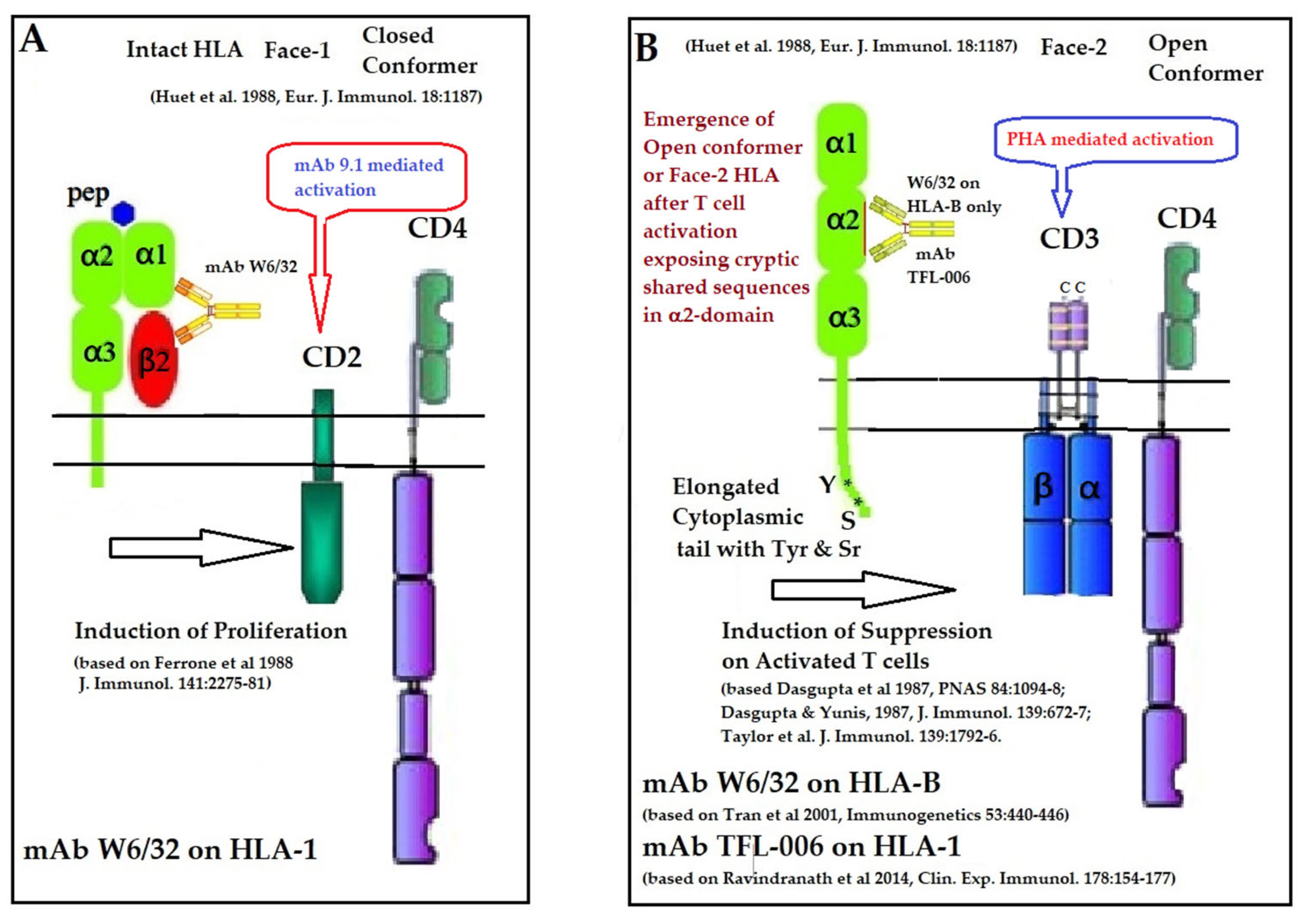
Ferrone and his team [125,126] observed that W6/32 enhanced the T cell proliferation induced by CD2-specific mAb 9.1 in a dose-dependent manner, which was not abrogated by the addition of exogenous IL-1 and IL-2. However, no such detectable enhancement was seen with CD2-induced stimulation by anti-β2m mAb NAMB-1. Ferrone’s team [127,128] also observed that W6/32 enhanced the proliferation of CD1- CD3+ CD4+ CD8+ T-prolymphocytic leukemic cells induced by CD2-specific mAb 9.1. However, W6/32 inhibited the proliferation induced by the CD3 binding mAb OKT3. These findings are supported by others [129,130,131]. Figure 10 summarizes the role of mAb W6/32 in enhancing the CD2- and CD3-mediated activation of T-cells.
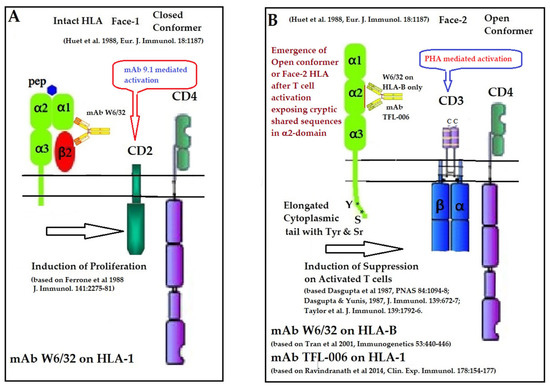
Figure 10.
Illustration of the role of mAb W6/32 in enhancing the CD2 and CD3 mediated activation of T-cells (A) as well as the role of HLA-I polyreactive mAbs TFL-006 and TFL-007 in suppression of (PHA)-CD3-mediated activation of T lymphocytes (B) [125,126,127,128,129,130,131].
It is known that HLA-I regulates T cell proliferation induced via both CD2 and CD3 pathways [132]. Evidently, the mAb W6/32 may ligate with both Face-1 and Face-2 of T-cells activated via CD2 and CD3 molecules on the T-cells. When the T-cells are activated by the ligation of the mAb 9.1 on CD2, the binding of W6/32 enhances proliferation. However, when the T-cells are activated via CD3 by the ligation of the mAb OKT3, the proliferation is suppressed. These findings suggest that the mAb W6/32 may influence HLA-I on the cell-surface differently, one upregulating intact HLA (Face-1) and another downregulating Face-2. Accordingly, W6/32 may enhance the proliferation of CD2-specific mAb-mediated activation of T-cells and suppress proliferation of CD3-mediated activation of T-cells. Examining HCs-polyreactive mAbs binding specifically to Face-2 may provide a better understanding of this aspect.
The findings of Tran et al. [103] clarified that β2m is not a prerequisite for W6/32 binding to HLA. However, it is still not clarified why that is applicable to β2m-free HCs of HLA-B and not applicable to β2m-free HCs of HLA-A or HLA-C. Since HLA-E, HLA-F and HLA-G were not tested, there is a long way to go to fully define the specificity of mAb W6/32.
5.2. The Immune-Reactive Potential of HLA-I Shared-Epitope Polyreactive mAbs (TFL-006/TFL-007) Directed against Anti-HLA Open Conformers (Face-2)
T-cells can also be activated by phytohemagglutinin (PHA), phorbol myristate acetate (PMA), anti-CD3 antibody, brefeldin A (PFA), and IFN-γ. Both CD2 and CD3 domains on T-cells may serve as receptors for several ligands such as PHA and get activated, resulting in proliferation. Such activated T-cells overexpress β2m-free HCs (Face-2) [12,15,16,17,18,19,20,24,25,26,55,56,57,58,92,93,95,96]. Upregulation of Face-2 upon activation of T-cells could also be related to CD3-mediated activation.
In naturally occurring β2m-free HCs (Face-2), cryptic epitopes hidden by β2m in Face-1 are exposed primarily in the α2 domain. This domain (Figure 2, Figure 3, Figure 4D–F Figure 5) contains several sequences (such as 141AYDGKDY147) shared by HLA-A, -B, -C, -E, -F and -G. (Table 3). The shared epitopes of all HLA isomers can be better appreciated when fitted on the same page, as shown in Figure 4.
Given the nature of the shared epitopes, mice were immunized with recombinant HCs of HLA-E and the Abs elicited after immunization were examined for binding to different alleles of different HLA isomers [104,105,106,107,108,109]. A profile of the antibody specificities of more than 200 mAbs are illustrated in the Table 6 and are described in detail elsewhere [104,105,106,107,108,109]. Distinctly, six mAbs were highly polyreactive to β2m-free HCs (Face-2) of all alleles of HLA isoforms. The shared epitopes were identified, and the synthetic amino acid sequences were used to inhibit the binding of the polyreactive mAbs [109,110,111,112,113]. Of these, TFL-006 ranks first and TFL-007 ranks second in their strength of polyreactivity as illustrated [68,110,112,113]. We will focus on their immunoreactive potential and compare it with (1) mAb W6/32 and (2) IVIg.
5.2.1. HLA-I Face-2 Polyreactive mAbs (TFL-006/TFL-007) Mimic IVIg HLA-I Reactivity and Regulate Activated Lymphocytes
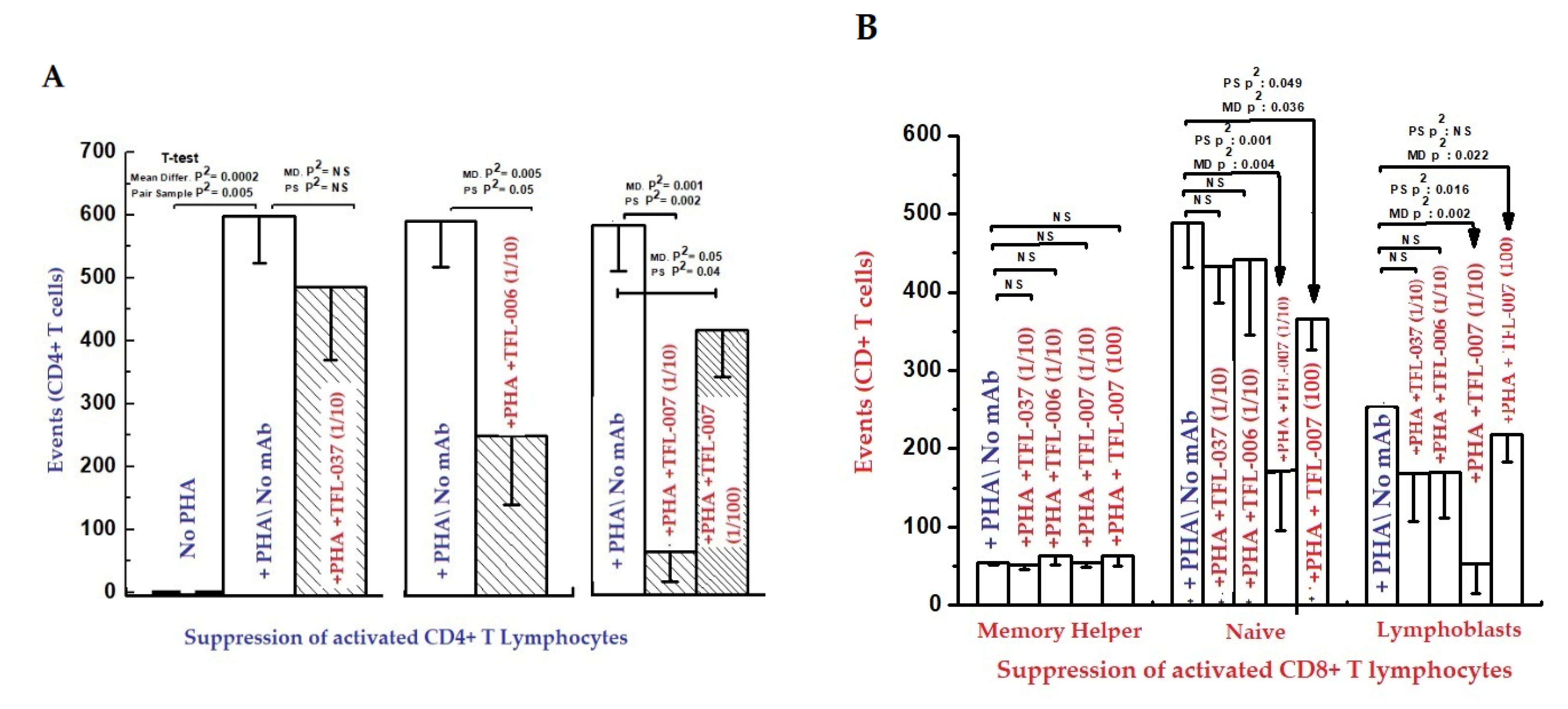
Immunologically relevant shared epitopes are located in the α2-domain of the heavy chain of HLA-I, as illustrated in Figure 2, Figure 3, Figure 4D,E, Figure 5 and Figure 6. These sequences are cryptic in Face-1 but exposed in Face-2 (Figure 5 and Figure 6). To determine how the recognition by mAbs of the shared epitopes of Face-2 impacts proliferation of activated cells, we have added mAbs TFL-006 and TFL-007 to human T-cells activated by PHA-P (PHA-Phaseolus) in vitro [133]. Two sets of experiments were done in triplicate on both CD4+ T-cells and CD8+ T-cells as follows:
- (i)
- The impact of the mAbs (TFL-006 and TFL-007) (Experimental) and controls (TFL-033 and TFL-037) (Controls) on the blastogenesis in PHA-treated and untreated T-cells was determined by counting lymphoblasts after culturing purified lymphocytes from donors for 72 h. Lymphoblasts were recognized by flow cytometry based on size (side-scatter) and granularity (forward scatter). The blastogenesis and suppression were compared with different commercial preparations of IVIg.
- (ii)
- The impact of the above listed mAbs was examined for enhancement or suppression of proliferation by labelling the purified lymphocytes during PHA activation with the intracellular fluorescent dye carboxyfluorescein succinimidyl ester (CFSE). Using flow cytometry, the mitotic activity was measured by the successive twofold reductions in fluorescent intensity of the T-cells placed in culture for 72 h. PHA-treated T-cells undergo, on average, four to six divisions by 72 h. The suppression of blastogenesis and proliferation can be visualized by cessation of progression of mitotic activity, as measured by the successive twofold reductions in the fluorescent intensity after 72 h of treatment.
The findings are elaborated in detail in previous reports [111,112,113]. Figure 11 and Figure 12, and Table 8 elaborate the results of dosimetric suppression of blastogenesis and proliferation of PHA-activation of both CD4+ and CD8+ T lymphocytes by anti-HLA-E mAbs (TFL-006 and TFL-007) that mimic the HLA class-Ia and-Ib reactivity of IVIg. The IVIg-mediated dosimetric suppression of blastogenesis and proliferation of PHA-activated T-cells was identical to that of the HLA-polyreactive mAbs, whereas the control anti-HLA-E mAbs (TFL-033 and TFL-037) did not have any impact on PHA-activated T-cells. Interestingly, the mAbs TFL-006 and TFL-007 are shown to be better suppressors of blastogenesis of the PHA-activated CD4+ T-cells than IVIg. The concentration of anti-HLA-E mAbs required for this suppression was as much as 150-fold lower than that of IVIg.
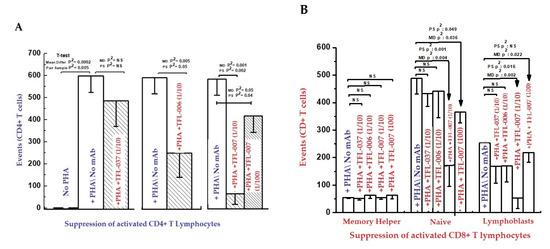
Figure 11.
Inhibition of Phytohaemagglutinin (PHA)-activated T-lymphocytes in vitro with two ((A) CD4+; (B) CD8+).
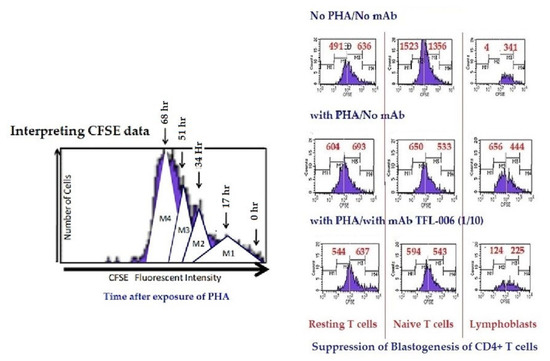
Figure 12.
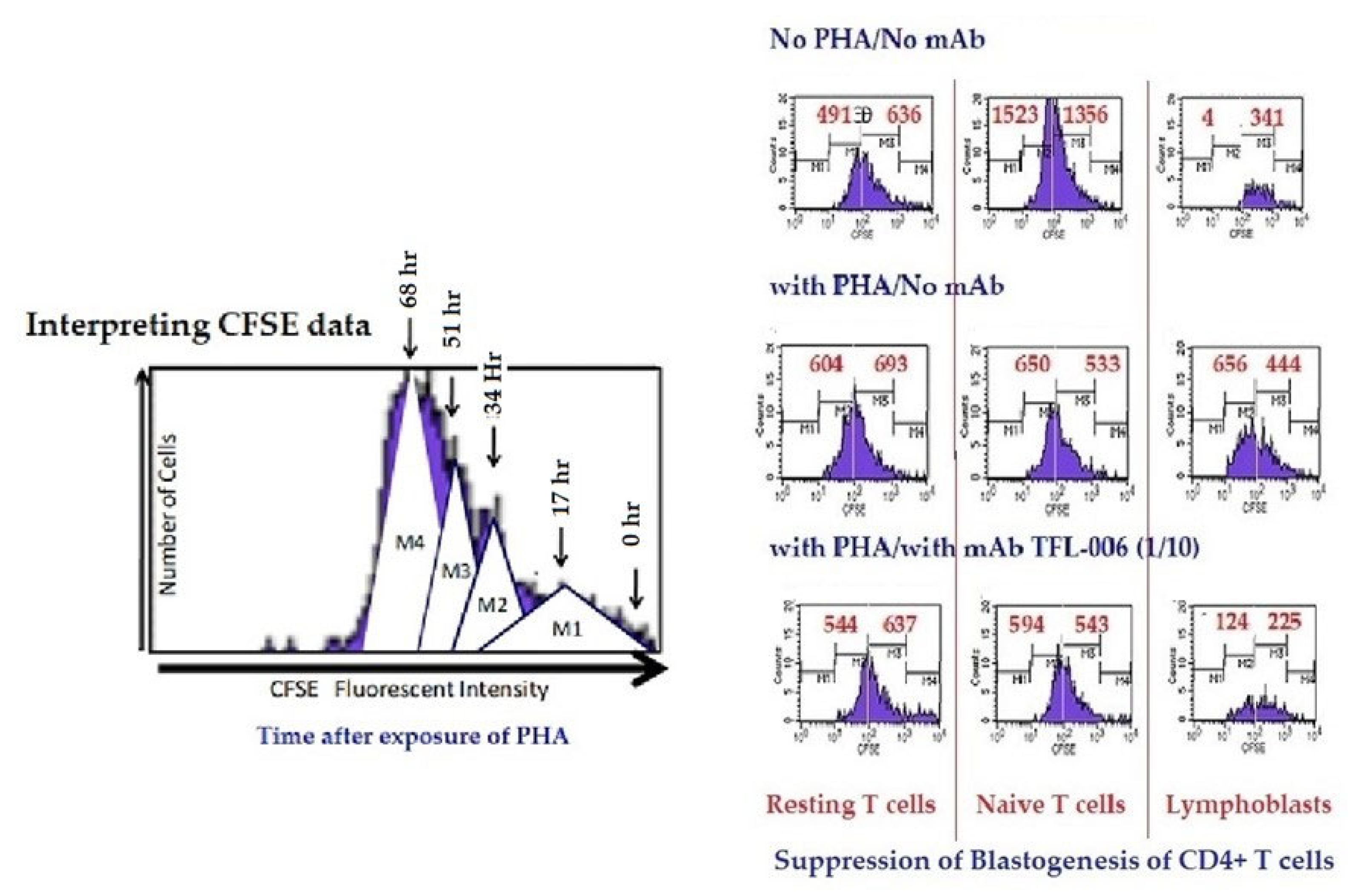
Suppression of in-vitro proliferation of PHA-activated CD4+ T lymphocytes by mAb TFL-006s (culture supernatant). The carboxyfluorescein succinimidyl ester (CFSE)-labelled lymphocytes were cultured with or without PHA or with PHA and mAb TFL-006s at 1/10 dilution. Three days after culture, cells were labelled with fluorescent dye-conjugated anti-CD4+ or anti-CD8+ Abs before analysis. CFSE labelling allowed us to gauge and show cell proliferation: when the CFSE-labelled cell population undergoes mitosis, after 72 h it has migrated from the right to the left side of each rectangular box (as shown in the top figure) depending on the number of mitoses. The distance moved shows the number of cell divisions. The effects of mAb TFL-006s on the proliferation of CD4+/CFSE+ T lymphocytes are shown in each box. After incubating cells with CFSE, the cells were treated as noted. Each box in the figure is divided by a vertical line into two sub-boxes, the right for mitoses 1 and 2 (M1/2) (parent lymphocytes) and the left for mitosis 3 to 5 (M3–5) (the progeny). The cell number after each treatment (including ‘no PHA’) of each T lymphocyte population was counted and compared, the number shown in each sub-box. Note that with no PHA the number of cells in the M3–5 sub-box is very meagre for all groups of CD4+ T lymphocytes. With PHA-only treatment, the very high number of cells for M3–5 indicates that proliferation has occurred in all three groups. The impact of treatment with PHA and TFL-006s is striking: the number of cells in the M3–5 sub-box is reduced, indicating suppression of proliferation. No such decrease was observed with resting or naive T lymphocytes.

Table 8.
Dose-dependent inhibition of blastogenesis and proliferation of activated lymphocytes.
HLA-I open conformer (Face-2) polyreactive monoclonal Abs (mAbs). The mAbs were generated following immunization with recombinant HCs of HLA-ER. The density of the population of CD4+ T cell (A) and CD8+ T cell (B) cultured with and without PHA, and after adding TFL-037s (control mAb) at 1/10 dilution, TFL-006s (1/10) and TFL-007s (1/10 and 1/100) to the PHA-treated CD4+ T-cells. The values are expressed as the mean of triplicate analyses +/− standard deviation (s.d.) and two-tailed p-values. The two-tailed p-values, if significant, are indicated by a horizontal line connecting the two groups. Note that the PHA-activated CD4+ T lymphocytes (A) remained unaffected by treatment with mAb TFL-037s, and that only lymphoblasts (not resting or naïve T-cells) decreased after treatment with mAb TFL-006s. Similarly, the lymphoblasts (not resting or naive T-cells) decreased significantly after treatment with mAb TFL-007s at 1/10, and after treatment with mAb TFL-007s at 1/100 dilution. Clearly, both anti-HLA-E mAbs TFL-006s and TFL-007s significantly suppress PHA-activated CD4+ T lymphoblasts, although the dosimetric suppression by TFL-007s is more striking. Similarly, the lymphoblasts of CD8+ Naïve T-cells (B) decreased after treatment with both mAbs but the decrease is significant with mAb TFL-007. Interestingly, mAb TFL-007 also suppressed PHA-activated naïve T-cells significantly. Note: “s” after mAb numericals (e.g., TFL-006s) refers to culture supernatant of the mAb.
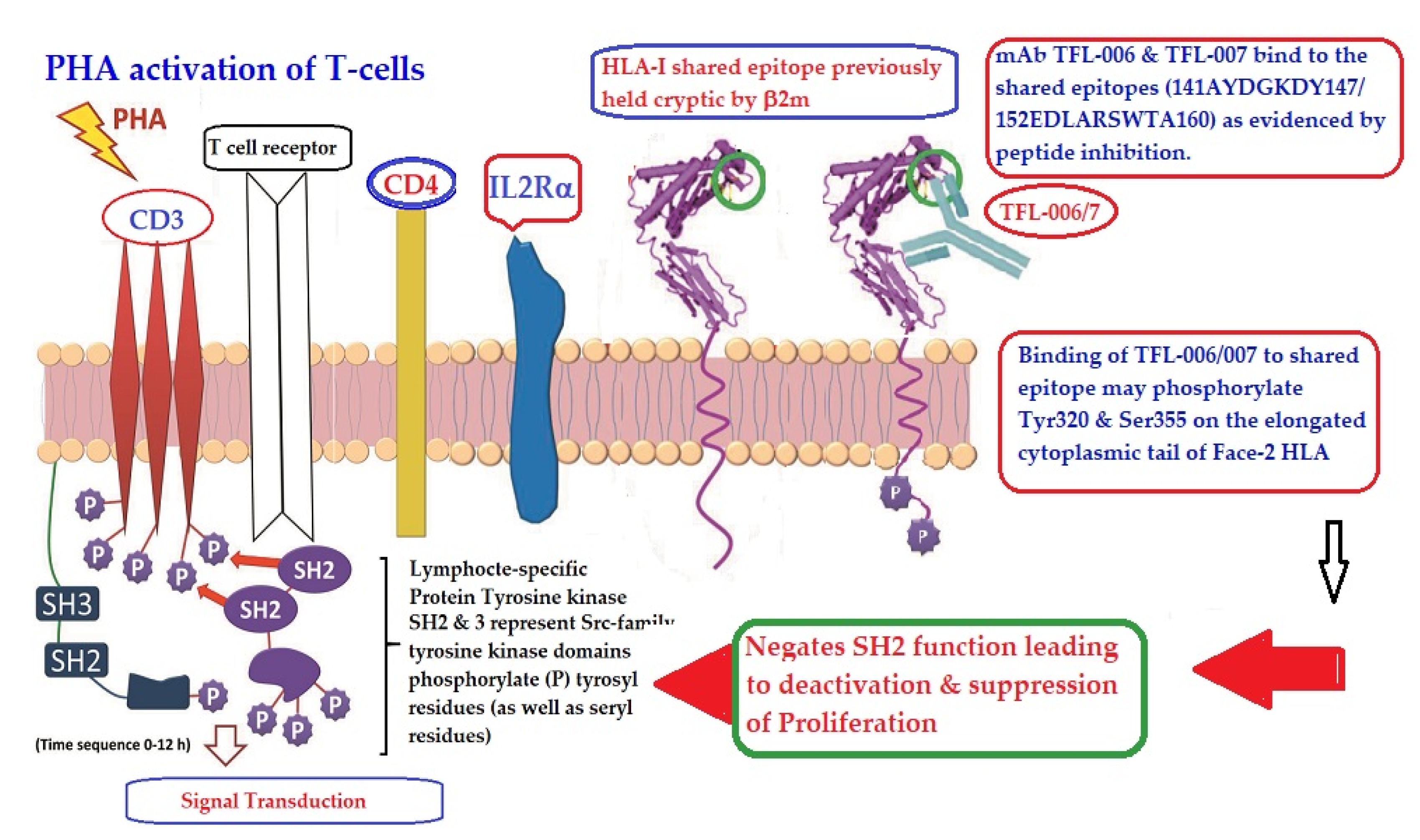
Figure 13 illustrates the possible mechanism of suppression of PHA activated T-cells mediated by mAbs directed against shared epitopes exposed in Face-2. The proposed mechanism is based on the Mustelin model for T-cell activation [134] and can be explained by the molecules shown on the cell surface. The CD3, CD4, IL-Rα and T cell receptor molecules are expressed on the cell surface. PHA interacts with surface ligands on CD3 and initiates phosphorylation of the cytoplasmic domain of CD3 leading to activation of transcription factors. Such a cascade of events results in the production of cell surface molecules such as IL-2Rα [135,136] and Face-2 [12,15,16,17]. Face-2 on the cell surface exposes the cryptic epitopes. The binding of HLA-I polyreactive Abs (e.g., mAbs TFL-006 & TFL-007) to the shared epitopes promotes the phosphorylation of Tyr 320 [137,138,139] and Ser 355 [140] in the cytoplasmic domain of Face-2. The phosphorylation mediated by the HLA-I polyreactive Abs leads to activation of phosphatases leading deactivation of transcription factors and abrogation of the synthesis of proteins involved in blastogenesis and mitosis [141,142,143,144], as portrayed in the Figure 13.
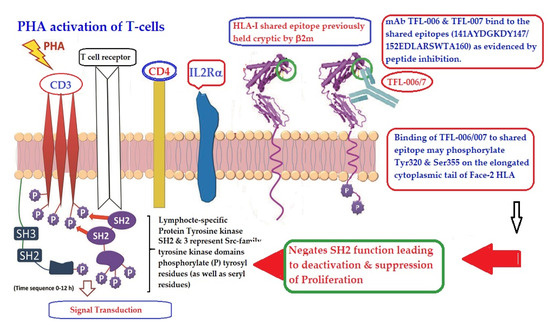
Figure 13.
An illustration of the possible mechanism of suppression of PHA activated T-cells mediated by mAbs directed against shared epitopes exposed in the Face-2 of almost all alleles of HLA-I. The figure shows CD3/T cell receptor (TCR)/CD4 on the bi-layered lipid membrane on the T-cells upon activation (Yellow complex arrow). Phosphorylation of the cytoplasmic domain of CD3 is induced by Lymphocyte-specific protein tyrosine kinase (LCK). SH represents Scr-family LCK. They phosphorylate the tyrosyl (even seryl) domain of the cytoplasmic tail of CD3. This event leads to activation of transcription factors and transcription cell-surface molecules such as IL2Rα and Face-2. Importantly, the HLA-I open conformers devoid of β2m (Face-2) expose unique epitopes shared by almost all HLA-I alleles. It is the site recognized by TFL-006 and TFL-007. It is postulated that the binding of these specific mAbs to the epitopes on the Face-2 may initiate phosphorylation of the elongated cytoplasmic tail. The elongated cytoplasmic tail results in the exposure of cryptic tyrosyl residue at position 320 [137,138,139] and serine at position 35 [140]. This leads to transduction that initiates dephosphorylation of the cytoplasmic domain of CD3, resulting in suppression of activation of T-cells. Evidently, the T-cell activating CD3-phosphorylation can be reversed.
5.2.2. HLA-I Face-2-Polyreactive mAbs (TFL-006/TFL-007) May Simulate IVIg in Suppressing allo-HLA Abs Produced by Human B Cells
IVIg, introduced as replacement therapy for patients with primary and secondary immune deficiencies [55,68], was extensively used for treatment of several autoimmune and systemic inflammatory diseases, acute and chronic relapsing diseases, and autoimmune disorders mediated by autoaggressive T-cells [35,36,37,38,39,40,41,42,43]. Additionally, IVIg modulates B-lineage cell antibody production. The use of IVIg for allograft recipients was approved by the US Food and Drug Administration (FDA) to reduce anti-HLA antibody levels prior to transplantation and to reverse humoral rejection [44,45,46,47,48,49,50,51,52,53,54].
All commercial IVIg preparations are reactive to almost all HLA-I antigens [55,145] (Figure 9), mimicking the HLA-I polyreactive mAbs (Table 7). Simulation of the antibody suppressive activity of IVIg by the polyreactive HLA mAbs was tested by comparing the efficacy of IVIg versus that of the mAb TFL-007 in suppressing the allo-HLA Abs production by hydridoma cell line (HML16), developed from a postpartum alloimmunized woman. The HML16 cell line produced anti-HLA class-I alloAbs with differing MFIs: high against B*07:02, B*81:01, B*67:01 and B*42:01; and low against B*27:08, B*27:05, B*55:01, B*56:01 and B*82:01.
The effects of two different therapeutic IVIg preparations (GamaSTAN and Gamunex) and the HLA-polyreactive mAb TFL-007 were tested on HML16 at different dilutions and protein concentrations and compared with the control media. Neither of the two IVIg preparations showed any significant suppression of the allo-HLA-B IgG production by HML16 cells. In contrast, the mAb TFL-007 showed a dose-dependent, (at 1:10 (100 μg/mL), 1:20, 1:40 and 1:80) significant reduction in the generation of Abs to both HLA-B*07:02 and B*81:01 [111], as compared with that of control media and mouse IgG control. The binding affinity of mAb TFL-007 is towards β2m-free HCs (Face-2) and not towards intact HLA (Face-1). This cannot be construed as a general effect since it involves specific binding of the mAbs to the cryptic domain exposed on Face-2. Since IVIg possess reactivity to several HLA antigens with a common shared sequence, IVIg may also bind to the shared epitopes to induce suppression of antibody production by the selected hybridoma.
5.2.3. The Polyreactive mAbs (TFL-006/TFL-007) Mimic IVIg in Upregulating CD4+/CD25+/Foxp3+ Regulatory T-Cells (T-Regs)
The CD4+CD25+(IL-2Rα)Foxp3+ regulatory T-cells (T-regs) in humans and mice, though different in their cell surface markers, are capable of suppressing proliferation and cytokine production by CD4+CD25− T-cells in response to a variety of stimuli in vivo [146]. Suppression of proliferation and activation of CD8+ T-cells in vitro by human T-regs is mediated by “a cell-contact-dependent cytokine-independent mechanism” [147,148]. IVIg is known to upregulate T-regs isolated from patients with Kawasaki disease and common variable immunodeficiency diseases but not those isolated from healthy individuals [149,150]. The ability of TFL-006 and TFL-007 to induce proliferation of CD4+CD25+Foxp3+ T-regs obtained from normal and healthy donors was assessed [113]. The impact of IVIg was compared with that of the mAb TFL-007 on untreated and PHA-treated isolated fractions of CD3+/CD4+ human T lymphocytes. A variety of cell-surface markers, including CD4, CD25 (IL-2Rα), CD45RA, and Foxp3, were monitored using their respective diagnostic mAbs. We have compared the efficacy of IVIg and anti-HLA-mAbs TFL-006 and TFL-007 to induce proliferation of CD4+CD25+ (IL-R2α), CD34RA and Foxp3 T-regs obtained from normal and healthy donors [134]. The effect of different commercial preparations (GamaSTAN, Octagam and Gamunex) of IVIg (at 1:10 and 1:80) and TFL-007 (at 1:10 and 1:60) were studied on the PHA-untreated T-regs obtained from a healthy non-alloimmunized volunteer.
Interestingly, all the three IVIg preparations did not significantly upregulate T-regs but in striking contrast, TFL-007 (at 1:10 but not at 1:60) significantly (p2: <0.001) upregulated T-reg cells. This finding once again resolves the role played by the polyreactive mAb binding to Face-2. Evidently, Face-2 is not overexpressed on normal T-cells but may be present at low levels, as observed for HLA-F and occasionally for HLA-B and HLA-C.
6. The Major Players in HLA-I-Mediated Cellular and Humoral Immune Homeostasis
6.1. HLA-I Variants as Receptors in Immune Regulation
HLA-I mediated immune homeostasis should be considered as a delicate and regulated balance of activation and suppression of immune and associated cells. It is also considered as a strategy to restore normalcy from alterations imposed by pathological conditions. Until 1989, β2m-associated HLAs (Face-1) were considered as the only functionally active cell-surface HLA class-I molecules. The Face-1 HLA-I molecules were known to present antigens derived from intracellular infection caused by pathogens to CD8+ cytotoxic T-cells (CTLs), which express TCRs specific for HLA-I. The T-cell co-receptor CD8 binds to the α3 domain of HCs of Face-1, while α1 and α2 domains of the HCs hold antigen peptides in the groove and present them to the receptor on T-cells (Figure 1). The antigen-presentation to CTLs initiates the cytolytic killing of the infected cells or even tumor cells. Since all human nucleated cells are potential targets for infections and malignancy, the HLA-I isomers are expressed on all such cells.
The discovery of naturally occurring HLA-I variants expands the functional role of HLA-I in inflammation, tissue injury, infection, malignancy, and end-stage organ diseases. Thus, the discovery of Face-2 in activated immune cells revolutionized concepts of HLA-mediated immune homeostasis. The Face-2 variant of HLA-I has exposed amino acid sequences known to be masked by β2m (Figure 5). Interestingly, most of these sequences (Table 3, Figure 2, Figure 3 and Figure 4D–F) are highly shared among all HLA-I alleles of different isomers, namely HLA-A, HLA-B, HLA-C, HLA-E, HLA-F and HLA-G.
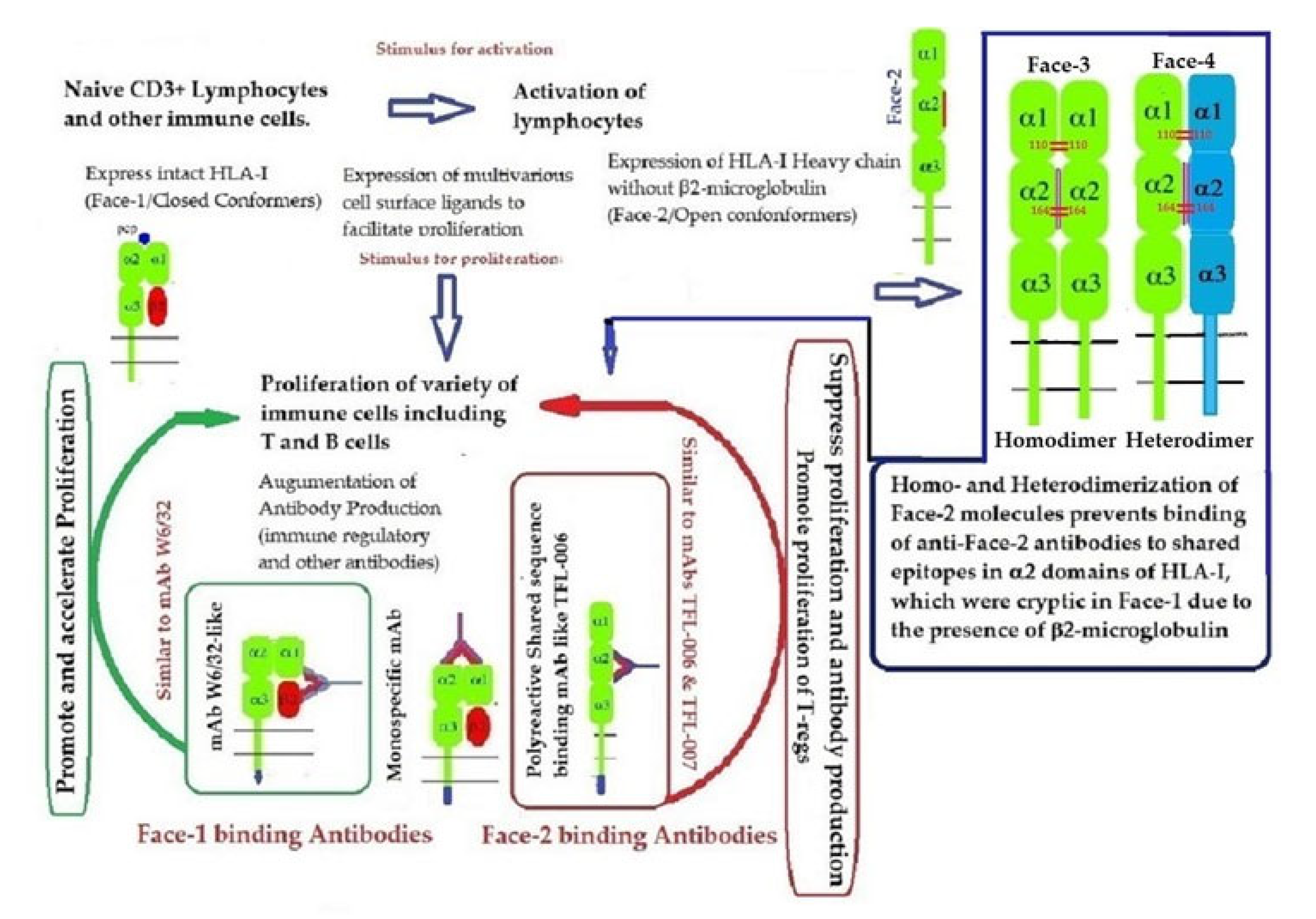
Recent reports on Face-2 prompted attention to other HLA variants (homodimers, Face-3, and heterodimers, Face-4) emanating from Face-2. The cysteine residues on the open heavy chain of Face-2 can interact with the cysteine residues (at positions 64, 101 and 164) on the other allele’s heavy chain resulting in the genesis of Face-3 (Figure 6). Face-2 conversion into Face-3 or Face-4 on the cell-surface has the functional significance of masking the shared epitopes exposed by the paucity of β2m. Since these shared epitopes play a major role as receptors for several ligands, including Abs that bind to them (such as the mAbs TFL-006 and TFL-007), the immunohomeostatic roles of these HLA-I structural variants (Face-2, Face-3, Face-4, including that of Face-1) become explicit. Thus, Face-3, the homodimeric variant of Face-2, and Face-4, the heterodimeric variant of Face-2, have become important factors that can prevent the exposure of cryptic domains. They perform the function previously performed by β2-m in Face-1 HLA-I (Figure 14). Masking of the exposed shared epitopes seems to be an important step to regulate the signal transduction leading to activation of immune cells. However, elucidating the roles of Face-2, Face-3, and Face-4 in different pathological conditions may unravel their delicate and regulated role in immune homeostasis. More focused investigations in this direction are needed to understand the immunohomeostatic role of different Faces of HLA-I.
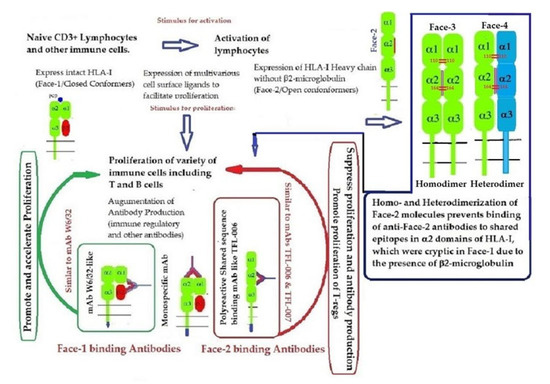
Figure 14.
Graphic overview of the roles played by HLA-I structural variants (Face-1, Face-2, Face-3, and Face-4) together with the Abs directed specifically against Face-1 (represented by the mAb W6/32) and/or the Abs directed specifically against Face-2 (exemplified by the mAbs TFL-006/007) in HLA-I mediated immune homeostasis.
6.2. Antibodies Formed against Face-1 and Face-2 as Major Players in HLA-Mediated Immune Homeostasis
6.2.1. Profiles of HLA Abs in Sera of Normal Females and Non-Alloimmunized Males
Both IgM and IgG occur naturally in the sera of normal healthy males and females (Table 4 and Figure 7A–C). These Abs are formed against almost all alleles of HLA-A, HLA-B, HLA-C, HLA-E, HLA-F and HLA-G. The profiles of HLA Abs differ among individuals. Some normal healthy individuals have Abs against almost all HLA alleles, while some have Abs against only a few alleles (Table 4). Figure 7A–C illustrate the relative proportions (measured as mean fluorescence intensity or MFI) of IgM and IgG reacting to HLA-E, HLA-F, and HLA-G in the unpurified sera of the normal individuals. Many Abs escape detection due to presence of either anti-idiotypic antibodies or other factors contributing to the formation of immune complexes, as evidenced from the eluates of sera after purifying the Abs with Protein-G affinity columns (Table 5). It is clarified from the works of Morales-Bunerostro et al. [32] that serum naturally occurring anti-HLA Abs react to both Face-1 and Face-2 of HLA-I. However, very little is known about the functional roles of these naturally occurring sera Abs. To elucidate the functions of such anti-HLA Abs, investigators have used most often the mAb directed against Face-1, namely W6/32. Since the natural anti-HLA-I Abs also react well with Face-2, there is a need to examine the functional potentials of Abs directed against Face-2.
6.2.2. Functional Potentials of Antibodies Directed against Face-2 as Evidenced from the Observations Made on IVIg
Anti-HLA-I Abs observed in IVIg, pooled and purified from the plasma of thousands of individuals, represent the Abs found in normal healthy individuals. The profiles of anti-HLA Abs in IVIg are not comparable to normal native sera but to the sera purified by Protein-G affinity column. Figure 9 illustrates the profile of HLA-Ia reactivity of a commercial preparation of IVIg. The Abs were monitored using three kinds of beadsets, namely regular beadsets coated with Face-1 admixed with Face-2, beadsets treated with a mild acid to expose cryptic epitopes resulting in only Face-2, and iBeads carrying only Face-1. It is evident from the figure that IVIg contains Abs reacting to both Face-1 and Face-2. When IVIg was used in vitro on activated T-cells, proliferation was not enhanced, as was observed with the mAb W6/32, but rather blastogenesis and proliferation were significantly suppressed. This finding prompted further investigation on the functional potential of mAbs directed against Face-2.
HLA variants may play a major role in conjunction with the Abs they generate. Face-2 variants of HLA-I can elicit different kinds of epitope-specific Abs, due to exposure of several cryptic epitopes. The different kinds of Abs may include different grades of HLA-I polyreactive Abs ranging from the lowest to the highest polyreactivity (Table 6), depending on the frequency of the respective epitopes among HLA-I alleles [104]. Typical example of highly polyreactive Abs include TFL-006 (Table 7) and TFL-007. In addition, Face-1 is also capable of generating Abs that can bind simultaneously to β2-m and α-HC of HLA-I. A typical and most popular example is mAb W6/32, although as noted above, Tran et al. [103] raised unique concerns about this mAb’s affinity to the HLA-B Face-2. Despite the concerns, the very ability of W6/32 to bind to both β2-m and α-HC of HLA-I clearly identifies as a Face-1 directed Ab, while TFL-006 and TFL-007 remain as Face-2 specific polyreactive Abs.
This review is restricted to one major issue of immune homeostasis related to activation CD3+/CD4+/CD8−, CD3+/CD4−/CD8+ and CD3+/CD4+/CD25+/Fox P3+ lymphocytes. An overview of HLA-I mediated immune homeostasis is diagrammatically illustrated in Figure 13 and Figure 14. They are enumerated as follows:
- (1)
- The difference in HLA variants found in normal and activated CD3+ lymphocytes. In normal healthy individuals, CD3+ lymphocytes express intact HLA-I (Face-1). The lymphocytes may get activated naturally upon inflammation and injury under different pathological conditions. In addition, other exogenous agents can induce activation, such as mAb 9.1 [125,126,127,128], phorbol myristate acetate (PMA), anti-CD3 antibody, phytohemagglutinin (PHA), brefeldin A (PFA), and IFN-γ [15]. Such activated T-cells initiate activation of transcription factors [64], production of cell-surface molecules such as IL-2Rα (Figure 13) [45,46,47], and Face-2 of HLA-I [12,15,16,17,115,116,117,118,119,120,121].
- (2)
- This unique shift in the cell-surface expression of Face-1 HLA-I on normal cells to Face-2 upon activation exposes epitopes that remain cryptic in Face-1 due to masking of amino acid sequences in the α2 domain by β2m.
- (3)
- This domain (Figure 2, Figure 3, Figure 4D–F Figure 5) exposes several sequences shared by HLA-A, -B. -C, -E, -F and -G. (Table 3). When all HLA alleles of all HLA isomers are seen on one screen, it is easy to pinpoint which sequences were cryptic and how they are shared by almost all alleles of all HLA isomers. The uniquely shared epitopes are important to understand and appreciate the role of HLA variants in immune homeostasis.
- (4)
- The exposure of cryptic epitopes may involve immediate immune recognition resulting in the formation of Abs against those shared and cryptic epitopes. This is exemplified by TFL-006 and TFL-007 and by their role in several immunoregulatory functions. Essentially, they are capable of suppressing proliferation of T and B-cells while promoting proliferation of T-regulatory cells (Figure 14).
- (5)
- Figure 13 clarifies the events that occur after the binding by mAbs TFL-006 and TFL-007 to Face-2 on the cell-surface of activated T-cells. The mAb binding to the exposed cryptic epitopes on Face-2 may involve a reversal of activation of T-cells mediated by signal transduction. In support of such an inference, it is documented that the elongation of the cytoplasmic tail of the HLA class-I open conformers will expose otherwise cryptic tyrosine320 [137,138,139] and serine335 [140] with a provision for phosphorylation (Figure 13). Although serine335 is generally considered the primary site of phosphorylation in this tail, phosphorylation of tyrosine320 has been documented by others [141,143]. Either way, what is most important is that the open conformers in activated normal human T-cells are associated with tyrosine phosphorylation and can enable cis interactions with cell-surface receptors or other signalling molecules [141,142,143,144]. The phosphorylation mediated by TFL mAbs may result in dephosphorylation of the cytoplasmic tails of CD3 molecules by activating phosphatases, leading to the arrest of transcription factors and synthesis of the proteins involved in blastogenesis and mitosis.
- (6)
- Again, a homeostatic function of the HLA variant Face-2 is the transient exposure of unique shared epitopes which is terminated by homo- and/or heterodimerization. Face-3, the homodimeric variant of Face-2, and Face-4, the heterodimeric variant of Face-2, recapitulate the function previously performed by β2-m in Face-1 HLA-I (Figure 14). The exposure of cryptic domains is blocked from receptor-ligand interactions and recognition by immunoregulatory Abs such as TFL-006 and TFL-007.
6.2.3. Are Immunoregulatory Roles Restricted Only to HLA-polyreactive mAbs?
HLA-I isoform reactivity of the monoclonal Abs generated from immunization with two alleles (HLA-ER107and HLA-EG107) of recombinant Face-2 resulted in several mAbs with a wide range of allelic reactivity against HLA-Ia and HLA-Ib isomers (Table 6). In contrast to polyreactive mAbs such as TFL-006 and TFL-007, highly monospecific mAbs against HLA-E were also observed. The recognition of HLA-E-specific amino acid sequences by mAbs is the defining criteria of the monospecificity. There is dosimetric inhibition of HLA-E binding of the monospecific mAb TFL-033 by HLA-E-restricted amino acid sequences (65RSARDT70 and 154AESADNSKQES144). Such monospecific anti-HLA-E mAb is highly valuable for immunodiagnosis of HLA-E [105,106] on human cancers which are known to upregulate HLA-E gene expression correlated with overexpression of cell-surface HLA-E [151,152,153,154,155,156,157,158,159,160]. HLA-E gene expression in some cancers (e.g., melanoma) is ranked 19th among the most overexpressed genes [153,154,155,156,157,158,159,160,161,162]. HLA-Ib overexpression, concomitant with loss of HLA-Ia molecules in cancer cells, is correlated with disease progression and poor survival of patients [163,164,165,166,167]. The disease progression is attributed to the suppression of the tumor-killing activity of CD8+ cytotoxic αβ T-cells (CTLs) and natural killer (NK)T-cells [168]. Binding of HLA-E to the inhibitory receptors CD94 and NKG2A on both CD8+ CTLs and NKT-cells hindered the cell-mediated anti-tumor activity. The interaction between HLA-E and the inhibitory receptors involves the binding of amino acids located on the α1 and α2 domains of HLA-E to specific amino acids on CD94 and NKG2A [168]. This interaction is attributed to the loss of the anti-tumor activity of CD8+ CTLs as well as that of NKT-cells [169].
The specific sequences located on the α1 and α2 domains of HLA-E are indeed the sequences recognized by the monospecific mAb TFL-033. The amino acids on the α1 and α2 domains of HLA-E—respectively, (R65D69) and (S152)—are essential for recognition by the inhibitory receptors CD94 and NKG2A present on the cell-surface of CD8+ T-cells and NK cells [105,106,107,108,109,134,168,169]. Therefore, the mAb TFL-033 has the potential to block the ligands of inhibitory receptors on the α1 and α2 helices of HLA-E [107,108,109]. When the mAb TFL-033 was tested in vitro on isolated CD3+/CD8+ T-cells both in the presence and absence of PHA, the mAb triggered proliferation of both non-activated and PHA-activated CD8+ T-cells [111]. Importantly, a significant increase in the number of CD8+ T lymphoblasts was observed even in the absence of an activating co-stimulant (PHA). This unique property of augmenting the proliferation of CD8+ T-cells, particularly when the classical HLA-Ia antigens are downregulated concomitant with the upregulation of HLA-E, suggests the unique immunoregulatory potential of the HLA-E monospecific mAb TFL-033.
These findings necessitate the need to further investigate the immunoregulatory potentials of other monospecific mAbs of HLA-F and HLA-G, after critical assessment of monospecificity by dosimetric peptide inhibtion studies. Such future investigations can expand the role of HLA further in immune homeostasis.
7. Conclusions
HLA class-I is long known to be a heterodimer with a polypeptide HC complexed with β2m on the surface of all nucleated cells. In the past three decades, it was discovered that there are three HLA-I variants (Face-2, Face-3 and Face-4) without β2m. Although the presence of these variants has been reported on activated immune cells, β2m-free HLA-I was observed in the monocytes of spondyloarthritis patients [170] and notably, the first trimester human endovascular trophoblast-cells [171] as well as on the extravillous cells of human placenta [172], which express both HLA-C and HLA-G. Siegel et al. [173] summarized reports on binding of Face-2 of HLA-C and HLA-F to activating and inhibiting receptors, such as KIR on NK cells. Increased frequency of HLA-C variants in rheumatoid arthritis (RA) is correlated with the strong association between RA and the KIR2DS2 activating receptors. They have also indicated that the homodimerized Face-2 (Face-3) can interact with the KIR receptors. HLA-C variants are also associated with increased susceptibility to systemic lupus erythematosus [174]. Activating KIR2DS4 was shown to specifically recognize Face-2 of HLA-F [20]. Infrequent reports of their presence of these variants in some, if not all, non-immune and malignant-cells have appeared [24,25,26,170]. It was unknown whether there are Abs directed against the three β2m-free HLA-I variants (Faces 2–3), until Morales-Bunerostro et al. [32] reported that the HLA-I Abs found in normal non-allo immunized males and females are against Face-2 variants of HLA-I. Indeed, many de novo donor-specific antibodies recognize β2m-free HLA but do not recognize intact HLA heterodimers [171].
Commonly used anti-HLA-I mAb W6/32, binds to both HLA-I HC and β2m. The assumption that the mAb W6/32 is specific for Face-1 was shattered when Tran et al. [103] reported that the mAb W6/32 recognizes an epitope preserved in free nonreduced αHCs of most HLA-B antigens but not in other HLA class-I α HCs. Essentially identical results were also obtained with another pan-HLA class-I mAb MEM-147 which cross-blocks W6/32. It is yet to be examined whether W6/32 recognizes an epitope on αHCs of HLA-Ib molecules.
The Face-2 HC-exposed epitopes that were previously masked by β2m are, interestingly, shared by all HLA-I isomers. Abs binding to these shared epitopes are considered as polyreactive mAbs. Immunizing the Face-2 of recombinant HLA-E elicited several Abs, some of them are truly polyreactive (TFL-006 and TFL-007). In contrast to mAb W6/32, they suppressed proliferation and blastogenesis of activated CD4+ T-cells and CD8+ T-cells but enhanced proliferation of T-regulatory cells (Figure 14). Face-2 also elicited Abs directed against specific domain of HLA-E not shared by any other HLA-Isomers. One such Ab is TFL-033, which has induced proliferation of both non-activated and activated CD8+ T-cells.
In summary, there exist several steps in HLA-I mediated immunohomeostasis.
- (1)
- Exposures of cryptic epitopes in β2-m free HC (Face-2) of HLA upon activation of cells. Such activation may occur during inflammation, injury, malignancy, and other pathological conditions, such as arthritis, autoimmune diseases and cancer.
- (2)
- Several of these exposed cryptic epitopes are shared by almost all HLA-I isomers.
- (3)
- Such exposure invites immune recognition by Abs and various other ligands.
- (4)
- It appears that such exposure may not last long, and it is prevented by homo- or heterodimerization of Face-2.
- (5)
- The Abs generated by the exposed cryptic epitopes are uniquely polyreactive with Face-2 of all HLA isomers since the epitopes are shared by all HLA isomers.
- (6)
- Binding of these Abs is capable of suppressing the proliferation and blastogenesis of CD4+ and CD8+ T-cells.
- (7)
- Binding of these Abs enhances the proliferation of T-regulatory cells, which are capable of several immunoregulatory functions.
- (8)
- The function of these Abs directed against cryptic epitopes on Face-2 is different from that of mAb W6/32 which is capable of binding to Face-1 of all loci (and Face-2 of B alleles) and promoting the proliferation of immune and non-immune cells.
This review highlights the importance of both polyreactive as well as the monospecific HLA-I mAbs as key elements for maintaining immune homeostasis. Confirming the epitope’s specificity of the mAbs by dosimetric peptide inhibition is a necessary prerequisite for understanding their precise roles. Examining the HLA alleles and their shared epitopes, as well as their corresponding Abs formed during specific pathological conditions, would enable developing therapeutic and diagnostic mAbs. These mAbs will have potential usefulness in specific immunodiagnosis and for developing active or passive immunotherapy for a variety of pathological conditions such as cancer, endstage organ diseases, and autoimmune diseases.
Author Contributions
Conceptualization: M.H.R., E.J.F., S.R.S., C.J.A.-M., F.E.H. and H.E.H.; Methodology: M.H.R.; Original Draft preparation: M.H.R. and F.E.H.; Critical review of the draft original and final versions: F.E.H., S.R.S., C.J.A.-M. and H.E.H.; Extensive Revision of the final version: E.J.F., F.E.H., S.R.S., H.E.H. and C.J.A.-M. All authors have read and agreed to the published version of the manuscript.
Funding
Funding is provided by a member of Terasaki Family Foundation, Professor Mark Terasaki from his private funds. There is no funding number available.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowlegments
This review is dedicated to the late Paul Terasaki for his invaluable guidance and for inculcating enthusiasm and passion to study the role of human Leukocyte antigens and their antibodies (specifically monoclonal antibodies) in immune homeostasis. The first author is highly indebted to Mark Terasaki for providing munificent funding, sustained support and encouragement, without which this project would not have come to light. Selvan contributed to this article in his personal capacity. The views expressed are his own and do not necessarily represent the views of the Food and Drug Administration or the United States Government.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marsh, S.G.E.; Parham, P.; Barber, L.D. The HLA, Facts Book; Facts Book Series; Academic Press: San Diego, CA, USA, 2000; ISBN 0-12-545025-7. [Google Scholar]
- Butler, E.A.; Flynn, F.V.; Harris, H.; Robson, E.B.A. A study of urine proteins by two-dimensional electrophoresis with special reference to the proteinuria of renal tubular disorders. Clin. Chim. Acta 1962, 7, 34–41. [Google Scholar] [CrossRef]
- Greeth, J.M.; Kekwick, R.A.; Flynn, F.V.; Harris, H.; Robson, E.B. An ultracentrifuge study of urine proteins with particular reference to the proteinuria of renal tubular disorders. Clin. Chim. Acta 1963, 8, 406414. [Google Scholar] [CrossRef]
- Berggård, I.; Bearn, A.G. Isolation and properties of a low molecular weight beta-2-globulin occurring in human biological fluids. J. Biol. Chem. 1968, 243, 4095–4103. [Google Scholar] [CrossRef]
- Cresswell, P.; Turner, M.J.; Strominger, J.L. 1973. Papain solubilized HLA antigens from cultured human lymphocytes contain two peptide fragments. Proc. Natl. Acad. Sci. USA 1973, 70, 1603–1607. [Google Scholar] [CrossRef]
- Tanigaki, N.; Nakamuro, K.; Appella, E.; Poulik, M.D.; Pressman, D. Identity of the HL-A common portion fragment and human beta2-microglobulin. Biochem. Biophys. Res. Commun. 1973, 55, 1234–1239. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 47, 45–148. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins—A tool for the selection of peptide antigens. Naturwissenschaft 1985, 72, 212–213. [Google Scholar] [CrossRef]
- Parker, J.M.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Ravindranath, N.M.; Selvan, S.R.; Filippone, E.J.; Amato-Menker, C.J.; El Hilali, F. Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines. Vaccines 2022, 10, 339. [Google Scholar] [CrossRef]
- Schnabl, E.; Stockinger, H.; Majdic, O.; Gaugitsch, H.; Lindley, I.J.; Maurer, D.; Hajek-Rosenmayr, A.; Knapp, W. Activated human T lymphocytes express MHC class I heavy chains not associated with beta 2-microglobulin. J. Exp. Med. 1990, 171, 1431–1442. [Google Scholar] [CrossRef]
- Lancet, D.; Parham, P.; Strominger, J.L. Heavy chain ofHLA-A and HLA-B antigens is conformationally labile: A possible role for beta2-microglobulin. Proc. Natl. Acad. Sci. USA 1979, 76, 3844–3848. [Google Scholar] [CrossRef]
- Raine, T.; Brown, D.; Bowness, P.; Hill Gaston, J.S.; Moffett, A.; Trowsdale, J.; Allen, R.L. Consistent patterns of expression of HLA class I free heavy chains in healthy individuals and raised expression in spondyloarthropathy patients point to physiological and pathological roles. Rheumatology 2006, 45, 1338–1344. [Google Scholar] [CrossRef]
- Demaria, S.; Schwab, R.; Bushkin, Y. The origin and fate of beta 2m-free MHC class I molecules induced on activated T-cells. Cell Immunol. 1992, 142, 103–113. [Google Scholar] [CrossRef]
- Aiuti, A.; Forte, P.; Simeoni, L.; Lino, M.; Pozzi, L.; Fattorossi, A.; Giacomini, P.; Ginelli, E.; Beretta, A.; Siccardi, A. Membrane expression of HLA-Cw4 free chains in activated T-cells of transgenic mice. Immunogenetics 1995, 42, 368–375. [Google Scholar] [CrossRef]
- Arosa, F.A.; Santos, S.G.; Powis, S.J. Open conformers: The hidden Face of MHC-I molecules. Trends Immunol. 2007, 28, 115–123. [Google Scholar] [CrossRef]
- Dangoria, N.S.; De Lay, M.L.; Kingsbury, D.J.; Mear, M.P.; Uchanska-Ziegler, B.; Ziegler, A.; Colbert, R.A. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J. Biol. Chem. 2002, 277, 23459–23468. [Google Scholar] [CrossRef]
- Allen, H.J.; Fraser, J.; Flyer, D.; Calvin, S.; Flavell, R. Beta 2-microglobulin is not required for cell-surface expression of the routine class I histocompatibility antigen H-2Db. Proc. Natl. Acad. Sci. USA 1986, 83, 7447–7451. [Google Scholar] [CrossRef]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty D., E. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J. Immunol. 2010, 184, 6199–6208. [Google Scholar] [CrossRef]
- Algarra, I.; Garcia-Lora, A.; Cabrera, T.; Ruiz-Cabello, F.; Garrido, F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: Implications for tumor immune escape. Cancer Immunol. Immunother. 2004, 53, 904–910. [Google Scholar] [CrossRef]
- Stam, N.J.; Spits, H.; Ploegh, H.L. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 1986, 137, 2299–2306. [Google Scholar] [PubMed]
- Grassi, F.; Meneveri, R.; Gullberg, M.; Lopalco, L.; Rossi, G.B.; Lanza, P.; De Santis, C.; Bratsand, G.; Butto, S.; Ginelli, E.; et al. Human immunodeficiency virus type 1 gpl20 mimics a hidden monomorphic epitope borne by class I major histocompatibility complex heavy chains. J. Exp. Med. 1990, 174, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Marozzi, A.; Meneveri, R.; Bunone, G.; De Santis, C.; Lopalco, L.; Beretta, A.; Siccardi, A.G.; Della Valle, G.; Ginelli, E. Expression of beta 2m-free HLA class I heavy chains in neuroblastoma cell lines. Scand. J. Immunol. 1993, 37, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Martayan, A.; Fiscella, M.; Setini, A.; Ciccarelli, G.; Gambari, R.; Feriotto, G.; Beretta, A.; Siccardi, A.G.; Appella, E.; Giacomini, P. Conformation and surface expression of free HLA-CW1 heavy chains in the absence of beta 2-microglobulin. Hum. Immunol. 1997, 53, 23–33. [Google Scholar] [CrossRef]
- Giacomini, P.; Beretta, A.; Nicotra, M.R.; Ciccarelli, G.; Martayan, A.; Cerboni, C.; Lopalco, L.; Bini, D.; Delfino, L.; Ferrara, G.B.; et al. HLA-C heavy chains free of β2-microglobulin: Distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-γ responsiveness. Tissue Antigens 1997, 50, 555–566. [Google Scholar] [CrossRef]
- Avrameas, S.; Guilbert, B.; Dighiero, B. Natural antibodies against tubulin. actin, myoglobin. Ihyroglobtilin. fetuin, albumin and transferin arc present in normal human sera, and monoclonal immunoglobulins from multiple myclomc and Waldenstrom macroglobulinemia may express similar antibody specificities. Ann. Immunol. 1981, 132, 103–113. [Google Scholar] [CrossRef]
- Collins, Z.V.; Arnold, P.F.; Peetoom, F.; Smith, G.S.; Walford, R.L. A naturally occurring monospecific anti-HLA8 isoantibody. Tissue Antigens 1973, 3, 358–363. [Google Scholar] [CrossRef]
- Tongio, M.M.; Falkenrodt, A.; Mitsuishi, Y.; Urlacher, A.; Bergerat, J.P.; North, M.L.; Mayer, S. Natural HLA antibodies. Tissue Antigens 1985, 26, 271–285. [Google Scholar] [CrossRef]
- Zhou, B.; Saito, S.; Nakazawa, Y.; Kobayashi, N.; Matsuda, M.; Matsumoto, Y.; Hosoyama, T.; Koike, K. Existence of an immunoglobulin G component of naturally occurring HLA class I antibodies that are not directed against self-antigens in human serum. Tissue Antigens 2008, 72, 98–104. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Terasaki, P.I.; Maehara, C.Y.; Jucaud, V.; Kawakita, S.; Pham, T.; Yamashita, W. Immunoglobulin (Ig)G purified from human sera mirrors intravenous Ig human leucocyte antigen (HLA) reactivity and recognizes one’s own HLA types, but may be masked by Fab complementarity-determining region peptide in the native sera. Clin. Exp. Immunol. 2015, 179, 309–328. [Google Scholar] [CrossRef]
- Morales-Buenrostro, L.E.; Terasaki, P.I.; Marino-Vazquez, L.A.; Lee, J.H.; El-Awar, N.; Alberú, J. “Natural” human leukocyte antigen antibodies found in nonalloimmunized healthy males. Transplantation 2008, 86, 1111–1115. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Jucaud, V.; Ferrone, S. Monitoring native HLA-I trimer specific antibodies in Luminex multiplex single antigen bead assay: Evaluation of beadsets from different manufacturers. J. Immunol. Methods 2017, 450, 73–80. [Google Scholar] [CrossRef]
- Jucaud, V.; Ravindranath, M.H.; Terasaki, P.I. Conformational Variants of the Individual HLA-I Antigens on Luminex Single Antigen Beads Used in Monitoring HLA Antibodies: Problems and Solutions. Transplantation 2017, 101, 764–777. [Google Scholar] [CrossRef]
- Sullivan, K.M.; Kopecky, K.J.; Jocom, J.; Fisher, L.; Buckner, C.D.; Meyers, J.D.; Counts, G.W.; Bowden, R.A.; Peterson, F.B.; Witherspoon, R.P.; et al. Immuno-modulatory and antimicrobial efficacy of intravenous immunoglobulin in bone marrow transplantation. N. Engl. J. Med. 1990, 323, 705–712. [Google Scholar] [CrossRef]
- Sullivan, K.M.; Storek, J.; Kopecky, K.J.; Jocom, J.; Longton, G.; Flowers, M.; Siadak, M.; Nims, J.; Witherspoon, R.P.; Anasetti, C.; et al. A controlled trial of long-term administration of intravenous immunoglobulin to prevent late infection and chronic graft-vs.-host disease after marrow transplantation: Clinical outcome and effect on subsequent immune recovery. Biol. Blood Marrow Transpl. 1996, 2, 44–53. [Google Scholar]
- Bayry, J.; Lacroix-Desmazes, S.; Kazatchkine, M.D.; Kaveri, S.V. Intravenous immunoglobulin for infectious diseases: Back to the pre-antibiotic and passive prophylaxis era? Trends Pharmacol. Sci. 2004, 25, 306–310. [Google Scholar] [CrossRef]
- Molica, S.; Musto, P.; Chiurazzi, F.; Specchia, G.; Brugiatelli, M.; Cicoira, L.; Levato, D.; Nobile, F.; Carotenuto, M.; Liso, V.; et al. Prophylaxis against infections with low-dose intravenous immunoglobulins (IVIg) in chronic lymphocytic leukemia. Results of a crossover study. Haematologica 1996, 81, 121–126. [Google Scholar]
- Elluru, S.R.; Kaveri, S.V.; Bayry, J. The protective role of immunoglobulins in fungal infections and inflammation. Semin. Immunopathol. 2015, 37, 187–197. [Google Scholar] [CrossRef]
- Dalakas, M.C. Intravenous immunoglobulin in autoimmune neuromuscular diseases. JAMA 2004, 291, 2367–2375. [Google Scholar] [CrossRef]
- Sibéril, S.; Elluru, S.; Graff-Dubois, S.; Negi, V.S.; Delignat, S.; Mouthon, L.; Lacroix-Desmazes, S.; Kazatchkine, M.D.; Bayry, J.; Kaveri, S.V. Intravenous immunoglobulins in autoimmune and inflammatory diseases: A mechanistic perspective. Ann. N. Y. Acad. Sci. 2007, 1110, 497–506. [Google Scholar] [CrossRef]
- Galeotti, C.; Kaveri, S.V.; Bayry, J. IVIG-mediated effector functions in autoimmune and inflammatory diseases. Int. Immunol. 2017, 29, 491–498. [Google Scholar] [CrossRef]
- Hoffmann, J.H.O.; Enk, A.H. High-Dose Intravenous Immunoglobulin in Skin Autoimmune Disease. Front. Immunol. 2019, 10, 1090. [Google Scholar] [CrossRef]
- Jordan, S.C.; Toyoda, M.; Vo, A.A. Regulation of immunity and inflammation by intravenous immunoglobulin: Relevance to solid organ transplantation. Expert Rev. Clin. Immunol. 2011, 7, 341–348. [Google Scholar] [CrossRef]
- Abu Jawdeh, B.G.; Cuffy, M.C.; Alloway, R.R.; Shields, A.R.; Woodle, E.S. Desensitization in kidney transplantation: Review and future perspectives. Clin. Transpl. 2014, 28, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Picascia, A.; Grimaldi, V.; Paolillo, R.; Vasco, M.; Casamassimi, A.; De Luca, F.P.; Cavalca, F.; Schiano, C.; Napoli, C. Intravenous human immunoglobulin treatment of serum from HLA-sensitized patients in kidney transplantation. Ren. Fail. 2014, 36, 585–588. [Google Scholar] [CrossRef]
- Kornberg, A. Intravenous immunoglobulins in liver transplant patients: Perspectives of clinical immune modulation. World J. Hepatol. 2015, 7, 1494–1508. [Google Scholar] [CrossRef]
- Tedla, F.M.; Roche-Recinos, A.; Brar, A. Intravenous immunoglobulin in kidney transplantation. Curr. Opin. Organ. Transpl. 2015, 20, 630–637. [Google Scholar] [CrossRef]
- Matignon, M.; Leibler, C.; Moranne, O.; Salomon, L.; Charron, D.; Lang, P.; Jacquelinet, C.; Suberbielle, C.; Grimbert, P. Anti-HLA sensitization after kidney allograft nephrectomy: Changes one year post-surgery and beneficial effect of intravenous immunoglobulin. Clin. Transpl. 2016, 30, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Matignon, M.; Pilon, C.; Commereuc, M.; Grondin, C.; Leibler, C.; Kofman, T.; Audard, V.; Cohen, J.; Canoui-Poitrine, F.; Grimbert, P. Intravenous immunoglobulin therapy in kidney transplant recipients with de novo DSA: Results of an observational study. PLoS ONE. 2017, 12, e0178572. [Google Scholar] [CrossRef]
- Glotz, D.; Rostaing, L.; Merville, P.; Squifflet, J.P.; Lebranchu, Y. Intravenous Immunoglobulins in the Prevention of Rejection of a Second or Third Kidney Graft. Transpl. Proc. 2018, 50, 70–71. [Google Scholar] [CrossRef]
- Bujnowska, A.; Michon, M.; Konopelski, P.; Hryniewiecka, E.; Jalbrzykowska, A.; Perkowska-Ptasinska, A.; Cieciura, T.; Zagozdzon, R.; Paczek, L.; Ciszek, M. Outcomes of Prolonged Treatment with Intravenous Immunoglobulin Infusions for Acute Antibody-mediated Rejection in Kidney Transplant Recipients. Transpl. Proc. 2018, 50, 1720–1725. [Google Scholar] [CrossRef]
- Hoang, J.; Krisl, J.; Moaddab, M.; Nguyen, D.T.; Graviss, E.A.; Hussain, I.; Kassi, M.; Yousefzai, R.; Kim, J.; Trachtenberg, B.; et al. Intravenous immunoglobulin in heart transplant recipients with mild to moderate hypogammaglobulinemia and infection. Clin. Transpl. 2022, 36, e14571. [Google Scholar] [CrossRef]
- Ulisses, L.R.S.; Paixão, J.O.; Agena, F.; Souza, P.S.; Paula, F.J.; Bezerra, G.; Rodrigues, H.; Panajotopolous, N.; David-Neto, E.; Castro, M.C.R. Desensitization using IVIG alone for living-donor kidney transplant: Impact on donor-specific antibodies. J. Bras. Nefrol. 2022, 8. [Google Scholar] [CrossRef]
- Vani, J.; Elluru, S.; Negi, V.S.; Lacroix-Desmazes, S.; Kazatchkine, M.D.; Bayry, J.; Kaveri, S.V. Role of natural antibodies in immune homeostasis: IVIg perspective. Autoimmun. Rev. 2008, 7, 440–444. [Google Scholar] [CrossRef]
- Sbrana, S.; Ruocco, L.; Vanacore, R.; Azzarà, A.; Ambrogi, F. In vitro effects of an immunoglobulin preparation for intravenous use (IVIG) on T-cells activation. Allergy Immunol. 1993, 25, 35–37. [Google Scholar]
- Hurez, V.; Kaveri, S.V.; Mouhoub, A.; Dietrich, G.; Mani, J.C.; Klatzmann, D.; Kazatchkine, M.D. Anti-CD4 activity of normal human immunoglobulin G for therapeutic use (intravenous immunoglobulin, IVIg). Ther. Immunol. 1994, 1, 269–277. [Google Scholar]
- Amran, D.; Renz, H.; Lack, G.; Bradley, K.; Gelfand, E.W. Suppression of cytokine-dependent human T-cell proliferation by intravenous immunoglobulin. Clin. Immunol. Immunopathol. 1994, 73, 180–186. [Google Scholar] [CrossRef]
- Andersson, J.; Skansén-Saphir, U.; Sparrelid, E.; Andersson, U. Intravenous immune globulin affects cytokine production in T lymphocytes and monocytes/macrophages. Clin. Exp. Immunol. 1996, 104 (Suppl. S1), 10–20. [Google Scholar] [CrossRef]
- Aktas, O.; Waiczies, S.; Grieger, U.; Wendling, U.; Zschenderlein, R.; Zipp, F. Polyspecific immunoglobulins (IVIg) suppress proliferation of human (auto) antigen-specific T-cells without inducing apoptosis. J. Neuroimmunol. 2001, 114, 160–167. [Google Scholar] [CrossRef]
- MacMillan, H.F.; Lee, T.; Issekutz, A.C. Intravenous immunoglobulin G-mediated inhibition of T-cell proliferation reflects an endogenous mechanism by which IgG modulates T-cell activation. Clin. Immunol. 2009, 132, 222–233. [Google Scholar] [CrossRef]
- Tha-In, T.; Metselaar, H.J.; Tilanus, H.W.; Boor, P.P.; Mancham, S.; Kuipers, E.J.; de Man, R.A.; Kwekkeboom, J. Superior immunomodulatory effects of intravenous immunoglobulins on human T-cells and dendritic cells: Comparison to calcineurin inhibitors. Transplantation 2006, 81, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Van Schaik, I.N.; Lundkvist, I.; Vermeulen, M.; Brand, A. Polyvalent immunoglobulin for intravenous use interferes with cell proliferation in vitro. J. Clin. Immunol. 1992, 12, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Prasad, N.K.; Papoff, G.; Zeuner, A.; Bonnin, E.; Kazatchkine, M.D.; Ruberti, G.; Kaveri, S.V. Therapeutic preparations of normal polyspecific IgG (IVIg) induce apoptosis in human lymphocytes and monocytes: A novel mechanism of action of IVIg involving the Fas apoptotic pathway. J. Immunol. 1998, 161, 3781–3790. [Google Scholar]
- Aubin, E.; Lemieux, R.; Bazin, R. Indirect inhibition of in vivo and in vitro T-cell responses by intravenous immunoglobulins due to impaired antigen presentation. Blood 2010, 115, 1727–1734. [Google Scholar] [CrossRef] [PubMed]
- Aubin, É.; Proulx, D.P.; Trépanier, P.; Lemieux, R.; Bazin, R. Prevention of T cell dependent native antigen presentation. Clin. Immunol. 2011, 141, 273–283. [Google Scholar] [CrossRef]
- Kaveri, S.; Vassilev, T.; Hurez, V.; Lengagne, R.; Lefranc, C.; Cot, S.; Pouletty, P.; Glotz, D.; Kazatchkine, M.D. Antibodies to a conserved region of HLA class I molecules, capable of modulating CD8 T cell-mediated function, are present in pooled normal immunoglobulin for therapeutic use. J. Clin. Investig. 1996, 97, 865–869. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Terasaki, P.I.; Pham, T.; Jucaud, V.; Kawakita, S. Therapeutic preparations of IVIg contain naturally occurring anti-HLA-E Abs that react with HLA-Ia (HLA-A/-B/-Cw) alleles. Blood 2013, 121, 2013–2028. [Google Scholar] [CrossRef]
- Cavallini, G.; Pozzan, T.; Selvatici, R.; Baricordi, R.; Mazzilli, C.; Gandini, E. Monoclonal antibodies against HLA class I antigens inhibit human lymphocyte proliferation but do not affect mitogen dependent second messenger generation. Biochem. Biophys. Res. Commun. 1988, 154, 712–718. [Google Scholar] [CrossRef]
- De Felice, M.; Turco, M.C.; Costanzo, F.; Corbo, L.; Ferrone, S.; Venuta, S. Inhibition by anti-HLA class I mAb of IL-2 and IL-2 receptor synthesis in lymphocytes stimulated with PHA-P. Cell Immunol. 1990, 126, 420–427. [Google Scholar] [CrossRef]
- Tanabe, M.; Sekimata, M.; Ferrone, S.; Takiguchi, M. Structural and functional analysis of monomorphic determinants recognized by monoclonal antibodies reacting with the HLA class I alpha 3 domain. J. Immunol. 1992, 148, 3202–3209. [Google Scholar]
- Smith, D.M.; Bluestone, J.A.; Jeyarajah, D.R.; Newberg, M.H.; Engelhard, V.H.; Thistlethwaite, J.R., Jr.; Woodle, E.S. Inhibition of T cell activation by a monoclonal antibody reactive against the alpha 3 domain of human MHC class I molecules. J. Immunol. 1994, 153, 1054–1067. [Google Scholar]
- Woodle, E.S.; Smith, D.M.; Zhou, N. Class I MHC mediates programmed cell death in human lymphoid cells. Transplantation 1997, 64, 140–146. [Google Scholar] [CrossRef]
- Woodle, E.S.; Smith, D.M.; Bluestone, J.A.; Kirkman, W.M., 3rd; Green, D.R.; Skowronski, E.W. Anti-human class I MHC antibodies induce apoptosis by a pathway that is distinct from the Fas antigen-mediated pathway. J. Immunol. 1997, 158, 2156–2164. [Google Scholar]
- Coupel, S.; Moreau, A.; Hamidou, M.; Horejsi, V.; Soulillou, J.-P.; Béatrice Charreau, B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 2007, 109, 2806–2814. [Google Scholar] [CrossRef]
- Harris, P.E.; Bian, H.; Reed, E.F. Induction of high-affinity fibroblast growth factor receptor expression and proliferation in human endothelial cells by anti-HLA antibodies: A possible mechanism for transplant atherosclerosis. J. Immunol. 1997, 159, 5697–5704. [Google Scholar]
- Bian, H.; Harris, P.E.; Mulder, A.; Reed, E.F. Anti-HLA antibody ligation to HLA class I molecules expressed by endothelial cells stimulates tyrosine phosphorylation, inositol phosphate generation, and proliferation. Hum. Immunol. 1997, 53, 90–97. [Google Scholar] [CrossRef]
- Bian, H.; Harris, P.E.; Reed, E.F. Ligation of HLA class I molecules on smooth muscle cells with anti-HLA antibodies induces tyrosine phosphorylation, fibroblast growth factor receptor expression and cell proliferation. Int. Immunol. 1998, 10, 1315–1323. [Google Scholar] [CrossRef]
- Bian, H.; Reed, E.F. Alloantibody-mediated class I signal transduction in endothelial cells and smooth muscle cells: Enhancement by IFN-γ and TNF-α. J. Immunol. 1999, 163, 1010–1018. [Google Scholar]
- Bian, H.; Reed, E.F. Anti-HLA antibodies transduce proliferative signals in endothelial cells and smooth muscle cells. Transpl. Proc. 1999, 31, 1924. [Google Scholar] [CrossRef]
- Nath, N.; Bian, H.; Reed, E.F.; Chellappan, S.P. HLA class I-mediated induction of cell proliferation involves cyclin E-mediated inactivation of Rb function and induction of E2F activity. J. Immunol. 1999, 162, 5351. [Google Scholar]
- Jin, Y.P.; Singh, R.P.; Du, Z.Y.; Rajasekaran, A.K.; Rozengurt, E.; Reed, E.F. Ligation of HLA class I molecules on endothelial cells induces phosphorylation of Src, paxillin, and focal adhesion kinase in an actin-dependent manner. J. Immunol. 2002, 168, 5415–5423. [Google Scholar] [CrossRef]
- Jin, Y.P.; Jindra, P.T.; Gong, K.W.; Lepin, E.J.; Reed, E.F. Anti-HLA class I antibodies activate endothelial cells and promote chronic rejection. Transplantation 2005, 15, S19–S21. [Google Scholar] [CrossRef]
- Jin, Y.P.; Fishbein, M.C.; Said, J.W.; Jindra, P.T.; Rajalingam, R.; Rozengurt, E.; Reed, E.F. Anti-HLA class I antibody-mediated activation of the PI3K/Akt signaling pathway and induction of Bcl-2 and Bcl-xL expression in endothelial cells. Hum. Immunol. 2004, 65, 291–302. [Google Scholar] [CrossRef]
- Jindra, P.T.; Zhang, X.; Mulder, A.; Claas, F.; Veale, J.; Jin, Y.P.; Reed, E.F. Anti-HLA antibodies can induce endothelial cell survival or proliferation depending on their concentration. Transplantation 2006, 82 (Suppl. S1), S33–S35. [Google Scholar] [CrossRef]
- Jindra, P.T.; Jin, Y.P.; Rozengurt, E.; Reed, E.F. HLA class I antibody-mediated endothelial cell proliferation via the mTOR pathway. J. Immunol. 2008, 180, 2357–2366. [Google Scholar] [CrossRef]
- Li, F.; Zhang, X.; Jin, Y.P.; Mulder, A.; Reed, E.F. Antibody ligation of human leukocyte antigen class I molecules stimulates migration and proliferation of smooth muscle cells in a focal adhesion kinase-dependent manner. Hum. Immunol. 2011, 72, 1150–1159. [Google Scholar] [CrossRef]
- Ziegler, M.E.; Souda, P.; Jin, Y.P.; Whitelegge, J.P.; Reed, E.F. Characterization of the endothelial cell cytoskeleton following HLA class I ligation. PLoS ONE 2012, 7, e29472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Valenzuela, N.M.; Reed, E.F. HLA class I antibody-mediated endothelial and smooth muscle cell activation. Curr. Opin. Organ Transpl. 2012, 17, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Lepin, E.J.; Bastin, J.M.; Allan, D.S.; Roncador, G.; Braud, V.M.; Mason, D.Y.; van der Merwe, P.A.; McMichael, A.J.; Bell, J.I.; Powis, S.H.; et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 2000, 30, 3552–3561. [Google Scholar] [CrossRef]
- Barnstable, C.J.; Bodmer, W.F.; Brown, G.; Galfre, G.; Milstein, C.; Williams, A.F.; Ziegler, A. Production of monoclonal antibodies to group A erythrocytes HLA and other human cell-surface antigens-new tools for genetic analysis. Cell 1978, 14, 9–20. [Google Scholar] [CrossRef]
- Parham, P.; Barnstable, C.J.; Bodmer, W.F. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A, B, C, antigens. J. Immunol. 1979, 123, 342–349. [Google Scholar]
- Brown, G.; Joshua, D.E.; Bastin, J.; Mason, D.Y.; Barnstable, C.J. Monoclonal antibodies to human cell-surface antigens. Haematol. Blood Transfus. 1979, 23, 365–368. [Google Scholar] [CrossRef]
- Smith, K.D.; Mace, B.E.; Valenzuela, A.; Vigna, J.L.; McCutcheon, J.A.; Barbosa, J.A.; Huczko, E.; Engelhard, V.H.; Lutz, C.T. Probing HLA-B7 conformational shifts induced by peptide-binding groove mutations and bound peptide with anti-HLA monoclonal antibodies. J. Immunol. 1996, 157, 2470–2478. [Google Scholar]
- Vigna, J.L.; Smith, K.D.; Lutz, C.T. Invariant chain association with MHC class I: Preference for HLA class I/beta 2-microglobulin heterodimers, specificity, and influence of the MHC peptide-binding groove. J. Immunol. 1996, 157, 4503–4510. [Google Scholar]
- Ivanyi, D.; van de Meugheuvel, W. A monomorphic HLA-specific monoclonal antibody, W6/32, reacts with the H-2Db molecule of normal mouse lymphocytes. Immunogenetics 1984, 20, 699–703. [Google Scholar] [CrossRef]
- Maziarz, R.T.; Fraser, J.; Strominger, J.L.; Burakoff, S.J. The human HLA-specific monoclonal antibody W6/32 recognizes a discontinuous epitope within the alpha 2 domain of murine H-2Db. Immunogenetics 1986, 24, 206–208. [Google Scholar] [CrossRef]
- Jefferies, W.A.; MacPherson, G.G. Expression of the W6/32 HLA epitope by cells of rat, mouse, human and other species: Critical dependence on the interaction of specific MHC heavy chains with human or bovine beta 2-microglobulin. Eur. J. Immunol. 1987, 17, 1257–1263. [Google Scholar] [CrossRef]
- Kahn-Perles, B.; Boyer, C.; Arnold, B.; Sanderson, A.R.; Ferrier, P.; Lemonnier, F.A. Acquisition of HLA class I W6/32 defined antigenic determinant by heavy chains from different species following association with bovine beta 2-microglobulin. J. Immunol. 1987, 138, 2190–2196. [Google Scholar]
- Kievits, F.; Ivanyi, P. Monomorphic anti-HLA monoclonal antibody (W6/32) recognizes polymorphic H-2 heavy-chain determinants exposed by association with bovine or human but not murine beta 2-microglobulin. Hum. Immunol. 1987, 20, 115–126. [Google Scholar] [CrossRef]
- Shields, M.J.; Ribaudo, R.K. Mapping of the monoclonal antibody W6/32: Sensitivity to the amino terminus of beta2-microglobulin. Tissue Antigens 1998, 51, 567–570. [Google Scholar] [CrossRef]
- Ladasky, J.J.; Shum, B.P.; Canavez, F.; Seuánez, H.N.; Parham, P. Residue 3 of beta2-microglobulin affects binding of class I MHC molecules by the W6/32 antibody. Immunogenetics 1999, 49, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.M.; Ivanyi, P.; Hilgert, I.; Brdicka, T.; Pla, M.; Breur, B.; Flieger, M.; Ivasková, E.; Horejsí, V. The epitope recognized by pan-HLA class I-reactive monoclonal antibody W6/32 and its relationship to unusual stability of the HLA-B27/beta2-microglobulin complex. Immunogenetics 2001, 53, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Ravindranath, N.M.; El Hilali, F.; Selvan, S.R.; Filippone, E.J. Ramifications of the HLA-I Allelic Reactivity of Anti-HLA-E*01:01 and Anti-HLA-E*01:03 Heavy Chain Monoclonal Antibodies in Comparison with Anti-HLA-I IgG Reactivity in Non-Alloimmunized Males, Melanoma-Vaccine Recipients, and End-Stage Renal Disease Patients. Antibodies 2022, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Terasaki, P.I. Diagnostic and Therapeutic Potential of HLA-E Monospecific Monoclonal IgG Antibodies Directed against Tumor Cell-surface and Soluble HLA-E. U.S. Patent 10656156, 5 July 2012. [Google Scholar]
- Sasaki, T.; Ravindranath, M.H.; Terasaki, P.I.; Freitas, M.C.; Kawakita, S.; Jucaud, V. Gastric cancer progression may involve a shift in HLA-E profile from an intact heterodimer to β2-microglobulin-free monomer. Int. J. Cancer 2014, 134, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Terasaki, P.I.; Pham, T.; Jucaud, V. The Monospecificity of Novel Anti-HLA-E Monoclonal Antibodies Enables Reliable Immunodiagnosis, Immunomodulation of HLA-E, and Upregulation of CD8+ T Lymphocytes. Monoclon. Antibodies Immunodiagn. Immunother. 2015, 34, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Hopfield, J.; Ferrone, S. HLA-E restricted monoclonal antibodies: Therapeutic potential as a double-edged sword against tumor progression. Intern. Rev. Med. 2017, 3, 1–49. [Google Scholar]
- Ravindranath, M.H.; El Hilali, F. Monospecific and Polyreactive Monoclonal Antibodies against human Leukocyte Antigen-E: Diagnostic and Therapeutic relevance. In Monoclonal Antibodies; Rezaei, N., Ed.; Intech Open: London, UK, 2021; ISBN 978-1-83968-369-5. [Google Scholar]
- Ravindranath, M.H.; Terasaki, P.I. Anti-HLA Class-IB Antibodies Mimic Immunoreactivity and Immunomodulatory Functions of Intravenous Immunoglobulin (IVIG) Useful as Therapeutic IVIG Mimetics and Methods of Their Use. International Patent WO 2013/106586 A2, 19 July 2013. [Google Scholar]
- Zhu, D.; Ravindranath, M.H.; Terasaki, P.I.; Miyazaki, T.; Pham, T.; Jucaud, V. Suppression of allo-human leucocyteantigen (HLA) Abs secreted by B memory cells in vitro: Intravenous immunoglobulin (IVIg) versus a monoclonal anti-HLA-E IgG that mimics HLA-I reactivities of IVIg. Clin. Exp. Immunol. 2014, 177, 464–477. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Zhu, D.; Pham, T.; Jucaud, V.; Hopfield, J.; Kawakita, S.; Terasaki, P.I. Anti-HLA-E monoclonal antibodies reacting with HLA-la and lb alleles like IVIg as potential IVIg-immunomimetics: An evolving therapeutic concept. Clin. Transpl. 2013, 2013, 293–305. [Google Scholar]
- Ravindranath, M.H. HLA Class Ia and Ib Polyreactive Anti-HLA-E IgG2a Monoclonal Antibodies (TFL-006 and TFL-007) suppress Anti-HLA IgG Production by CD19+ B Cells and Proliferation of CD4+ T-cells While upregulating T-regs. J. Immunol. Res. 2017, 2017, 3475926. [Google Scholar] [CrossRef]
- Majdic, O.; Schnabl, E.; Stockinger, H.; Gadd, S.; Maurer, D.; Radaszkiewics, T. LA45, an activation-induced human lymphocyte antigen with strong homology to MHC class I molecules. In Leukocyte Typing IV; Oxford University Press: Oxford, UK, 1989; pp. 8–511. [Google Scholar]
- Schumacher, T.N.; Heemels, M.T.; Neefjes, J.J.; Kast, W.M.; Melief, C.J.; Ploegh, H.L. Direct binding of peptide to empty MHC class I molecules on intacT-cells and in vitro. Cell 1990, 62, 563–567. [Google Scholar] [CrossRef]
- Benjamin, R.J.; Madrigal, J.A.; Parham, P. Peptide binding to empty HLA-B27 molecules of viable human cells. Nature 1991, 351, 74–77. [Google Scholar] [CrossRef]
- Madrigal, J.A.; Belich, M.P.; Benjamin, R.J.; Little, A.M.; Hildebrand, W.H.; Mann, D.L.; Parham, P. Molecular definition of a polymorphic antigen (LA45) of free HLA-A and -B heavy chains found on the surfaces of activated B and T-cells. J. Exp. Med. 1991, 174, 1085–1095. [Google Scholar] [CrossRef]
- Demaria, S.; Bushkin, Y. CD8 and beta 2microglobulin-free MHC class I molecules in T cell immunoregulation. Int. J. Clin. Lab. Res. 1993, 23, 61–69. [Google Scholar] [CrossRef]
- Capps, G.G.; Robinson, B.E.; Lewis, K.D.; Zúñiga, M.C. In vivo dimeric association of class I MHC heavy chains. Possible relationship to class I MHC heavy chain-beta 2microglobulin dissociation. J. Immunol. 1993, 151, 159–169. [Google Scholar]
- Matko, J.; Bushkin, Y.; Wei, T.; Edidin, M. Clustering of class I HLA molecules on the surfaces of activated and transformed human cells. J. Immunol. 1994, 152, 3353–3360. [Google Scholar]
- Khare, S.D.; Hansen, J.; Luthra, H.S.; David, C.S. HLA-B27 heavy chains contribute to spontaneous inflammatory disease in B27/human beta2microglobulin (beta2m) double transgenic mice with disrupted mouse beta2m. J. Clin. Investig. 1996, 98, 2746–2755. [Google Scholar] [CrossRef]
- Narayanan, K.; Jendrisak, M.D.; Phelan, D.L.; Mohanakumar, T. HLA class I antibody mediated accommodation of endothelial cells via the activation of PI3K/cAMP dependent PKA pathway. Transpl. Immunol. 2006, 15, 187–197. [Google Scholar] [CrossRef]
- Reznik, S.I.; Jaramillo, A.; Zhang, L.; Patterson, G.A.; Cooper, J.D.; Mohanakumar, T. Anti-HLA antibody bindng to HLA class I molecules induces proliferation of airway epithelial cells: A potential mechanism for bronchiolitis obliterans syndrome. J. Thorac. Cardiovasc. Surg. 2000, 119, 39–45. [Google Scholar] [CrossRef]
- Jaramillo, A.; Smith, C.R.; Maruyama, T.; Zhang, L.; Patterson, G.A.; Mohanakumar, T. Anti-HLA class I antibody binding to airway epithelial cells induces production of fibrogenic growth factors and apoptotic cell death: A possible mechanism for bronchiolitis obliterans syndrome. Hum. Immunol. 2003, 64, 521–529. [Google Scholar] [CrossRef]
- Turco, M.C.; De Felice, M.; Corbo, L.; Giarrusso, P.C.; Yang, S.Y.; Ferrone, S.; Venuta, S. Enhancing effect of anti-HLA class I monoclonal antibodies on T cell proliferation induced via CD2 molecule. J. Immunol. 1988, 141, 2275–2281. [Google Scholar]
- Santoli, D.; Radka, S.F.; Kreider, B.L.; Ferrone, S. Regulatory role of monomorphic and polymorphic determinants of HLA class I antigens in the proliferation of T-cells stimulated by autologous and allogeneic B and T lymphoid cells. Cell Immunol. 1988, 116, 263–273. [Google Scholar] [CrossRef]
- Gilliland, L.K.; Norris, N.A.; Grosmaire, L.S.; Ferrone, S.; Gladstone, P.; Ledbetter, J.A. Signal transduction in lymphocyte activation through crosslinking of HLA class I molecules. Hum. Immunol. 1989, 25, 269–289. [Google Scholar] [CrossRef]
- Turco, M.C.; De Felice, M.; Alfinito, F.; Lamberti, A.; Costanzo, F.; Giordano, M.; Martinelli, V.; Rotoli, B.; Ferrone, S.; Venuta, S. Proliferative pathways in CD1- CD3+ CD4+ CD8+ T-prolymphocytic leukemic cells: Analysis with monoclonal antibodies and cytokines. Blood 1989, 74, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.S.; Nowell, P.C.; Kornbluth, J. Anti-HLA class I antibodies inhibit the T cell-independent proliferation of human B lymphocytes. J. Immunol. 1987, 139, 1792–1796. [Google Scholar] [PubMed]
- De Felice, M.; Turco, M.C.; Giarrusso, P.C.; Corbo, L.; Pizzano, R.; Martinelli, V.; Ferrone, S.; Venuta, S. Differential regulatory role of monomorphic and polymorphic determinants of histocompatibility leukocyte antigen class I antigens in monoclonal antibody OKT3-induced T cell proliferation. J. Immunol. 1987, 139, 2683–2689. [Google Scholar] [PubMed]
- De Felice, M.; Turco, M.C.; Corbo, L.; Carandente Giarrusso, P.; Lamberti, A.; Valerio, G.; Temponi, M.; Costanzo, F.; Ferrone, S.; Venuta, S. Lack of a role of monocytes in the inhibition by monoclonal antibodies to monomorphic and polymorphic determinants of HLA class I antigens of PHA-P-induced peripheral blood mononuclear cell proliferation. Cell Immunol. 1989, 122, 164–177. [Google Scholar] [CrossRef]
- Huet, S.; Boumsell, L.; Dausset, J.; Degos, L.; Bernard, A. The required interaction between monocytes and peripheral blood T lymphocytes (T-PBL) upon activation via CD2 or CD3. Role of HLA class I molecules from accessory cells and the differential response of T-PBL subsets. Eur. J. Immunol. 1988, 18, 1187–1194. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Terasaki, P.I.; Pham, T.; Jucaud, V.; Kawakita, S. Suppression of blastogenesis and proliferation of activated CD4(+) T-cells: Intravenous immunoglobulin (IVIg) versus novel anti-human leucocyte antigen (HLA)-E mAbs mimicking HLA-I reactivity of IVIg. Clin. Exp. Immunol. 2014, 178, 154–177. [Google Scholar] [CrossRef]
- Mustelin, T.; Vang, T.; Bottini, T.N. Protein tyrosine phosphatases and the immune response. Nat. Rev. Immunol. 2005, 5, 43–57. [Google Scholar] [CrossRef]
- Modai, D.; Berman, S.; Sheleg, Y.; Cohn, M.; Weissgarten, J.; Averbukh, Z. Interleukin-2 receptor is similarly expressed by activated lymphocytes from patients on chronic hemodialysis and healthy subjects. Clin. Immunol. Immunopathol. 1990, 55, 237–241. [Google Scholar] [CrossRef]
- Stentz, F.B.; Kitabchi, A.E. Transcriptome and proteome expression in activated human CD4 and CD8 T-lymphocytes. Biochem. Biophys. Res. Commun. 2004, 324, 692–696. [Google Scholar] [CrossRef]
- Le Gall, S.; Erdtmann, L.; Benichou, S.; Berlioz-Torrent, C.; Liu, L.; Benarous, R.; Heard, J.M.; Schwartz, O. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 1998, 8, 483–495. [Google Scholar] [CrossRef]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef]
- Santos, S.G.; Antoniou, A.N.; Sampaio, P.; Powis, S.J.; Arosa, F.A. Lack of tyrosine 320 impairs spontaneous endocytosis and enhances release of HLA-B27 molecules. J. Immunol. 2006, 176, 2942–2949. [Google Scholar] [CrossRef]
- Guild, B.; Strominger, J.L. Human and murine class I MHC antigens share conserved serine 335, the site of HLA phosphorylation in vivo. J. Biol. Chem. 1984, 259, 9235–9240. [Google Scholar] [CrossRef]
- Arosa, F.A.; de Jesus, O.; Porto, G.; Carmo, A.M.; de Sousa, M. Calreticulin is expressed on the cell-surface of activated human peripheral blood T lymphocytes in association with major histocompatibility complex class I molecules. J. Biol. Chem. 1999, 274, 16917–16922. [Google Scholar] [CrossRef]
- Allen, R.L.; Trowsdale, J. Recognition of classical and heavy chain forms of HLA-B27 by leukocyte receptors. Curr. Mol. Med. 2004, 4, 59–65. [Google Scholar] [CrossRef]
- Jones, D.C.; Kosmoliaptsis, V.; Apps, R.; Lapaque, N.; Smith, I.; Kono, A.; Chang, C.; Boyle, L.H.; Taylor, C.J.; Trowsdale, J.; et al. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J. Immunol. 2011, 186, 2990–2997. [Google Scholar] [CrossRef]
- Tamir, I.; Cambier, J.C. Antigen receptor signaling: Integration of protein tyrosine kinase functions. Oncogene 1988, 17, 135364. [Google Scholar] [CrossRef]
- EL Hilali, F.; Jucaud, V.; EL Hilali, H.; Bhuiyan, M.H.; Mancuso, A.; LiuSullivan, N.; Elidrissi, A.; Mazouz, H. Characterization of the Anti-HLA Class I and II IgG Antibodies in Moroccan IVIg Using Regular Beads and Ibeads in Luminex Multiplex Single Antigen Immunoassay. Int. J. Immunol. 2017, 5, 53–65. [Google Scholar] [CrossRef][Green Version]
- Hara, M.; Kingsley, C.I.; Niimi, M.; Read, S.; Turvey, S.E.; Bushell, A.R.; Morris, P.J.; Powrie, F.; Wood, K.J. IL-10 is required for regulatory T-cells to mediate tolerance to alloantigens in vivo. J. Immunol. 2001, 166, 3789–3796. [Google Scholar] [CrossRef]
- Levings, M.; Sangregorio, R.; Roncarolo, M.G. Human CD25+CD4+ T regulatory cells suppress naive and memory T-cell proliferation and can be expanded in vitro without loss of function. J. Exp. Med. 2001, 193, 1295–1301. [Google Scholar] [CrossRef]
- Van Maurik, A.; Wood, K.J.; Jones, N. Cutting edge: CD4+CD25+ alloantigen-specific immunoregulatory cells that can prevent CD8+ T-cell-mediated graft rejection: Implications for anti-CD154 immunotherapy. J. Immunol. 2002, 169, 5401–5404. [Google Scholar] [CrossRef]
- Jia, S.; Li, C.; Wang, G.; Yang, J.; Zu, Y. The T helper type 17/regulatory T cell imbalance in patients with acute Kawasaki disease. Clin. Exp. Immunol. 2010, 162, 131–137. [Google Scholar] [CrossRef]
- Kasztalska, K.; Ciebiada, M.; Cebula-Obrzut, B.; Górski, P. Intravenous immunoglobulin replacement therapy in the treatment of patients with common variable immunodeficiency disease: An open-label prospective study. Clin. Drug Investig. 2011, 31, 299–307. [Google Scholar] [CrossRef]
- Levy, E.M.; Bianchini, M.; Von Euw, E.M.; Barrio, M.M.; Bravo, A.I.; Furman, D.; Domenichini, E.; Macagno, C.; Pinsky, V.; Zucchini, C.; et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int. J. Oncol. 2008, 32, 633–641. [Google Scholar] [CrossRef]
- Gonçalves, M.A.; Le Discorde, M.; Simões, R.T.; Rabreau, M.; Soares, E.G.; Donadi, E.A.; Carosella, E.D. Classical and non-classical HLA molecules and p16 (INK4a) expression in precursors’ lesions and invasive cervical cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 141, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Hyodo, H.; Fujii, T.; Nagamatsu, T.; Matsumoto, J.; Kawana, K.; Yamashita, T.; Yasugi, T.; Kozuma, S.; Taketani, Y. Effect of progesterone on HLA-E gene expression in JEG-3 choriocarcinoma cell line. Am. J. Reprod. Immunol. 2009, 61, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Shen, Y.; Xia, M.; Xu, L.; Pan, N.; Yin, Y.; Miao, F.; Shen, C.; Xie, W.; Zhang, J. Expression of the nonclassical HLA class I and MICA/B molecules in human hepatocellular carcinoma. Neoplasma 2011, 58, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Kren, L.; Slaby, O.; Muckova, K.; Lzicarova, E.; Sova, M.; Vybihal, V.; Svoboda, T.; Fadrus, P.; Lakomy, R.; Vanhara, P.; et al. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: An unexpected prognostic significance? Neuropathology 2011, 31, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Kren, L.; Fabian, P.; Slaby, O.; Janikova, A.; Soucek, O.; Sterba, J.; Krenova, Z.; Michalek, J.; Kral, Z. Multifunctional immune-modulatory protein HLA-E identified in classical Hodgkin lymphoma: Possible implications. Pathol. Res. Pract. 2012, 208, 45–49. [Google Scholar] [CrossRef]
- Tremante, E.; Ginebri, A.; Lo Monaco, E.; Benassi, B.; Frascione, P.; Grammatico, P.; Cappellacci, S.; Catricalà, C.; Arcelli, D.; Natali, P.G.; et al. A melanoma immune response signature including Human Leukocyte Antigen-E. Pigment. Cell Melanoma Res. 2014, 27, 103–112. [Google Scholar] [CrossRef]
- De Kruijf, E.M.; Sajet, A.; van Nes, J.G.; Natanov, R.; Putter, H.; Smit, V.T.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.; Kuppen, P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef]
- Bossard, C.; Bezieau, S.; Matysiak-Budnik, T.; Volteau, C.; Laboisse, C.L.; Jotereau, F.; Mosnier, J.-F. HLA-E/beta2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int. J. Cancer 2012, 131, 855–863. [Google Scholar] [CrossRef]
- Dutta, N.; Majumder, D.; Gupta, A.; Mazumder, D.N.; Banerjee, S. Analysis of human lymphocyte antigen class I expression in gastric cancer by reverse transcriptase-polymerase chain reaction. Hum. Immunol. 2005, 66, 164–169. [Google Scholar] [CrossRef]
- Van Esch, E.M.; Tummers, B.; Baartmans, V.; Osse, E.M.; Ter Haar, N.; Trietsch, M.D.; Hellebrekers, B.W.; Holleboom, C.A.; Nagel, H.T.; Tan, L.T.; et al. Alterations in classical and nonclassical HLA expression in recurrent and progressive HPV-induced usual vulvar intraepithelial neoplasia and implications for immunotherapy. Int. J. Cancer 2014, 135, 830–842. [Google Scholar] [CrossRef]
- Bianchini, M.; Levy, E.; Zucchini, C.; Pinski, V.; Macagno, C.; De Sanctis, P.; Valvassori, L.; Carinci, P.; Mordoh, J. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int. J. Oncol. 2006, 29, 83–94. [Google Scholar] [CrossRef]
- Dutta, N.; Gupta, A.; Mazumder, D.N.; Banerjee, S. Down-regulation of locus-specific human lymphocyte antigen class I expression in Epstein-Barr virus-associated gastric cancer: Implication for viral-induced immune evasion. Cancer 2006, 106, 1685–1693. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, A.; Zhang, J.G.; Bao, W.G.; Xu, D.P.; Ruan, Y.Y.; Yan, W.H. Alteration of HLA-F and HLA I antigen expression in the tumor is associated with survival in patients with esophageal squamous cell carcinoma. Int. J. Cancer 2013, 132, 82–89. [Google Scholar] [CrossRef]
- Ashrafi, G.H.; Haghshenas, M.R.; Marchetti, B.; O’Brien, P.M.; Campo, M.S. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int. J. Cancer 2005, 113, 276–283. [Google Scholar] [CrossRef]
- Näsman, A.; Andersson, E.; Nordfors, C.; Grün, N.; Johansson, H.; Munck-Wikland, E.; Massucci, G.; Dalianis, T.; Ramqvist, T. MHC class I expression in HPV positive and negative tonsillar squamous cell carcinoma in correlation to clinical outcome. Int. J. Cancer 2013, 132, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Campoli, M.; Ferrone, S. HLA antigen changes in malignanT-cells: Epigenetic mechanisms and biologic significance. Oncogene 2008, 27, 5869–5885. [Google Scholar] [CrossRef] [PubMed]
- Petrie, E.J.; Clements, C.S.; Lin, J.; Sullivan, L.C.; Johnson, D.; Huyton, T.; Heroux, A.; Hoare, H.L.; Beddoe, T.; Reid, H.H.; et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J. Exp. Med. 2008, 205, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Schell, T.D.; Mylin, L.M.; Tevethia, S.S.; Joyce, S. The assembly of functional beta(2)-microglobulin-free MHC class I molecules that interact with peptides and CD8(+) T lymphocytes. Int. Immunol. 2002, 14, 775–782. [Google Scholar] [CrossRef]
- Ding, J.; Feng, Y.; Zheng, Z.H.; Li, X.Y.; Wu, Z.B.; Zhu, P. Increased expression of human leucocyte antigen class I free heavy chains on monocytes of patients with spondyloarthritis and cells transfected with HLA-B27. Int. J. Immunogenet. 2015, 42, 4–10. [Google Scholar] [CrossRef]
- Pröll, J.; Blaschitz, A.; Hutter, H.; Dohr, G. First trimester human endovascular trophoblasT-cells express both HLA-C and HLA-G. Am. J. Reprod. Immunol. 1999, 42, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Blaschitz, A.; Hutter, H.; Dohr, G. HLA Class I protein expression in the human placenta. Early Pregnancy 2001, 5, 67–69. [Google Scholar] [PubMed]
- Siegel, R.J.; Bridges, L., Jr.; Ahmed, S. HLA-C: An Accomplice in Rheumatic Diseases. ACR Open Rheumatol. 2019, 1, 571–579. [Google Scholar] [CrossRef]
- Gambino, C.M.; Di Bona, D.; Aiello, A.; Carru, C.; Duro, G.; Guggino, G.; Ferrante, A.; Zinellu, A.; Caruso, C.; Candore, G.; et al. HLA-C1 ligands are associated with increased susceptibility to systemic lupus erythematosus. Hum. Immunol. 2018, 79, 172–177. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).