Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
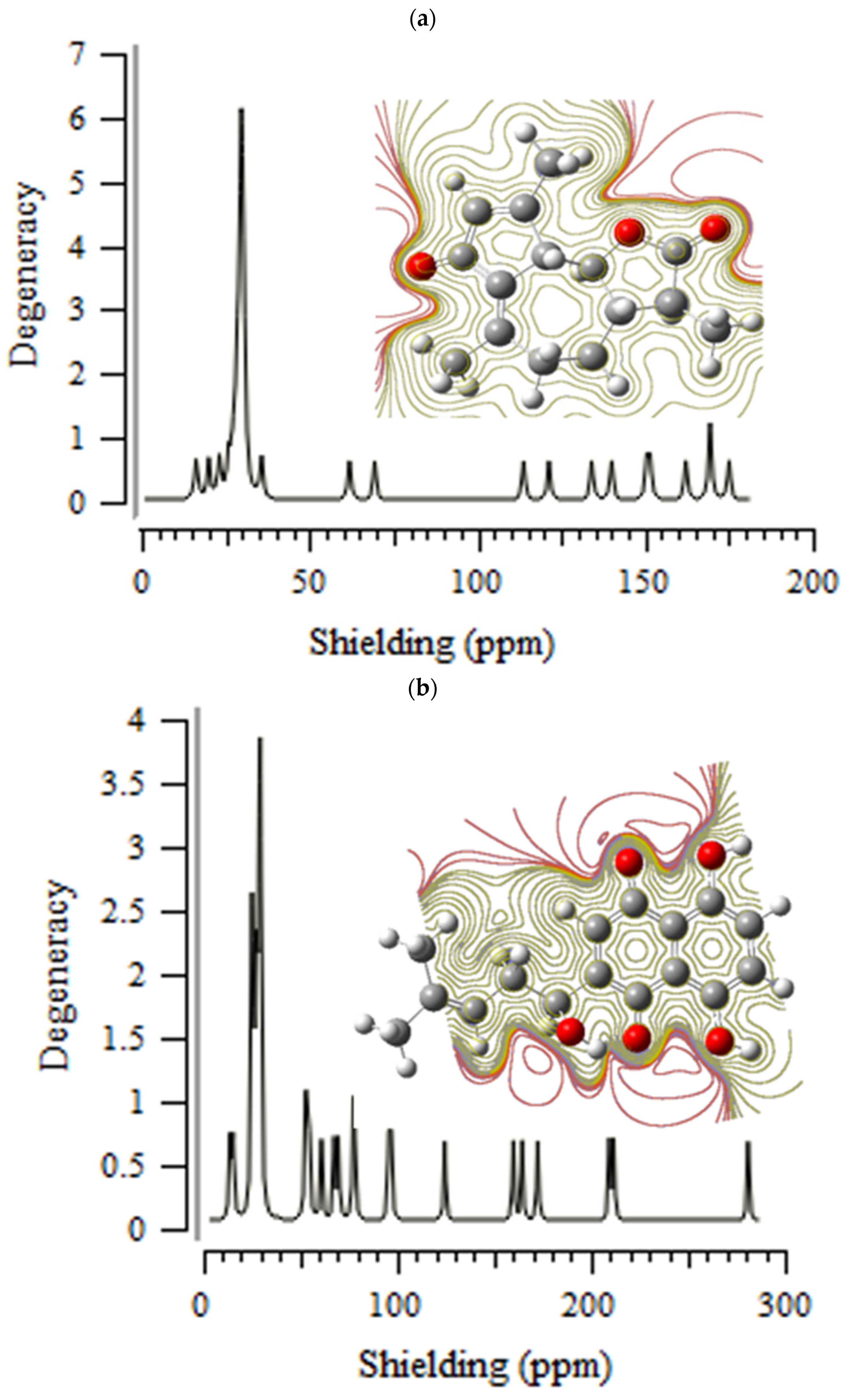
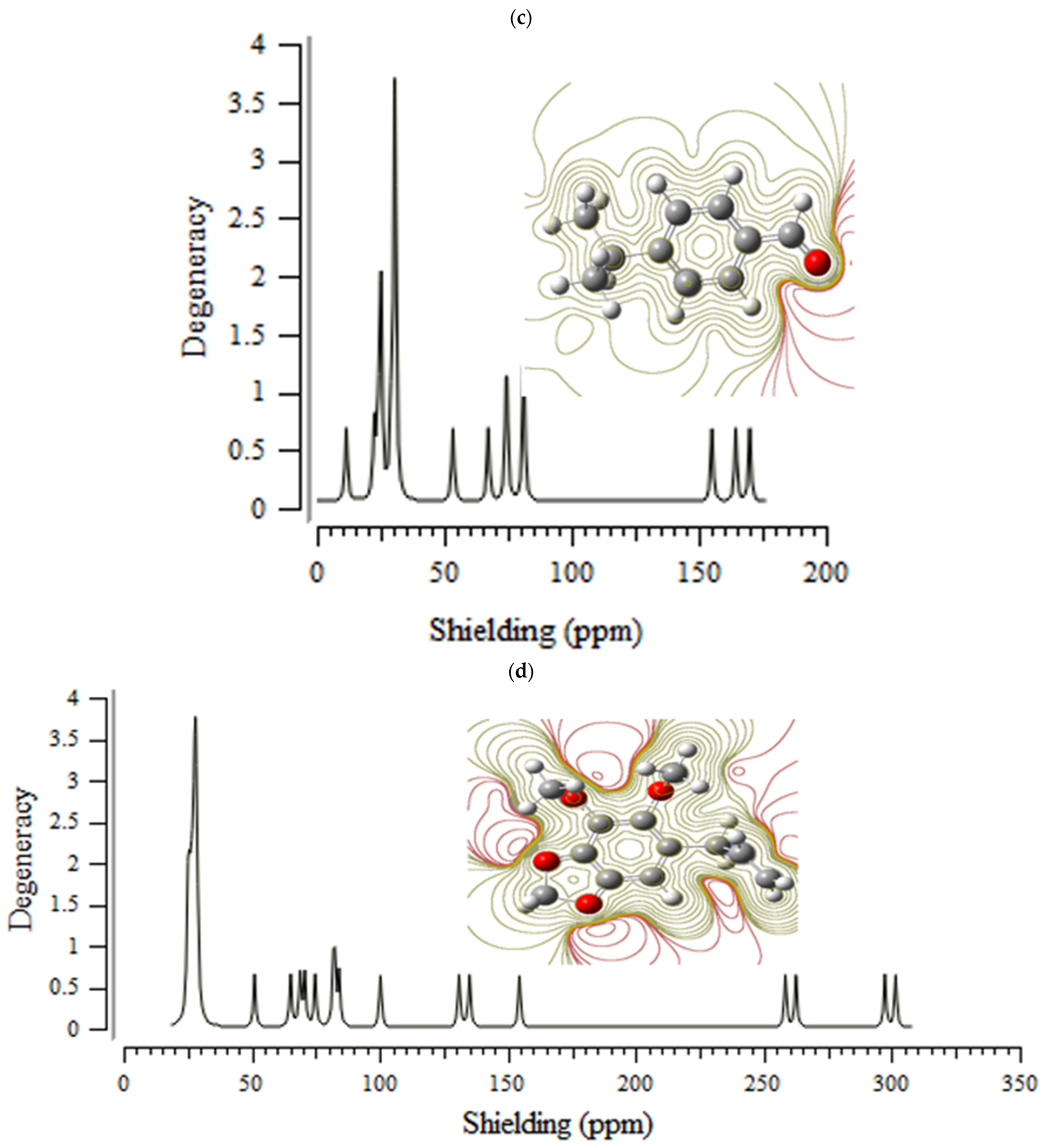
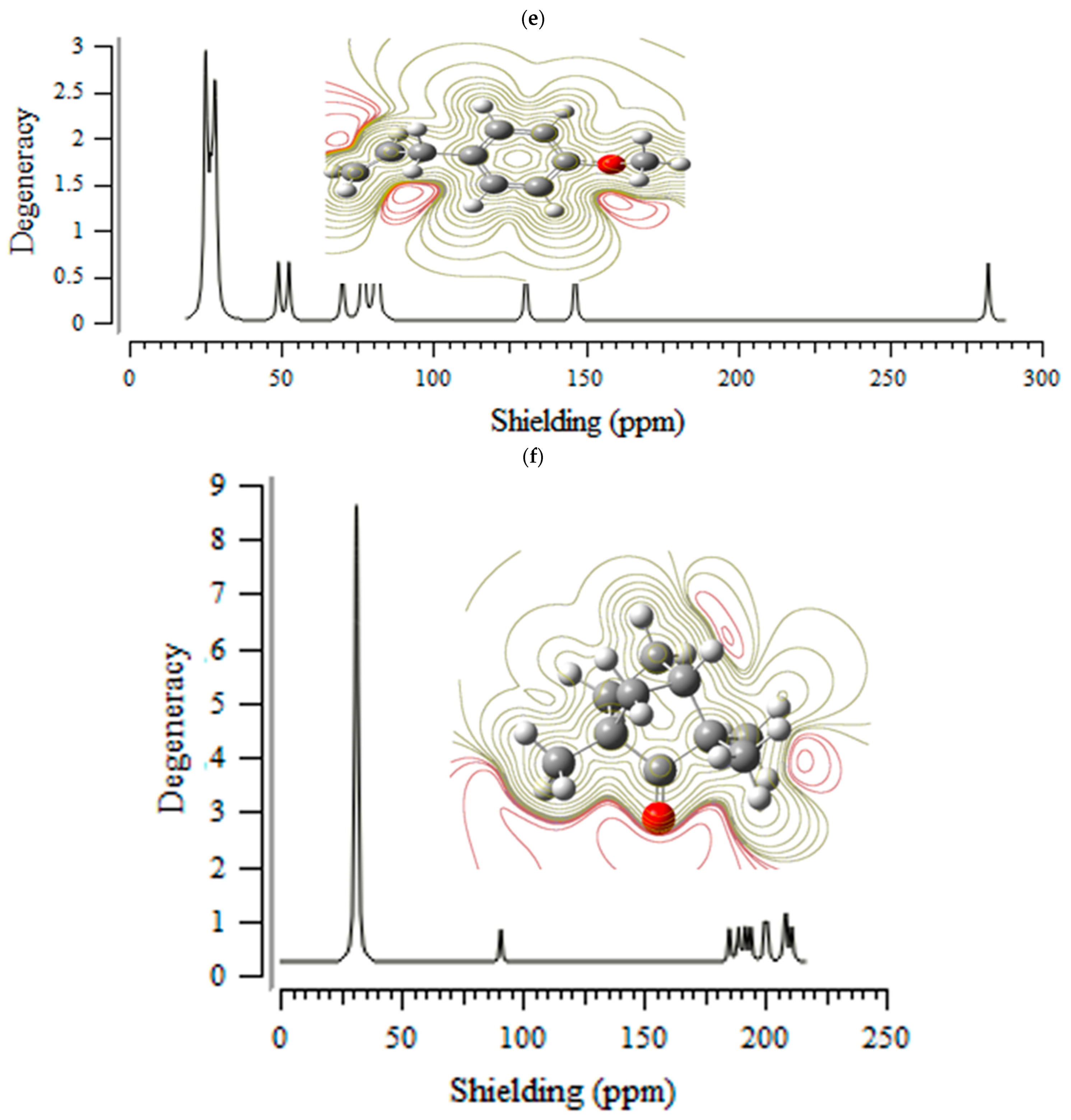
3.1. Nuclear Magnetic Resonance (NMR) Analysis
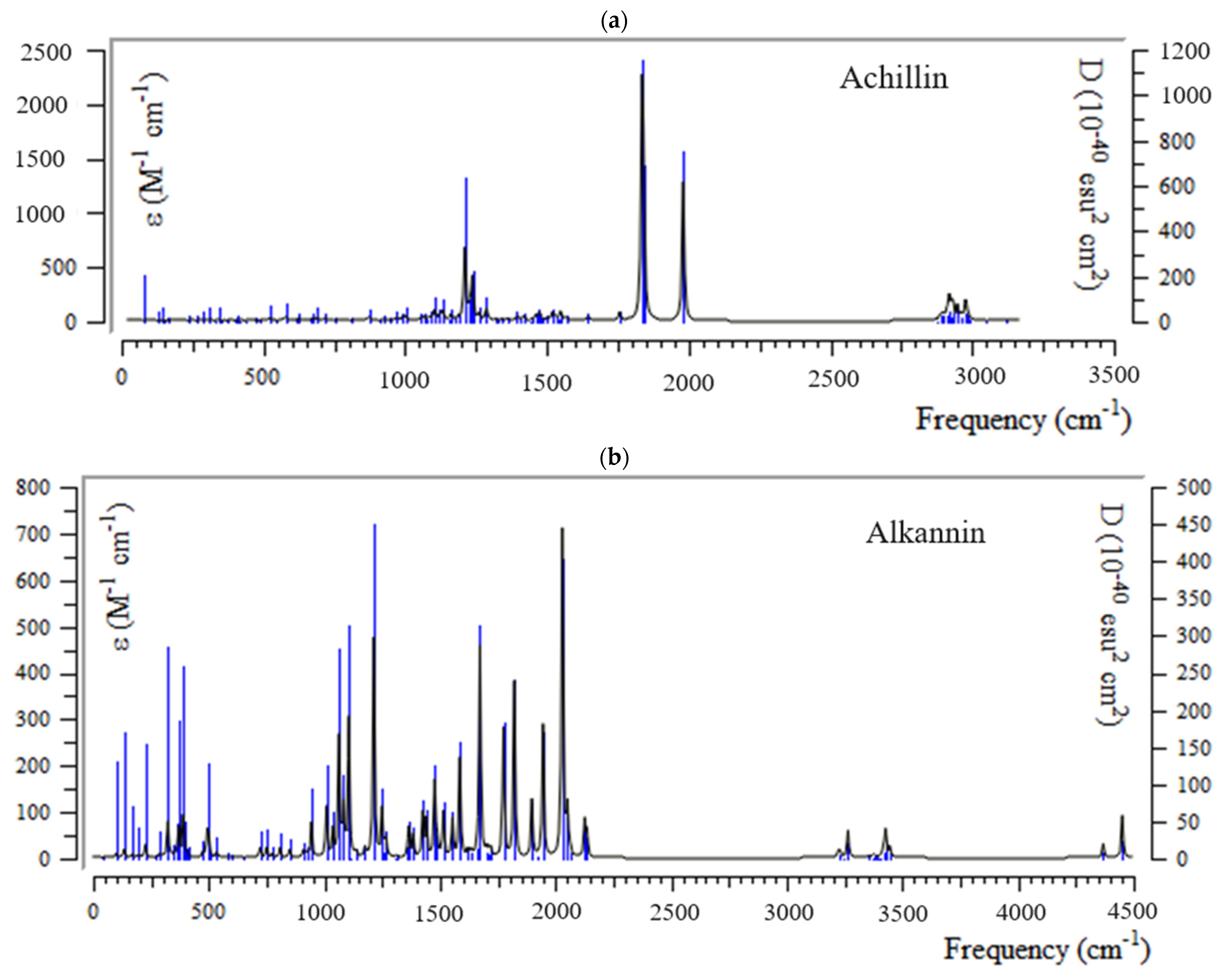
3.2. Infrared (IR) Spectra Analysis and Thermodynamic Properties
3.3. Charge Distribution
3.4. HOMO and LUMO: Frontier Orbitals
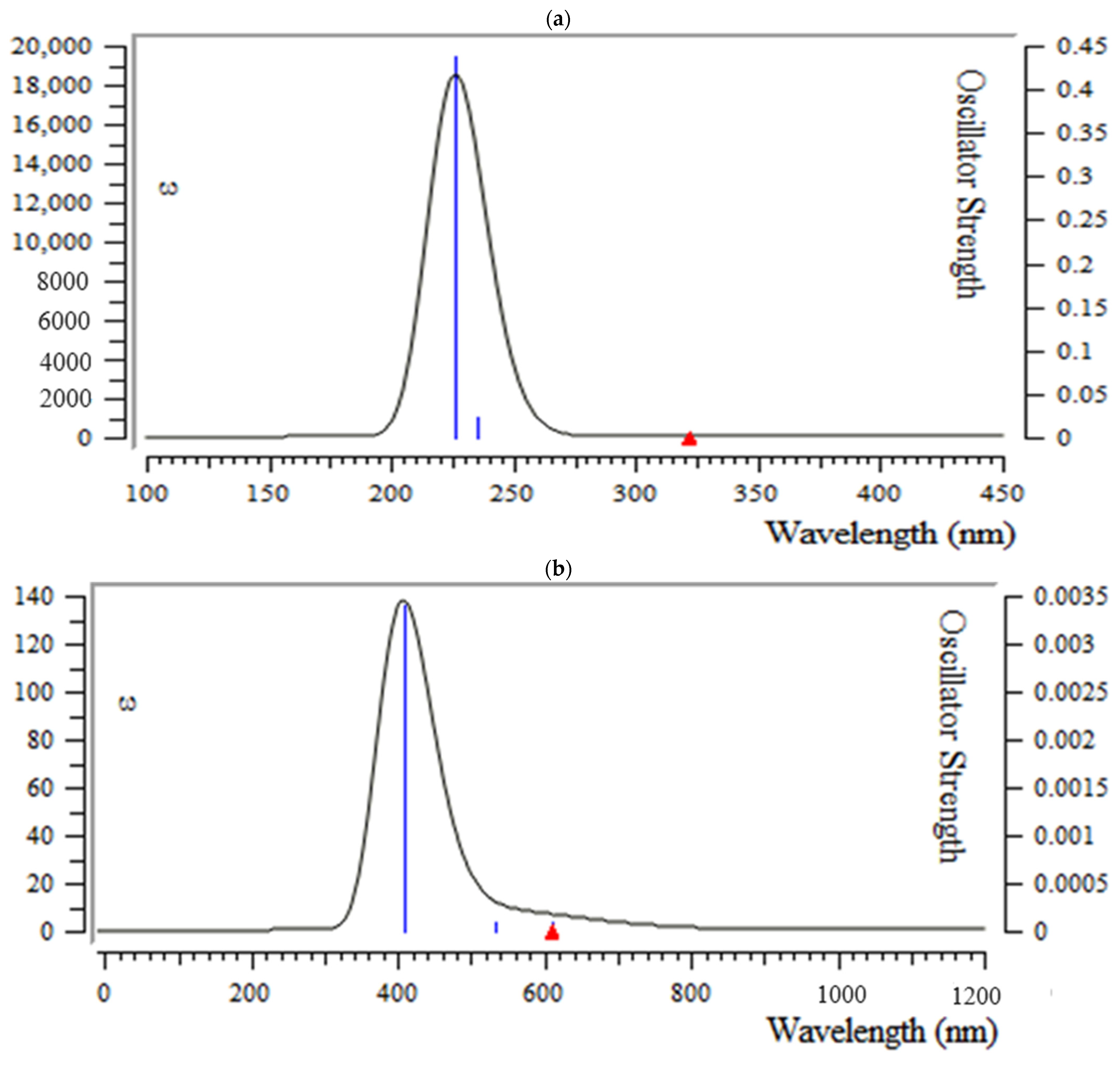
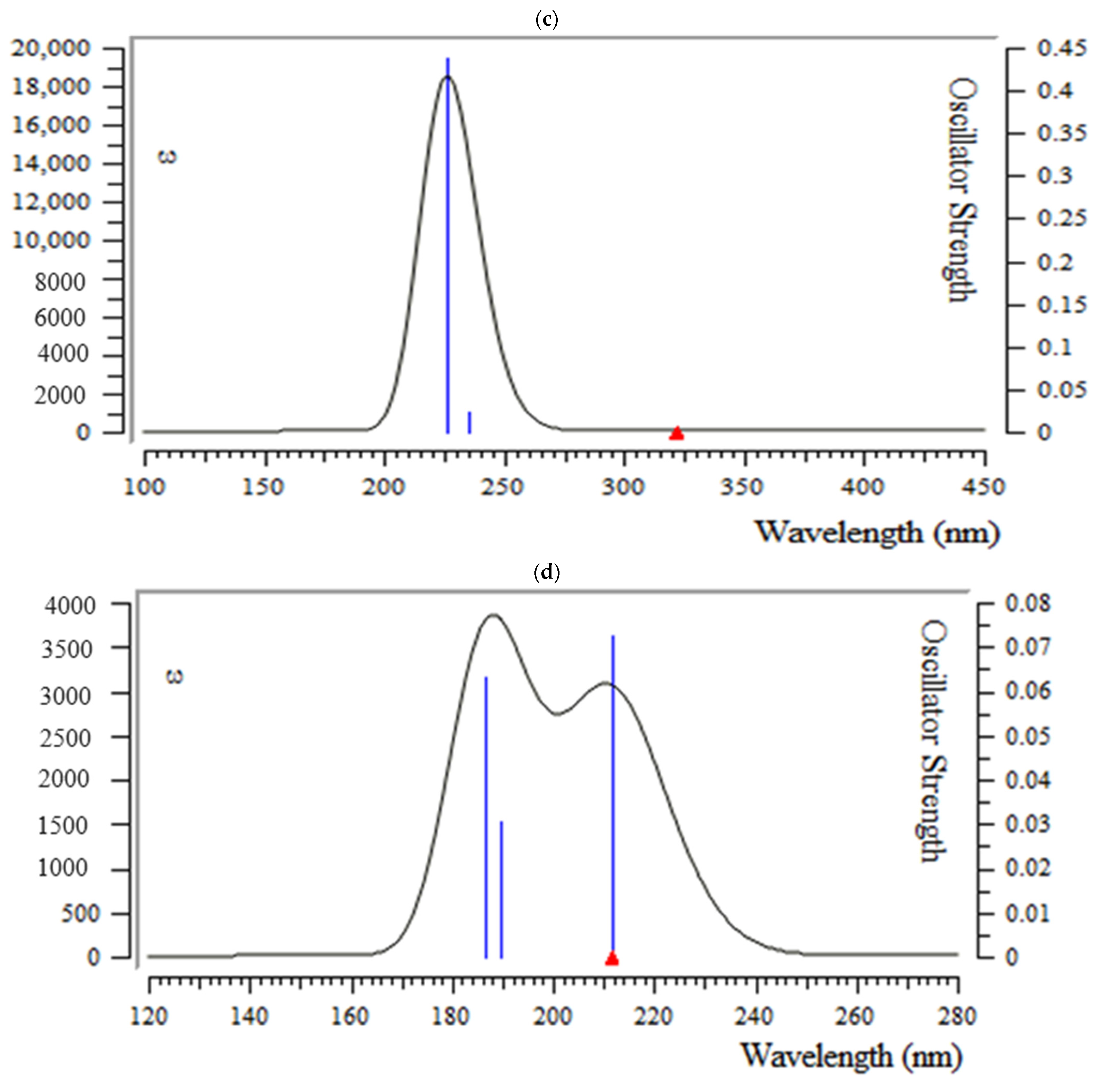
3.5. Analysis of UV-VIS Spectroscopy
4. Conclusions
 H] and [O
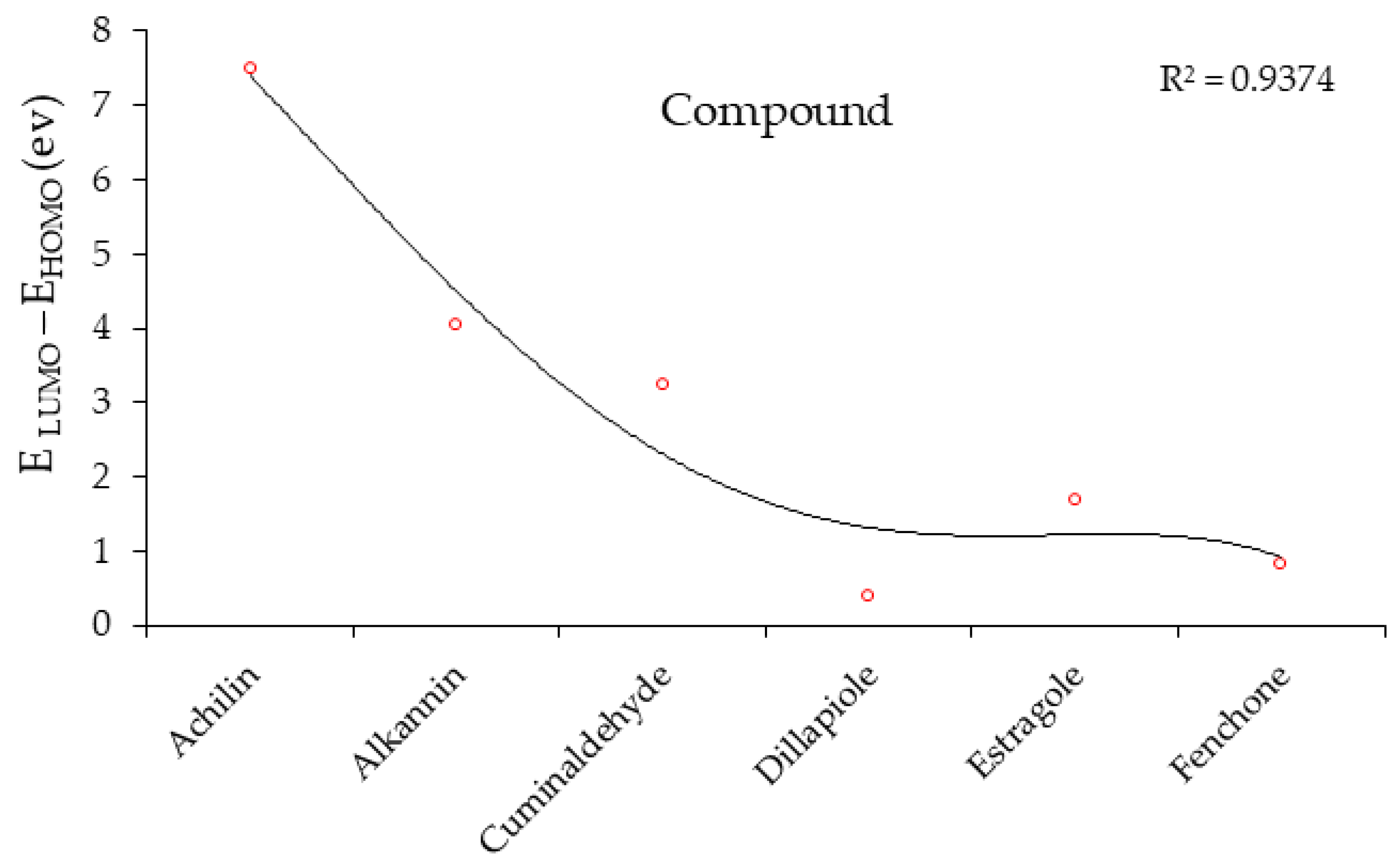
H] and [O H] with distinct atomic charges have been inquired using “IR” approaches. Therefore, the thermodynamic attributes of Gibbs free energy, enthalpy of formation, Electronic Energy, and Core–Core interaction can authorize the consistency of anti-COVID-19 due to hydrogen bonding foundation using the drug design framework. Moreover, the lowering of the energy gap [∆E = ELUMO –EHOMO] has illustrated the charge transfer interactions taking place within achillin, alkannin, cuminaldehyde, dillapiole, estragole, and fenchone. The atomic charges have donated the proper perception of molecular theory and the energies of fundamental molecular orbitals.
H] with distinct atomic charges have been inquired using “IR” approaches. Therefore, the thermodynamic attributes of Gibbs free energy, enthalpy of formation, Electronic Energy, and Core–Core interaction can authorize the consistency of anti-COVID-19 due to hydrogen bonding foundation using the drug design framework. Moreover, the lowering of the energy gap [∆E = ELUMO –EHOMO] has illustrated the charge transfer interactions taking place within achillin, alkannin, cuminaldehyde, dillapiole, estragole, and fenchone. The atomic charges have donated the proper perception of molecular theory and the energies of fundamental molecular orbitals.Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alomair, L.; Mustafa, S.; Jafri, M.S.; Alharbi, W.; Aljouie, A.; Almsned, F.; Alawad, M.; Bokhari, Y.A.; Rashid, M. Molecular Dynamics Simulations to Decipher the Role of Phosphorylation of SARS-CoV-2 Nonstructural Proteins (nsps) in Viral Replication. Viruses 2022, 14, 2436. [Google Scholar] [CrossRef]
- Plavec, Z.; Domanska, A.; Liu, X.; Laine, P.; Paulin, L.; Varjosalo, M.; Auvinen, P.; Wolf, S.G.; Anastasina, M.; Butcher, S.J. SARS-CoV-2 Production, Purification Methods and UV Inactivation for Proteomics and Structural Studies. Viruses 2022, 14, 1989. [Google Scholar] [CrossRef]
- Monajjemi, M.; Shahriari, S.; Mollaamin, F. Evaluation of Coronavirus Families & COVID-19 Proteins: Molecular Modeling Study. Biointerface Res. Appl. Chem. 2020, 10, 6039–6057. [Google Scholar] [CrossRef]
- Yarovaya, O.I.; Shcherbakov, D.N.; Borisevich, S.S.; Sokolova, A.S.; Gureev, M.A.; Khamitov, E.M.; Rudometova, N.B.; Zybkina, A.V.; Mordvinova, E.D.; Zaykovskaya, A.V.; et al. Borneol Ester Derivatives as Entry Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses. Viruses 2022, 14, 1295. [Google Scholar] [CrossRef] [PubMed]
- Shahriari, S.; Monajjemi, M.; Mollaamin, F. Determination of proteins specification with SARS-COVID-19 based ligand designing. J. Chil. Chem. Soc. 2022, 67, 5468–5476. [Google Scholar] [CrossRef]
- Majeed, A.; Zhang, X. On the Adoption of Modern Technologies to Fight the COVID-19 Pandemic: A Technical Synthesis of Latest Developments. COVID 2023, 3, 90–123. [Google Scholar] [CrossRef]
- Bonaccorsi, G.; Pierri, F.; Cinelli, M.; Flori, A.; Galeazzi, A.; Porcelli, F.; Schmidt, A.L.; Valensise, C.M.; Scala, A.; Quattrociocchi, W.; et al. Economic and social consequences of human mobility restrictions under COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 15530–15535. [Google Scholar] [CrossRef]
- Barakat, A.; Mostafa, A.; Ali, M.; Al-Majid, A.M.; Domingo, L.R.; Kutkat, O.; Moatasim, Y.; Zia, K.; Ul-Haq, Z.; Elshaier, Y.A.M.M. Design, Synthesis and In Vitro Evaluation of Spirooxindole-Based Phenylsulfonyl Moiety as a Candidate Anti-SAR-CoV-2 and MERS-CoV-2 with the Implementation of Combination Studies. Int. J. Mol. Sci. 2022, 23, 11861. [Google Scholar] [CrossRef] [PubMed]
- Mollaamin, F.; Monajjemi, M. Thermodynamic research on the inhibitors of coronavirus through drug delivery method. J. Chil. Chem. Soc. 2021, 66, 5195–5205. [Google Scholar] [CrossRef]
- Sardar, T.; Nadim, S.S.; Rana, S.; Chattopadhyay, J. Assessment of lockdown effect in some states and overall India: A predictive mathematical study on COVID-19 outbreak. Chaos Solitons Fract. 2020, 139, 110078. [Google Scholar] [CrossRef]
- Mollaamin, F.; Shahriari, S.; Monajjemi, M. Treating omicron BA.4 & BA.5 via herbal antioxidant asafoetida: A DFT study of carbon nanocarrier in drug delivery. J. Chil. Chem. Soc. 2023, 68, 5781–5786. [Google Scholar] [CrossRef]
- Zeng, F.; Huang, Y.; Guo, Y.; Yin, M.; Chen, X.; Xiao, L.; Deng, G. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int. J. Infect. Dis. 2020, 96, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Mollaamin, F. Physicochemical investigation of anti-COVID19 drugs using several medicinal plants. J. Chil. Chem. Soc. 2022, 67, 5537–5546. [Google Scholar] [CrossRef]
- Jamal, Q.M.S. Antiviral Potential of Plants against COVID-19 during Outbreaks—An Update. Int. J. Mol. Sci. 2022, 23, 13564. [Google Scholar] [CrossRef]
- Remali, J.; Aizat, W.M. A review on plant bioactive compounds and their modes of action against coronavirus infection. Front. Pharmacol. 2021, 11, 589044. [Google Scholar] [CrossRef]
- Singh, S.; Sk, M.F.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via rna-dependent RNA polymerase (rdrp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2021, 39, 6249–6264. [Google Scholar] [CrossRef] [PubMed]
- Capell, T.; Twyman, R.M.; Armario-Najera, V.; Ma, J.K.-C.; Schillberg, S.; Christou, P. Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 2020, 25, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Mollaamin, F. Function of anti-cov structure using inh [1-6]-tyr160-met161-his162 complex. Biointerface Res. Appl. Chem. 2021, 11, 14433–14450. [Google Scholar] [CrossRef]
- Bibi, S.; Khan, M.S.; El-Kafrawy, S.A.; Alandijany, T.A.; El-Daly, M.M.; Yousafi, Q.; Fatima, D.; Faizo, A.A.; Bajrai, L.H.; Azhar, E.I. Virtual screening and molecular dynamics simulation analysis of Forsythoside A as a plant-derived inhibitor of SARS-CoV-2 3clpro. Saudi Pharm. J. 2022, 30, 979–1002. [Google Scholar] [CrossRef]
- Raudone, L.; Vilkickyte, G.; Marksa, M.; Radusiene, J. Comparative Phytoprofiling of Achillea millefolium Morphotypes: Assessing Antioxidant Activity, Phenolic and Triterpenic Compounds Variation across Different Plant Parts. Plants 2024, 13, 1043. [Google Scholar] [CrossRef]
- Faran, M.; Tcherni, A. Medicinal Herbs in Modern Medicine (Ṣimḥei Marpé Bir’fū’ah ha-Modernīt); Akademon (Hebrew University of Jerusalem): Jerusalem, Israel, 1997; Volume 1, p. 242. ISBN 965-350-068-6. (In Hebrew) [Google Scholar]
- Martin, C. How to Grow Perennial Vegetables; Green Books: Newark, NJ, USA, 2012; ISBN 978-1-900322-84-3. [Google Scholar]
- Rojas-Martínez, R.; Arrieta, J.; Cruz-Antonio, L.; Arrieta-Baez, D.; Velázquez-Méndez, A.M.; Sánchez-Mendoza, M.E. Dillapiole, Isolated from Peperomia pellucida, Shows Gastroprotector Activity against Ethanol-Induced Gastric Lesions in Wistar Rats. Molecules. 2013, 18, 11327–11337. [Google Scholar] [CrossRef]
- Schepetkin, I.A.; Özek, G.; Özek, T.; Kirpotina, L.N.; Klein, R.A.; Khlebnikov, A.I.; Quinn, M.T. Composition and Biological Activity of the Essential Oils fromWild Horsemint, Yarrow, and Yampah from Subalpine Meadows in Southwestern Montana: Immunomodulatory Activity of Dillapiole. Plants 2023, 12, 2643. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.M. Artemisia dracunculus. In Flora of North America North of Mexico (FNA); Flora of North America Editorial Committee, Ed.; eFloras.org: New York, NY, USA; Oxford, UK; Missouri Botanical Garden: St. Louis, MO, USA; Harvard University Herbaria: Cambridge, MA, USA, 2006; Volume 19. [Google Scholar]
- Zeller, A.; Rychlik, M. Impact of estragole and other odorants on the flavour of anise and tarragon. Flavour Fragr. J. 2007, 22, 105–113. [Google Scholar] [CrossRef]
- Ribeiro-Santos, R.; Andrade, M.; Sanches-Silva, A.; de Melo, N.R. Essential Oils for Food Application: Natural Substances with Established Biological Activities. Food Bioprocess Technol. 2017, 11, 43–71. [Google Scholar] [CrossRef]
- Badgujar, S.B.; Patel, V.V.; Bandivdekar, A.H. Foeniculum vulgare Mill: A Review of Its Botany, Phytochemistry, Pharmacology, Contemporary Application, and Toxicology. BioMed Res. Int. 2014, 2014, 842674. [Google Scholar] [CrossRef] [PubMed]
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: Promising natural compounds against viral infections. Arch. Virol. 2017, 162, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Seema, T.M.; Thyagarajan, S.P. Pa-9: A flavonoid extracted from plectranthus amboinicus inhibits HIV-1 protease. Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 1020–1024. [Google Scholar]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef]
- Dhama, K.; Natesan, S.; Iqbal Yatoo, M.; Patel, S.K.; Tiwari, R.; Saxena, S.K.; Harapan, H. Plant-based vaccines and antibodies to combat COVID-19: Current status and prospects. Hum. Vaccines Immunother. 2020, 16, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Nawrot-Hadzik, I.; Zmudzinski, M.; Matkowski, A.; Preissner, R.; Kęsik-Brodacka, M.; Hadzik, J.; Drag, M.; Abel, R. Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study. Pharmaceuticals 2021, 4, 742. [Google Scholar] [CrossRef]
- Dwarka, D.; Agoni, C.; Mellem, J.J.; Soliman, M.E.; Baijnath, H. Identification of potential SARS-CoV-2 inhibitors from South African medicinal plant extracts using molecular modelling approaches. S. Afr. J. Bot. 2020, 133, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.A.; Nagarajan, S.K.; Ramesh, V.; Palaniyandi, V.; Selvam, S.P.; Madhavan, T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J. Mol. Struct. 2020, 1221, 128823. [Google Scholar] [CrossRef] [PubMed]
- Shree, P.; Mishra, P.; Selvaraj, C.; Singh, S.K.; Chaube, R.; Garg, N.; Tripathi, Y.B. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants—Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)—A molecular docking study. J. Biomol. Struct. Dyn. 2020, 40, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, J.; Gornowicz-Porowska, J.; Budzianowski, J.; Nowak, G.; Schroeder, G.; Kurczewska, J. Medicinal Herbs in the Relief of Neurological, Cardiovascular, and Respiratory Symptoms after COVID-19 Infection A Literature Review. Cells 2022, 11, 1897. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865. [Google Scholar] [CrossRef] [PubMed]
- Bakhshi, K.; Mollaamin, F.; Monajjemi, M. Exchange and correlation effect of hydrogen chemisorption on nano V(100) surface: A DFT study by generalized gradient approximation (GGA). J. Comput. Theor. Nanosci. 2011, 8, 763–768. [Google Scholar] [CrossRef]
- Monajjemi, M.; Mahdavian, L.; Mollaamin, F.; Khaleghian, M. Interaction of Na, Mg, Al, Si with carbon nanotube (CNT): NMR and IR study. Russ. J. Inorg. Chem. 2009, 54, 1465–1473. [Google Scholar] [CrossRef]
- Mollaamin, F.; Shahriari, S.; Monajjemi, M. Drug design of medicinal plants as a treatment of omicron variant (COVID-19 variant B.1.1.529). J. Chil. Chem. Soc. 2022, 67, 5562–5570. [Google Scholar] [CrossRef]
- Monajjemi, M.; Noei, M.; Mollaamin, F. Design of fMet-tRNA and Calculation of its Bonding Properties by Quantum Mechanics. Nucleosides Nucleotides Nucleic Acids 2010, 29, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Mollaamin, F.; Monajjemi, M. Harmonic Linear Combination and Normal Mode Analysis of Semiconductor Nanotubes Vibrations. J. Comput. Theor. Nanosci 2015, 12, 1030–1039. [Google Scholar] [CrossRef]
- Khaleghian, M.; Zahmatkesh, M.; Mollaamin, F.; Monajjemi, M. Investigation of Solvent Effects on Armchair Single-Walled Carbon Nanotubes: A QM/MD Study. Fuller. Nanotub. Carbon Nanostruct. 2011, 19, 251–261. [Google Scholar] [CrossRef]
- Sarasia, E.M.; Afsharnezhad, S.; Honarparvar, B.; Mollaamin, F.; Monajjemi, M. Theoretical study of solvent effect on NMR shielding tensors of luciferin derivatives. Phys. Chem. Liquids 2011, 49, 561–571. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. In Situ Ti-Embedded SiC as Chemiresistive Nanosensor for Safety Monitoring of CO, CO2, NO, NO2: Molecular Modelling by Conceptual Density Functional Theory. Russ. J. Phys. Chem. B 2024, 18, 49–66. [Google Scholar] [CrossRef]
- Tahan, A.; Mollaamin, F.; Monajjemi, M. Thermochemistry and NBO analysis of peptide bond: Investigation of basis sets and binding energy. Russ. J. Phys. Chem. A 2009, 83, 587–597. [Google Scholar] [CrossRef]
- Mollaamin, F.; Shahriari, S.; Monajjemi, M. Monkeypox disease treatment by tecovirimat adsorbed onto single-walled carbon nanotube through drug delivery method. J. Chil. Chem. Soc. 2023, 68, 5796–5801. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. Molecular modelling framework of metal-organic clusters for conserving surfaces: Langmuir sorption through the TD-DFT/ONIOM approach. Mol. Simul. 2023, 49, 365–376. [Google Scholar] [CrossRef]
- Shahriari, S.; Mollaamin, F.; Monajjemi, M. Increasing the Performance of {[(1-x-y) LiCo0.3Cu0.7] (Al and Mg doped)] O2}, xLi2MnO3, yLiCoO2 Composites as Cathode Material in Lithium-Ion Battery: Synthesis and Characterization. Micromachines 2023, 14, 241. [Google Scholar] [CrossRef]
- McArdle, S.; Mayorov, A.; Shan, X.; Benjamin, S.; Yuan, X. Digital quantum simulation of molecular vibrations. Chem. Sci. 2019, 10, 5725–5735. [Google Scholar] [CrossRef]
- Monajjemi, M.; Baie, M.T.; Mollaamin, F. Interaction between threonine and cadmium cation in [Cd(Thr)] (n = 1–3) complexes: Density functional calculations. Russ Chem Bull. 2010, 59, 886–889. [Google Scholar] [CrossRef]
- Zadeh, M.A.A.; Lari, H.; Kharghanian, L.; Balali, E.; Khadivi, R.; Yahyaei, H.; Mollaamin, F.; Monajjemi, M. Density functional theory study and anti-cancer properties of shyshaq plant: In view point of nano biotechnology. J. Comput. Theor. Nanosci. 2015, 12, 4358–4367. [Google Scholar] [CrossRef]
- Ciliberto, G.; Cardone, L. Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discov. Today 2020, 25, 946–948. [Google Scholar] [CrossRef]
- Hatada, R.; Okuwaki, K.; Mochizuki, Y.; Handa, Y.; Fukuzawa, K.; Komeiji, Y.; Okiyama, Y.; Tanaka, S. Fragment molecular orbital based interaction analyses on COVID-19 main protease-inhibitor N3 complex (PDB ID: 6LU7). J. Chem. Inf. Model. 2020, 60, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Peele, K.A.; Chandrasai, P.; Srihansa, T.; Krupanidhi, S.; Sai, A.V.; Babu, D.J.; Indira, M.; Reddy, A.R.; Venkateswarulu, T. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Inform. Med. Unlocked 2020, 19, 100345. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Zhang, H.; Ji, H.-F.; Chen, Q. Computational view toward the inhibition of SARS-CoV-2 spike glycoprotein and the 3CL protease. Computation 2020, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Pitsillou, E.; Karagiannis, C.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: Molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput. Biol. Chem. 2020, 87, 107292. [Google Scholar] [CrossRef]
- Mollaamin, F. Conocimiento de enfermedades virales terapéuticas: Aplicación de SWCNT en la administración de fármacos. Rev. Colomb. Quim. 2023, 52, 28–35. [Google Scholar] [CrossRef]
- Monajjemi, M.; Mollaamin, F.; Shojaei, S. An overview on coronaviruses family from past to COVID-19: Introduce some inhibitors as antiviruses from Gillan’s plants. Biointerface Res. Appl. Chem. 2020, 3, 5575–5585. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. Adsorption ability of Ga5N10 nanomaterial for removing metal ions contamination from drinking water by DFT. Int. J. Quantum Chem. 2024, 124, e27348. [Google Scholar] [CrossRef]
- Wang, S. Efficiently Calculating Anharmonic Frequencies of Molecular Vibration by Molecular Dynamics Trajectory Analysis. ACS Omega 2019, 4, 9271–9283. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M.; Salemi, S.; Baei, M.T. A Dielectric Effect on Normal Mode Analysis and Symmetry of BNNT Nanotube. Fuller. Nanotub. Carbon Nanostruct. 2011, 19, 182–196. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. Trapping of toxic heavy metals from water by GN–nanocage: Application of nanomaterials for contaminant removal technique. J. Mol. Struct. 2024, 1300, 137214. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. Transition metal (X = Mn, Fe, Co, Ni, Cu, Zn)-doped graphene as gas sensor for CO2 and NO2 detection: A molecular modeling framework by DFT perspective. J. Mol. Model. 2023, 29, 119. [Google Scholar] [CrossRef] [PubMed]
- Aji, G.K.; Hatou, K.; Morimoto, T. Modeling the Dynamic Response of Plant Growth to Root Zone Temperature in Hydroponic Chili Pepper Plant Using Neural Networks. Agriculture 2020, 10, 234. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. Tailoring and functionalizing the graphitic-like GaN and GaP nanostructures as selective sensors for NO, NO2, and NH3 adsorbing: A DFT study. J. Mol. Model. 2023, 29, 170. [Google Scholar] [CrossRef]
- Khalili Hadad, B.; Mollaamin, F.; Monajjemi, M. Biophysical chemistry of macrocycles for drug delivery: A theoretical study. Russ. Chem. Bull. 2022, 60, 238–241. [Google Scholar] [CrossRef]
- Mollaamin, F.; Ilkhani, A.; Sakhaei, N.; Bonsakhteh, B.; Faridchehr, A.; Tohidi, S.; Monajjemi, M. Thermodynamic and solvent effect on dynamic structures of nano bilayer-cell membrane: Hydrogen bonding study. J. Comput. Theor. Nanosci. 2015, 12, 3148–3154. [Google Scholar] [CrossRef]
- Monajjemi, M.; Khaleghian, M.; Tadayonpour, N.; Mollaamin, F. The effect of different solvents and temperatures on stability of single-walled carbon nanotube: A QM/MD study. Int. J. Nanosci. 2010, 9, 517–529. [Google Scholar] [CrossRef]
- Aihara, J. Reduced HOMO−LUMO Gap as an Index of Kinetic Stability for Polycyclic Aromatic Hydrocarbons. J. Phys. Chem. A 1999, 103, 7487–7495. [Google Scholar] [CrossRef]
- Mollaamin, F.; Monajjemi, M. In Silico-DFT Investigation of Nanocluster Alloys of Al-(Mg,Ge,Sn) Coated by Nitrogen Heterocyclic Carbenes as Corrosion Inhibitors. J. Clust. Sci. 2023, 34, 2901–2918. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute Hardness: Companion Parameter to Absolute Electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Politzer, P.; Abu-Awwad, F. A comparative analysis of Hartree-Fock and Kohn-Sham orbital energies. Theor. Chem. Acc. 1998, 99, 83–87. [Google Scholar] [CrossRef]
- Mollaamin, F.; Shahriari, S.; Monajjemi, M.; Khalaj, Z. Nanocluster of Aluminum Lattice via Organic Inhibitors Coating: A Study of Freundlich Adsorption. J. Clust. Sci. 2023, 34, 1547–1562. [Google Scholar] [CrossRef]




| Compound | Molecular Structure | Sources | Applied Symptom |
|---|---|---|---|
| Achillin | 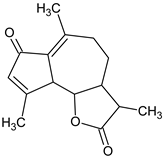 | Achillea millefolium (Yarrow) | weakness, cough, sore throat, nausea-vomiting |
| Alkannin | 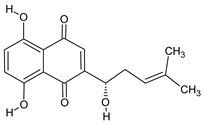 | Alkanet | skin rash, diarrhea |
| Cuminaldehyde | 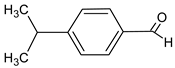 | Rumex patientia (Patience dock) | skin rash, sore throat, fever |
| Dillapiole |  | Dill | anorexia |
| Estragole | 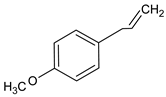 | Tarragon | fever, muscle-joint pain, anorexia |
| Fenchone | 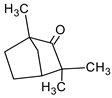 | Sweet fennel | shortness of breath |
| Medicinal Extracts—COVID-19 Active Area | Bond Length | (Å) | Bond/Torsion Angle | (°) |
|---|---|---|---|---|
| Achillin | N67-H68 | 1.036 | N67-H68-O15 | 176.181 |
| H68-O15 | 0.9974 | |||
| O15-C13 | 1.4123 | N67-H68-O15-C13 | 178.492 | |
| Cuminaldehyde | N61-H62 | 1.0351 | N61-H62-O9 | 179.192 |
| H62-O9 | 0.9966 | |||
| O9-C7 | 1.4150 | N61-H62-O9-C7 | 31.2731 | |
| Dillapiole | N78-H79 | 1.0295 | N78-H79-C13 | 179.216 |
| H79-C13 | 1.1193 | |||
| C13-O12 | 1.4131 | N78-H79-C13-O12 | 55.0114 | |
| Estragole | N71-H72 | 1.0358 | N71-H72-C11 | 179.208 |
| H72-C11 | 1.1244 | |||
| C11-O10 | 1.4099 | N71-H72-C11-O10 | 106.924 |
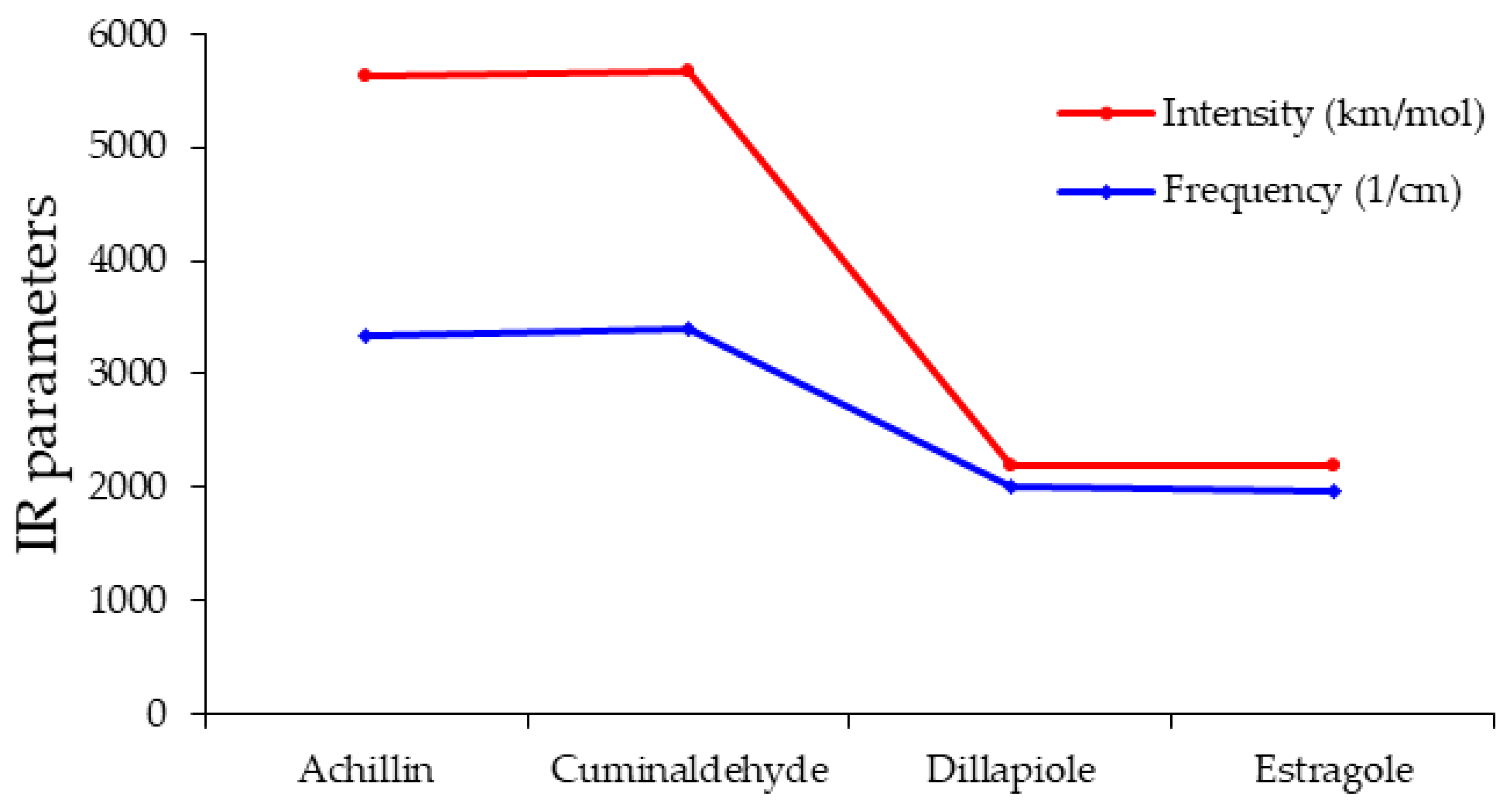
| Inhibitor | Normal Mode | Frequency (1/cm) | Intensity (km/mol) |
|---|---|---|---|
| Achillin | 275 | 3336.01 | 2292.987 |
| Cuminaldehyde | 236 | 3395.38 | 2270.866 |
| Dillapiole | 205 | 1998.66 | 202.722 |
| Estragole | 185 | 1971.26 | 226.961 |
| Plant Component—Active Site | ∆G × 10−4 (kcal/mol) | ∆S (kcal/K.mol) | Eelectronic × 10−4 (kcal/mol) | Ecore-core × 10−4 (kcal/mol) |
|---|---|---|---|---|
| Achillin | −18.2174 | 607.2787 | −214.3615 | 196.1441 |
| Alkannin | −19.6971 | 656.5647 | −232.3721 | 212.6750 |
| Cuminaldehyde | −15.2850 | 509.7272 | −165.8901 | 150.6051 |
| Dillapiole | −17.8553 | 595.2878 | −198.4780 | 180.6227 |
| Estragole | −15.2109 | 507.3701 | −160.6792 | 145.4682 |
| ∆HTMH × 10−4 25.8242 (kcal/mol) | ∆H Achillin | ∆H(Achillin - active site) | ∆HF × 10−4 = ∆H (Achillin - active site) – (∆H Achillin + ∆H active site) |
| −76.2424 | 9.5452 | −25.8156 | |
| ∆H Alkannin | ∆H(Alkannin - active site) | ∆HF × 10−4 = ∆H (Alkannin - active site) – (∆H Alkannin + ∆H active site) | |
| −80.8417 | −1.3898 | −25.8162 | |
| ∆H Cuminaldehyde | ∆H(Cuminaldehyde - active site) | ∆HF × 10−4 = ∆H (Cuminaldehyde - active site) – (∆H Cuminaldehyde +∆H active site) | |
| −3.6680 | 67.8448 | −25.8170 | |
| ∆H Dillapiole | ∆H(Dillapiole - active site) | ∆HF × 10−4 = ∆H(Dillapiole - active site) – (∆H Dillapiole + ∆H active site) | |
| −31.3428 | 33.0993 | −25.8177 | |
| ∆H Estragole | ∆H (Estragole - active site) | ∆HF × 10−4 =∆H (Estragole - active site) – (∆H Estragole + ∆Hactive site) | |
| 101.5614 | 14.9017 | −25.8328 |
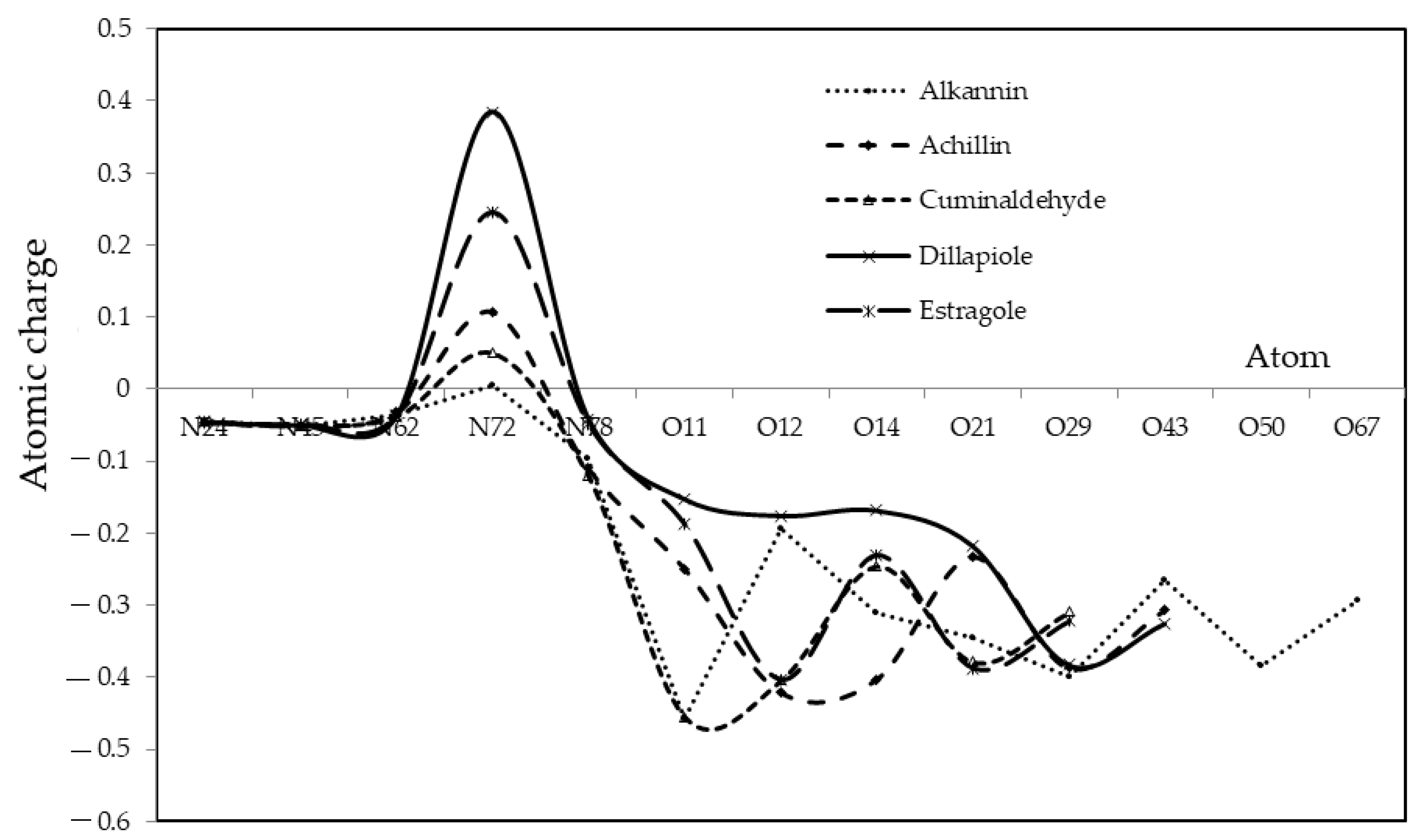
| Achillin | Q | Alkannin | Q | Cuminaldehyde | Q | Dillapiole | Q | Estragole | Q |
|---|---|---|---|---|---|---|---|---|---|
| N19 | −0.0463 | N24 | −0.0442 | N13 | −0.0450 | N30 | −0.0455 | N23 | −0.0463 |
| N40 | −0.0506 | N45 | −0.0513 | N34 | −0.0503 | N51 | −0.0501 | N44 | −0.0482 |
| N57 | −0.0323 | N62 | −0.0368 | N51 | −0.0375 | N68 | −0.0377 | N61 | −0.0367 |
| N67 | 0.1071 | N72 | 0.0057 | N61 | 0.0505 | N78 | 0.3846 | N71 | 0.2459 |
| N73 | −0.1103 | N78 | −0.0961 | N67 | −0.1211 | N84 | −0.0413 | N77 | −0.0487 |
| O14 | −0.2489 | O11 | −0.4572 | O9 | −0.4548 | O9 | −0.1526 | O10 | −0.1878 |
| O15 | −0.4204 | O12 | −0.1939 | O18 | −0.4041 | O10 | −0.1758 | O28 | −0.4031 |
| O24 | −0.4027 | O14 | −0.3115 | O32 | −0.2455 | O12 | −0.1684 | O42 | −0.2305 |
| O38 | −0.2326 | O21 | −0.3452 | O39 | −0.3786 | O49 | −0.2174 | O49 | −0.3875 |
| O45 | −0.3872 | O29 | −0.4001 | O56 | −0.3090 | O56 | −0.3826 | O66 | −0.3223 |
| O62 | −0.3070 | O43 | −0.2657 | O73 | −0.3254 | ||||
| O50 | −0.3842 | ||||||||
| O67 | −0.2939 |
| Molecule | ELUMO (a.u.) | EHOMO (a.u.) | ∆E = ELUMO – EHOMO (eV) | ||
|---|---|---|---|---|---|
| Achilin | 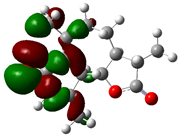 | −0.0266 | 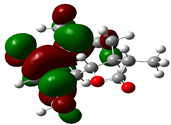 | −0.3019 | 7.4899 |
| Alkannin | 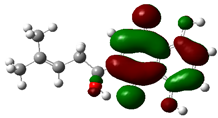 | −0.0283 |  | −0.1767 | 4.0381 |
| Cuminaldehyde |  | −0.0887 |  | −0.2075 | 3.2327 |
| Dillapiole | 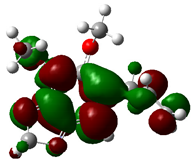 | −0.1575 | 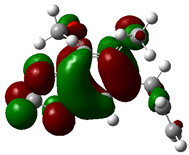 | −0.1717 | 0.3864 |
| Estragole | 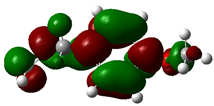 | −0.1523 |  | −0.2141 | 1.6816 |
| Fenchone | 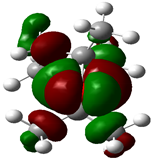 | −0.1605 | 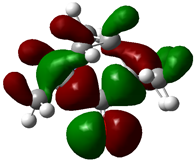 | −0.1907 | 0.8218 |
| Compounds | µ = (EHOMO + ELUMO)/2 | χ = –(EHOMO + ELUMO)/2 | η = (ELUMO–EHOMO)/2 | ζ = 1/(2η) | ψ = µ2/(2η) |
|---|---|---|---|---|---|
| Achilin | −4.4694 | 4.4694 | 3.74495 | 0.1335 | 2.6670 |
| alkannin | −2.7891 | 2.7891 | 2.01905 | 0.2476 | 1.9264 |
| cuminaldehyde | −4.0300 | 4.0300 | 1.61635 | 0.3093 | 5.0239 |
| dillapiole | −4.4790 | 4.4790 | 0.1932 | 2.5880 | 51.9188 |
| estragole | −4.9851 | 4.9851 | 0.8408 | 0.5947 | 14.7783 |
| fenchone | −4.7783 | 4.7783 | 0.8408 | 0.5947 | 13.5776 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mollaamin, F. Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms. Antibodies 2024, 13, 38. https://doi.org/10.3390/antib13020038
Mollaamin F. Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms. Antibodies. 2024; 13(2):38. https://doi.org/10.3390/antib13020038
Chicago/Turabian StyleMollaamin, Fatemeh. 2024. "Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms" Antibodies 13, no. 2: 38. https://doi.org/10.3390/antib13020038
APA StyleMollaamin, F. (2024). Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms. Antibodies, 13(2), 38. https://doi.org/10.3390/antib13020038






